Abstract
Backyard gardens, dump heaps, and kitchen middens are thought to have provided important venues for early crop domestication via generation of hybrids between otherwise isolated plant species. However, this process has rarely been demonstrated empirically. For the majority of polyploid crops, it remains uncertain to what extent hybridization and polyploidization preceded domestication or were precipitated by human activities. Using archaeological, ethnobotanical, geographical, and genetic data, we investigate the extent and significance of predomestication cultivation, backyard sympatry, and spontaneous hybridization for the Mimosoid legume tree Leucaena, which is used as a food crop throughout south-central Mexico. We show that predomestication cultivation was widespread, involved numerous independent transitions from the wild to cultivation, and resulted in extensive artificial sympatry of 2–6 species locally and 13 species in total. Using chloroplast and rapidly evolving nuclear-encoded DNA sequences, we demonstrate that hybridization in Leucaena has been extensive and complex, spawning a diverse set of novel hybrids as a result of juxtaposition of species in cultivation. The scale and complexity of hybridization in Leucaena is significantly greater than that documented for any other Mexican plant domesticates so far. However, there are striking parallels between Leucaena and the other major Mexican perennial domesticates Agave and Opuntia, which show very similar domestication via backyard hybridization pathways. Our results suggest that backyard hybridization has played a central role in Mesoamerican crop domestication and demonstrate that the simple step of bringing species together in cultivation can provide a potent trigger for domestication.
Keywords: domestication, Leguminosae, Leucaena, Mesoamerica
Hybridization between plant species is frequent in many contemporary contexts where species have been brought together in cultivation (1–4). It is also clear that hybridization and polyploidy have played a central role in the origins of many of the world's crops (5–7). Pinpointing where, when, and how hybridization occurred and assessing the extent to which hybrids and polyploids can be attributed to movement of wild and incipiently domesticated species by early proto-agriculturalists are central to understanding the role of hybridization in crop domestication and the processes that prompted and facilitated early domestication more generally. Hexaploid breadwheat and octaploid strawberry provide well documented examples of crops, which are unknown in the wild and are thought to have arisen via spontaneous hybridization in cultivation (7). In contrast, allotetraploid cotton is thought to have originated well before the arrival of humans in the New World (8). However, for the majority of polyploid crops, such as bananas, citrus, potatoes, kiwi fruit, oca, and peanuts, it remains uncertain to what extent hybridization and polyploidization preceded domestication or were precipitated by human activities (9).
To investigate the role of hybridization in crop domestication, we focus on Mesoamerica, which is one of the most important regions where crops were domesticated (10–15). Recent studies pinpoint where, when, how many times, and from what progenitors the mainstream crops of this region, such as maize, beans, and squash, were domesticated (11–15). However, the origins of most other Mesoamerican domesticates remain poorly understood (16). Among these is a set of important polyploid crops in the genera Agave, Opuntia, and Leucaena for which there is preliminary evidence to suggest that hybridization between species in cultivation may have been important in driving domestication (2, 17, 18). For all three genera, archaeobotanical data provide evidence of use over the last 6,000–9,000 years (17, 19, 20); utilization involves more than one, and sometimes numerous, species in any one area; for each there are finely differentiated cognitive systems indicative of sophisticated comparative knowledge of species traits (21–23); present-day use suggests that each genus presents a complex mosaic of wild, managed, and cultivated backyard populations often involving species in sympatry; and finally, hybridization and polyploidy are frequent in all three genera.
To gain insight into hybrid origins and the extent to which these can be attributed to predomestication cultivation, we examine one of these domesticates, the Mimosoid legume tree Leucaena, which comprises 22 species of small trees distributed from the southern United States to Peru (24). There is clear evidence to suggest that interspecific hybridization has been important in Leucaena. Five tetraploid species have been documented (25), and there is preliminary evidence that two of these, L. leucocephala and L. confertiflora, are allopolyploids (26). Artificial crossing experiments have shown that cross-ability among species is high (27). In addition, two named hybrids, L. × mixtec, a putative sterile triploid between tetraploid L. leucocephala and diploid L. esculenta, and L. × spontanea, a putative fertile hybrid between tetraploid L. leucocephala and L. diversifolia, have been proposed (26, 28, 29). Furthermore, several other putative hybrids found growing in backyards in south-central (S-C) Mexico (Fig. 1), are investigated here. Previous attempts to disentangle divergent from reticulate relationships in Leucaena have relied on chloroplast restriction fragment length polymorphism (30) and internal transcribed spacer (ITS) sequence data (26). These analyses provided evidence for three robustly supported clades of diploid species but lack of resolution within clades, and the occurrence of potential ITS pseudogene sequences limited the insights gained into hybrid and polyploid origins from these data (26). To address this lack of resolution we developed a sequence-characterized amplified region-based technique to identify rapidly evolving DNA sequence loci (31) that can generate gene trees with the resolution and support required (31, 32). Here, we present a densely sampled and well resolved gene tree that includes all of the known species of Leucaena and a set of putative backyard hybrids for one of these rapidly evolving loci, the nuclear-encoded locus, 23L (Fig. 2), alongside a three-gene chloroplast sequence data set [supporting information (SI) Fig. 3].
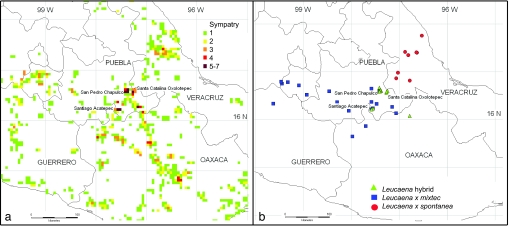
Fig. 1.
Sympatry and hybridization among Leucaena species in S-C Mexico. (a) Map showing the number of co-occurring species of Leucaena in 3-min grid cells. Backyard sympatry attributable to cultivation between pairs of species is common across this part of S-C Mexico (see also SI Figs. 4 and 5). Mixtures of three species are common in northern Oaxaca and southern Puebla. Higher levels of sympatry with four to six co-occurring species are found in villages on the fringes of the upper Tehuacán Valley. (b) The distribution of backyard hybrids in S-C Mexico coincides with these sympatry hotspots.
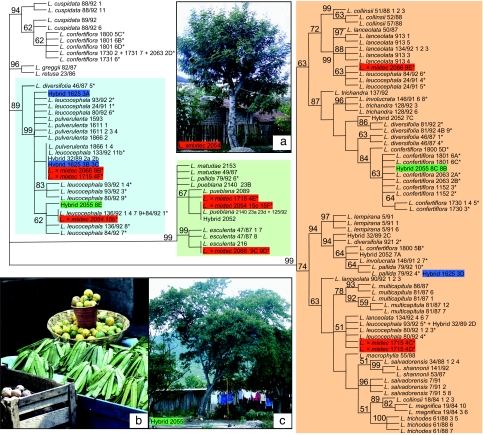
Fig. 2.
Biparentally inherited nuclear 23L gene tree for Leucaena. Tetraploid species names are indicated by *. The major clades (color-shaded) are congruent with previous studies (26, 30, 31) and the chloroplast gene tree (SI Fig. 3). Highlighted terminals show divergent placements of sequence types recovered from L. × mixtec (red), hybrid 2055 (green) between the tetraploids L. leucocephala and L. confertiflora, and hybrid 1625 (blue) between the tetraploids L. leucocephala and L. pallida. Strict consensus of 168 equally parsimonious trees is shown. Length = 268 steps; consistency index = 0.66; retention index = 0.93. The 23L matrix comprises 909 aligned characters with 130 parsimony informative substitutions and 30 informative gaps. Numbers at nodes are bootstrap values. (a–c) Photos illustrate backyard hybrids and marketing of Leucaena pods. (a) Sterile triploid L. × mixtec in a backyard in San Pedro Chapulco, Puebla. (b) Pods of L. leucocephala, the most popular and widespread species for food use, for sale in San Cristobal de las Casas, Chiapas. (c) Hybrid 2055 (highlighted in green) between L. leucocephala and L. confertiflora in a backyard in San Pedro Chapulco, Puebla.
Establishing whether the hybrids involved in crop domestication resulted from early cultivation requires knowledge about the identities of their parents and evidence of predomestication cultivation and translocation that brought those parents together. Obtaining this evidence is rarely straightforward. First, hybrid origins of crop plants can be difficult to unravel (9). Hybrid systems involving polyploidy are more tractable than homoploid hybrids, but can still be highly complex (9). Second, archaeological evidence for early transitions from the wild to cultivation is often lacking. Finally, the distributions of domesticates and their wild progenitors are often poorly known and obscured by a long history of cultivation, selective breeding, and introgression. Investigation of early-stage domesticates rather than mainstream crops may avoid some of these difficulties. For these early-stage domesticates, predomestication cultivation and the products of incipient domestication can often be directly surveyed in extant traditional agroecosystems providing unique insights into these processes (16). To test the hypothesis that Leucaena hybrids were precipitated by cultivation, we have assembled extensive ethnobotanical and geographical data to establish the extent of predomestication cultivation, translocation, and sympatry. Together with the phylogenetic results from chloroplast and nuclear-encoded gene trees, these studies provide the most complete picture of the degree and primary causes of hybridization in Leucaena to date to our knowledge.
Results and Discussion
Predomestication Cultivation and Sympatry.
Seeds of 13 species of Leucaena are used for food across S-C Mexico (SI Table 1). Food use is widespread and intensive in Chiapas, Oaxaca, Puebla, Guerrero, and Morelos and more sporadic further north, but unknown further south despite the availability of native species of Leucaena. Present-day food use varies from gathering of pods from local free-living populations for home consumption to intensive harvesting of commercial quantities from cultivated trees and transportation of seeds to regional markets (SI Table 1). This spectrum of increasing human intervention involves transitions from wild to managed to cultivated, from home consumption to local to wider regional marketing, and from very local to regional to much wider translocation of species (SI Table 1).
At one extreme, pods of L. cuspidata are gathered from natural populations close to settlements and consumed and marketed locally with only sporadic cultivation in adjacent backyards. Wider cultivation and limited translocation are apparent for L. confertiflora and L. collinsii. Zárate (33) noted what he termed incipient domestication of L. confertiflora in backyards in San Pedro Chapulco, Puebla, and determined from local informants that cultivated trees were derived from nearby natural populations. Since then, similar occurrences of cultivated L. confertiflora said by local informants to be derived from local native stands have been located in other villages across Puebla and Oaxaca. It is clear that L. confertiflora has been brought into cultivation independently from nearby natural stands in several places. Three species, L. esculenta, L. pallida, and L. leucocephala are much more widely and intensively used. Pods of these species are harvested commercially and marketed regionally and in cities several 100 km away (Fig. 2b). All three species are widely cultivated in backyards and orchards, sometimes on high-quality agricultural land. Indeed, these species are among the most widely cultivated trees throughout S-C Mexico (33). All three species have been widely introduced into new areas in Mexico.
Widespread cultivation makes it difficult to discern the native distribution of species with certainty, but the broad patterns in Leucaena are clearly apparent. Archaeobotanical data from sites across S-C Mexico (19, 34–36) suggest that Leucaena seeds have provided a minor, but constant, food source in this region for the last 6,000 years and provide evidence for early cultivation and translocation. Seed remains in caves in Tamaulipas and Oaxaca, and the earlier remains from Tehuacán, correspond to L. pulverulenta, L. trichandra, and L. pueblana,¶ respectively (19) that grow naturally, but not in cultivation, in the vicinity of these caves today, suggesting that these resulted from archaic foraging in free-living populations (35, 36). The first appearance of seeds and pod fragments of L. esculenta from ≈3,200 B.P. in the Coxcatlán and Purrón caves has been interpreted as marking the start of cultivation of Leucaena in the Tehuacán Valley (19). Seeds of L. leucocephala, the most widely cultivated species today, were first identified in the Coxcatlán and Purrón sequences between 2,300 B.P. and 1,200 B.P. (19). These two species are abundantly cultivated in the Tehuacán Valley today, but there is no evidence of natural populations to suggest that either species is native there (19). These records are compatible with field observations suggesting that the present-day distribution of L. esculenta is much wider than the original native range (19, 24). Archaeological evidence thus suggests that Leucaena was brought into cultivation soon after the wider transition from incipient cultivation to village agriculture that started once fusion of the Mesoamerican mainstream crop complex was established ≈4,000 B.P. (11, 37). This sequence fits with the idea that tree cultivation lags behind domestication of annual crops because of longer life cycles and the need for sedentary agriculture before tree cultivation (38).
A total of 1,631 3-min grid cells were recorded containing Leucaena from the United States to Peru. Of these, 1,474 contain just a single species, reflecting the predominance of allopatric species distributions (SI Figs. 4 and 5). Backyard sympatry between pairs of species is common across S-C Mexico (Fig. 1a and SI Figs. 4 and 5) with 31 of the 120 possible species combinations (of the 16 species in that area) detected. Although the widely cultivated L. esculenta, L. leucocephala, and L. pallida dominate sympatry, all 13 species used for food were found in sympatry somewhere (SI Table 1). Mixtures of three species occur commonly in northern Oaxaca and southern Puebla and sporadically in Chiapas, Morelos, and Veracruz. Higher levels of sympatry with four to six co-occurring species are found in villages around the upper Tehuacán Valley (Fig. 1a). The substantially lower levels of sympatry found when records representing cultivated trees are excluded (SI Fig. 4) demonstrate that sympatry in Leucaena is predominantly artificial and attributable to cultivation. Cultivation of multiple species together has been driven by pursuit of diverse seed and pod qualities and production seasons and the potential for year-round pod production (SI Table 1).
The overall geography of Leucaena food use and the concentration of sympatry and spontaneous hybrids in the upper Tehuacán Valley (Fig. 1 and SI Fig. 5) are consistent with the archaeological data suggesting early cultivation in that area and with the emerging focus on S-C Mexico as a center for early agricultural development (11, 37). After initial cultivation, it is clear that Leucaena has been brought into cultivation many times independently, at scattered localities across a broad swathe of S-C Mexico, involving at least nine species and, for three species, extensive translocation. Cultivation in the absence of domestication is not unusual among Neotropical fruit trees (16, 39). For Leucaena, predomestication cultivation was extensive and involved numerous repeated and temporally and spatially isolated wild to cultivated transitions of a range of species.
Spontaneous Hybridization.
The phylogenetic analyses suggest that all five polyploid species, L. confertiflora, L. diversifolia, L. involucrata, L. leucocephala, and L. pallida may be allopolyploids (Fig. 2) and provide hypotheses for their parentage. Robustly supported placement of divergent 23L sequences from polyploid individuals with specific diploids suggest L. cuspidata and L. trichandra as parents of L. confertiflora, L. pulverulenta and L. lanceolata are parents of L. leucocephala, and L. pulverulenta and L. trichandra are probable parents of L. diversifolia (Fig. 2). With the exception of the evidence for a hybrid origin of L. diversifolia and multiple independent origins of L. leucocephala, the results are consistent with previous data (26, 30) and a chloroplast DNA gene tree (SI Fig. 3). Anthropogenic origins of two of the five allopolyploid species, L. leucocephala and L. pallida, are suggested by several lines of evidence, and notably by the lack of known natural populations of these species. The case for L. leucocephala is particularly compelling. Despite numerous field collection efforts for this species, no natural populations (apart from weedy occurrences in Yucatan) have been located. Year-round pod production, abundant pod set, seed set on isolated trees caused by self-compatibility, and sweet seeds make L. leucocephala outstanding for food production compared with other species (Fig. 2b). These attributes mean that L. leucocephala is now the most widely cultivated species of Leucaena throughout Mexico. Despite this present-day abundance in cultivation and a 6,000-year archaeological record of Leucaena seed fragments, seeds of L. leucocephala first appeared only ≈2,300 years ago (19). Attempts to establish L. leucocephala at or beyond its drought and cold tolerance limits using irrigation or even potted trees are evidence of the continued drive to cultivate this species. A spontaneous origin of L. leucocephala would have been immediately noticed and seized on by Leucaena-conscious farmers. Such a scenario is compatible with the lack of natural populations, low levels of genetic variation among Mexican accessions of the widely cultivated subsp. glabrata favored for food use (40), and a possible origin in central Veracruz where L. leucocephala occurs sympatrically with sporadically cultivated trees of L. pulverulenta in areas where L. lanceolata is abundant.
The analyses also provide evidence for the parentage of a number of individuals that were identified as putative morphological hybrids (Fig. 2). Notably, three 23L sequence types placed in strongly supported divergent clades were recovered from putative L. × mixtec hybrids (1715 and 2066 in Fig. 2). This finding is as expected for triploids between tetraploid L. leucocephala, itself with two divergent sequence types, and one of two diploid species from the L. esculenta clade (26, 28). Spontaneous backyard hybridization in Leucaena was first suggested by the sporadic, but widespread, occurrence of this hybrid in six states in S-C Mexico (28) (Figs. 1b and 2a). As these hybrids are sterile and Leucaena is only propagated by seed and not vegetatively, all individuals must represent independent F1 hybrids and all are found as backyard trees. Leucaena × mixtec is recognized as the “guaje macho” because of its lack of pods but remains relatively common despite culling of trees, probably caused by how frequently this hybrid arises and inadvertent cultivation. Many L. × mixtec trees are reported to have grown from seed purchased as “guaje verde” (L. leucocephala), which were either sown deliberately or discarded in backyards. This finding is in line with evidence that most L. × mixtec individuals had L. leucocephala as the female parent (28). Although it is of no use for food, this hybrid provides tangible evidence of spontaneous backyard hybridization and the complex and serendipitous pathways by which such spontaneous hybrids may be disseminated, often unwittingly, via backyards and kitchen middens.
Individuals 2055 and 1625 (Fig. 2) are hybrids between tetraploid L. leucocephala and the tetraploids L. confertiflora and L. pallida, respectively, in line with sympatric occurrences of these species in backyards in San Pedro Chapulco, Santiago Acatepec, and Santa Catalina Oxolotepec where they are found (Fig. 1b). That these hybrids have L. leucocephala as one parent reflects the very widespread cultivation of this species. Current evidence suggests that these hybrids are sporadic and uncommon. However, field survey of Leucaena hybrids has only scratched the surface so far; only four villages were surveyed in detail for hybrids in this study. Furthermore, backyard plants are often neglected by botanical collectors, and it is likely that backyard Leucaena hybrids are more common than current collections suggest. Evidence for hybrids arising in natural populations of Leucaena is lacking, with the exception of putative hybrids between L. collinsii and L. magnifica in southeast Guatemala (26) (SI Fig. 6).
Conclusions
Edgar Anderson (41, 42) was among the first to point out the importance of disturbed sites such as kitchen middens and backyard gardens as suitable habitats for spontaneous hybridization, where otherwise isolated plant species were brought into sympatry in cultivation. Anderson (42) suggested that these sites were important venues for the generation of hybrids and polyploids involved in crop domestication, but this process has rarely been demonstrated in practice. For Leucaena, we have shown just how prevalent and influential the bringing together of species, casually or intentionally, in cultivation in backyards can be. Predomestication cultivation of Leucaena species has resulted in extensive artificial sympatry and a complex series of geographically dispersed spontaneous hybrids and at least one and possibly more polyploids. The scale and complexity of spontaneous hybridization in Leucaena is far greater than that documented for any other Mexican plant domesticate so far (2) to our knowledge.
Two other genera that have produced important Mexican plant domesticates, Agave and Opuntia, show striking similarities to Leucaena. Although genetic studies of hybrids and polyploids in Agave and Opuntia and surveys of their cultivation are patchy, the evidence for high levels of sympatry in cultivation and spontaneous interspecific hybridization for both these genera is nonetheless compelling (17, 20). For example, in the Bajío region of Guanajuato fruits and nopales are harvested from 16 species of Opuntia that are wild, tolerated in fields, encouraged in “nopalera,” and cultivated in gardens (43). At least five species are widely cultivated alongside other minor cultivated and wild species (20). Ease of hybridization and vegetative propagation are well known in Opuntia (18), and it has been suggested that the most widely cultivated domesticate, Opuntia ficus-indica, an octoploid, arose as a spontaneous hybrid in cultivation (18). The so-called Man-Agave symbiosis (17) is similar. Ease of vegetative propagation and a long archaeological record suggest that Agaves were among the earliest cultivated plants in Mexico. At least 15 species of Agave are widely used and cultivated, and high levels of sympatry in cultivation are common across S-C Mexico. A suite of putative hybrids has been identified (17), including possible spontaneous hybrid origins for the three polyploid domesticates Agave fourcroydes, Agave sisalana, and Agave tequilana after translocation and cultivation (17, 44).
Flannery (45) suggested Agave, cactus fruits, and legume seeds as three of six fundamental procurement systems involved in archaic foraging in Mesoamerica. Taken together Leucaena, Agave, and Opuntia comprise three of the dominant perennial plants cultivated in S-C Mexico today. In all three genera, domestication has apparently been facilitated by spontaneous hybridization after extensive predomestication cultivation. In each case there is evidence that the prominent species in cultivation, L. leucocephala, O. ficus-indica and A. sisalana/A. fourcroydes/A. tequilana, have hybrid origins most likely after cultivation. There is also evidence to suggest that hybridization has been important in many other Mesoamerican crops, such as Cucurbita, Phaseolus, Capsicum, Lepidium, Hyptis, and Panicum (2, 46). It seems that, as suggested by Anderson (42), the simple step of bringing species together, consciously or casually, in dump heaps and informal backyard orchards has played a central role in Mesoamerican crop domestication. As we unravel the domestication history of other crops in Mesoamerica and elsewhere, we can expect other examples where hybridization triggered by early cultivation has been instrumental in generating much of the diversity that we see today.
Methods
Ethnobotanical Survey.
We assessed present-day use, marketing, cultivation, and translocation of Leucaena throughout its native range from Peru to the United States from published studies (22, 23, 33) and new data assembled from interviews with farmers and people harvesting, processing, and selling pods during 14 field collection expeditions that have allowed us to observe all 22 species and hybrids (SI Table 1). In addition, four villages in southern Puebla, San Pedro Chapulco, Santa Catalina Oxolotepec, Santiago Acatepec, and Santa Ana Teloxtoc, where high levels of sympatry were found, were surveyed for hybrids in detail in 2001 and 2003.
Geographic Data.
Present-day species and hybrid distributions were mapped from specimens in 21 herbaria geo-referenced where necessary from 1:250,000 maps and supplemented by records of species occurrences observed by the authors. These provided a total of 2,617 data points for the 22 species and two named hybrids. Further details on each specimen are available at http://herbaria.plants.ox.ac.uk/bol/?leucaena. A further 332 herbarium records were discarded because of the lack of detailed locality data. Of the 2,617 records, 802 represented trees in cultivation based on conservative assessment of field observations, herbarium specimen notes, and data from local informants. Although distribution maps inevitably reflect collection effort, this problem is less serious for Leucaena where the very large number of records for a genus of this size reflect intensive collecting of Leucaena as forage genetic resources (47). Nevertheless, high levels of sympatry among species and the occurrence of hybrids are likely to be more frequent in S-C Mexico than currently depicted in Fig. 1. Taxonomy follows Hughes (24). Sympatry among species was mapped by using a circular neighborhood with a radius of 5 km to assign records to a grid cell by exporting data points from BRAHMS (www.brahmsonline.com) to the DIVA-GIS software (www.diva-gis.org). This approach produces a smoother grid that is less biased by the origin of the grid and less sensitive to errors in coordinate data than standard grid-cell approaches (www.diva-gis.org). This neighborhood size reflects estimated distances for pollen flow facilitated by Xylocopa bees that visit Leucaena flowers and groom for pollen. Separate maps were assembled in this way including (Fig. 1a and SI Fig. 5) and excluding (SI Fig. 4) cultivated records.
Sequence Data and Phylogenetic Analysis.
Seventy-nine accessions representing all diploid and tetraploid species (most with multiple accessions), 15 putative hybrids, and two outgroups (chloroplast data only) were sampled. Three noncoding chloroplast loci (psbA-trnH and the two trnK introns) were sequenced by using standard protocols. In addition, one anonymous nuclear DNA sequence locus, termed 23L, was selected as the most informative of a set of sequence characterized amplified region-based loci developed for Leucaena (31). For finding resolution within Leucaena this approach worked well (31), but with the limitation that 23L fails to amplify in outgroup taxa. The 23L gene tree is thus rooted internally (Fig. 2 and SI Fig. 6). The major clades highlighted in Fig. 2 and SI Fig. 6 are consistent with previous data (26, 30, 31). For all hybrid, polyploid, and the majority of diploid accessions, 23L PCR products were cloned by using standard protocols to sample intraindividual sequence diversity attributable to hybridization or heterozygosity. Voucher details and GenBank accession numbers are provided in SI Table 2. Numbers of distinct sequence types recovered from individuals are generally in line with expectations for different ploidy levels (SI Fig. 6). With two notable exceptions, both probably caused by hybridization, sequences of diploids group according to species (SI Fig. 6). Extensive cloning and sampling in the 23L gene tree and congruence with previous internal transcribed spacer and chloroplast restriction fragment length polymorphism gene trees (26) provide no indications of underlying paralogy problems attributable to multiple copies of the 23L locus (SI Fig. 6). The assumption in phylogenetic analyses of multiple DNA alignments that all sites share the same evolutionary history is violated if genomic or PCR-mediated recombination has occurred among sequences, causing errors in tree estimation (48, 49). We identified five potential recombinant sequences, all from polyploid or hybrid accessions, based on excess homoplasy on terminal branches (50) and the φ test (51). These were excluded from the analysis. Sequences were aligned by using CLUSTALX and manually adjusted. Indels were coded by using a simple gap-coding method (52). Separate parsimony analyses of the chloroplast and 23L data sets were conducted with NONA (www.cladistics.com) by using 1,000 random addition sequences, tree bisection and reconnection, holding 100 trees per replication and swapping to completion.
Supplementary Material
Acknowledgments
We thank Rob Forrest, Helga Ochoterena, Mario Sousa, Jose Luis Contreras, and Jose Dimas Rodriguez for help with fieldwork, Sergio Zárate and Alejandro Casas for sharing ideas on Leucaena domestication, and John Pannell and two anonymous reviewers for helpful and constructive comments. This work was supported by the Leverhulme Trust, National Science Foundation Grant EF-0542228 (to C.D.B.), the Royal Society, and the United Kingdom Department for International Development.
Abbreviation
- S-C
south-central.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EF643842–EF644103 and EF682017–EF682031).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702193104/DC1.
References
- 1.Abbott RJ. Trends Ecol Evol. 1992;7:401–405. doi: 10.1016/0169-5347(92)90020-C. [DOI] [PubMed] [Google Scholar]
- 2.Bye R. In: Biological Diversity of Mexico: Origins and Distribution. Ramamoorthy TP, Bye R, Lot A, Fa J, editors. New York: Oxford Univ Press; 1993. pp. 707–731. [Google Scholar]
- 3.Ellstrand NC, Prentice HC, Hancock JF. Annu Rev Ecol Syst. 1999;30:539–563. [Google Scholar]
- 4.Ellstrand NC, Schierenbeck KA. Proc Natl Acad Sci USA. 2000;97:7043–7050. doi: 10.1073/pnas.97.13.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doebley J. In: Molecular Systematics of Plants. Soltis PS, Soltis DE, Doyle JJ, editors. New York: Chapman & Hall; 1992. pp. 202–222. [Google Scholar]
- 6.Smartt J, Simmonds NW, editors. Evolution of Crop Plants. New York: Longman Scientific; 1995. [Google Scholar]
- 7.Hancock JF. Plant Evolution and the Origin of Crop Species. Wallingford, UK: CABI; 2004. [Google Scholar]
- 8.Wendel JF, Cronn RC. Adv Agron. 2003;78:139–186. [Google Scholar]
- 9.Emshwiller E. In: Documenting Domestication: New Genetic and Archaeological Paradigms. Zeder MA, Bradley DG, Emshwiller E, Smith BD, editors. Berkeley: Univ California Press; 2006. pp. 153–168. [Google Scholar]
- 10.Smith BD. The Emergence of Agriculture. New York: Scientific American Library; 1998. [Google Scholar]
- 11.Smith BD. Proc Natl Acad Sci USA. 2001;98:1324–1326. doi: 10.1073/pnas.98.4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith BD. Science. 1997;276:932–934. [Google Scholar]
- 13.Piperno DR, Flannery KV. Proc Natl Acad Sci USA. 2001;98:2101–2103. doi: 10.1073/pnas.98.4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuoka Y, Vigouroux Y, Goodman MM, Sanches J, Buckler E, Doebley J. Proc Natl Acad Sci USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L. Proc Natl Acad Sci USA. 2002;99:535–540. doi: 10.1073/pnas.012577299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller A, Schaal B. Proc Natl Acad Sci USA. 2005;102:12801–12806. doi: 10.1073/pnas.0505447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentry HS. Agaves of Continental North America. Tucson: Univ Arizona Press; 1982. [Google Scholar]
- 18.Griffith MP. Am J Bot. 2004;91:1915–1921. doi: 10.3732/ajb.91.11.1915. [DOI] [PubMed] [Google Scholar]
- 19.Zárate PS. Econ Bot. 2000;54:477–499. [Google Scholar]
- 20.Casas A, Barbera G. In: Cacti: Biology and Uses. Nobel PS, editor. Berkeley: Univ California Press; 2002. pp. 143–162. [Google Scholar]
- 21.Casas A, Caballero J. Econ Bot. 1996;50:167–181. [Google Scholar]
- 22.Berlin B., Breedlove DE, Raven PH. Principles of Tzeltal Plant Classification. New York: Academy Press; 1974. [Google Scholar]
- 23.Casas A, Viveros JL, Caballero J. Etnobotánica Mixteca: Sociedad, Cultura y Recursos Naturales en la Montana de Guerrero. Mexico D.F., Mexico: Consejo Nac Cultura Artes; 1994. [Google Scholar]
- 24.Hughes CE. Syst Bot Monogr. 1998;55:1–244. [Google Scholar]
- 25.Cardoso MB, Schifino-Wittmann MT, Bodanse-Zanettini MH. Bot J Linn Soc. 2000;134:549–556. [Google Scholar]
- 26.Hughes CE, Bailey CD, Harris SA. Am J Bot. 2002;89:1057–1073. doi: 10.3732/ajb.89.7.1057. [DOI] [PubMed] [Google Scholar]
- 27.Sorensson CT, Brewbaker JL. Am J Bot. 1994;81:240–247. [Google Scholar]
- 28.Hughes CE, Harris SA. Plant Syst Evol. 1994;192:177–197. [Google Scholar]
- 29.Hughes CE, Harris SA. Plant Syst Evol. 1998;212:53–77. [Google Scholar]
- 30.Harris SA, Hughes CE, Ingram R, Abbott RJ. Plant Syst Evol. 1994;191:1–26. [Google Scholar]
- 31.Bailey CD, Hughes CE, Harris SA. Syst Bot. 2004;29:4–14. [Google Scholar]
- 32.Linder CR, Rieseberg LH. Am J Bot. 2004;91:1700–1708. [PMC free article] [PubMed] [Google Scholar]
- 33.Zárate PS. J Ethnobiol. 1999;19:1–23. [Google Scholar]
- 34.MacNeish RS. Am Philos Soc Trans. 1958;48:1–210. [Google Scholar]
- 35.Byers DS, editor. The Prehistory of the Tehuacan Valley: Environment and Subsistence. Austin: Univ Texas Press; 1967. [Google Scholar]
- 36.Flannery KV. Guilá Naquitz: Archaic Foraging and Early Agriculture in Oaxaca, Mexico. New York: Academy Press; 1986. [Google Scholar]
- 37.Smith BD. Latin Am Antiquity. 1997;8:342–383. [Google Scholar]
- 38.Zohary D, Hopf M. Domestication of Plants in the Old World. Oxford: Oxford Univ Press; 1988. [Google Scholar]
- 39.Clement CR. Econ Bot. 1999;53:188–202. [Google Scholar]
- 40.Harris SA, Hughes CE, Abbot RJ, Ingram R. Silvae Genet. 1994;43:159–167. [Google Scholar]
- 41.Anderson E. Introgressive Hybridization. New York: Wiley; 1949. [Google Scholar]
- 42.Anderson E. Plants, Man, and Life. London: Melrose; 1954. [Google Scholar]
- 43.Colunga G-M, Hernández XE, Castillo MA. Agrociencia. 1986;65:7–49. [Google Scholar]
- 44.Wienk JF. In: Evolution of Crop Plants. Smartt J, Simmonds NW, editors. New York: Longman Scientific; 1995. pp. 4–8. [Google Scholar]
- 45.Flannery KV. In: Anthropological Archaeology in the Americas. Meggers BJ, editor. Washington, DC: Anthropological Society; 1968. pp. 67–87. [Google Scholar]
- 46.Felger RS, Wilson MF. Biodiversity and Management of the Madrean Archipelago: The Ski Islands of the Southwestern United States and Northwestern Mexico. Washington, DC: US Department of Agriculture; 1994. General Technical Report RM-GTR-264. [Google Scholar]
- 47.Hughes CE. Leucaena, A Genetic Resources Handbook. Oxford, UK: Oxford Forestry Institute; 1998. [Google Scholar]
- 48.Posada D, Crandall KA. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cronn R, Cedroni M, Haselkorn T, Grover C, Wendel JF. Theor Appl Genet. 2002;104:482–489. doi: 10.1007/s001220100741. [DOI] [PubMed] [Google Scholar]
- 50.Maynard Smith J, Smith NH. Mol Biol Evol. 1998;15:590–599. doi: 10.1093/oxfordjournals.molbev.a025960. [DOI] [PubMed] [Google Scholar]
- 51.Bruen TC, Philippe H, Bryant D. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simmons MP, Ochoterena H. Syst Biol. 2000;49:369–381. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.