Abstract
Cytolethal distending toxins (CDTs) are inhibitory cyclomodulins, which block eukaryotic cell proliferation and are produced by a diverse group of Gram-negative bacteria, including Escherichia coli strains associated with intestinal and extraintestinal infections. However, the mode of transmission of the toxin gene clusters among diverse bacterial pathogens is unclear. We found that Cdt-I produced by enteropathogenic E. coli strains associated with diarrhea is encoded by a lambdoid prophage, which is inducible and infectious. The genome of Cdt-I converting phage (CDT-1Φ) comprises 47,021 nucleotides with 60 predicted ORFs organized into six genomic regions encoding the head and tail, virulence, integrase, unknown functions, regulation, and lysis. The genomic organization of CDT-1Φ is similar to those of SfV, a serotype-converting phage of Shigella flexneri, and UTI89, a prophage identified in uropathogenic E. coli. Besides the cdtI gene cluster, the virulence region of CDT-1Φ genome contains sequences homologous to a truncated cycle inhibiting factor and a type 3 effector protein. Mutation analysis of susceptible E. coli strain C600 suggested that the outer membrane protein OmpC is a putative receptor for CDT-1Φ. CDT-1Φ genome was also found to integrate into the host bacterial chromosome forming lysogens, which produced biologically active Cdt-I. Furthermore, phage induction appeared to cause enhanced toxigenicity of the E. coli strains carrying lysogenic CDT-1Φ. Our results suggest that CDT-1Φ is the latest member of a growing family of lambdoid phages encoding bacterial cyclomodulins and that the phage may have a role in horizontal transfer of these virulence genes.
Keywords: bacterial genotoxin, converting phage, cyclomodulins
Cytolethal distending toxins (CDTs) constitute a family of secreted bacterial protein toxins characterized by their capacity to inhibit proliferation of mammalian cells by inducing an irreversible cell-cycle block (1). CDT production has been reported for diverse species of pathogenic bacteria, including Escherichia coli, Actinobacillus actinomycetemcomitans, Haemophilus ducreyi, Campylobacter spp., Shigella spp., and Helicobacter spp (2). CDT has been shown to arrest the tumor-derived HeLa cell line in the G2/M transition phase of the cell cycle (1). Thus, CDT inhibits cell proliferation and induces the formation of enlarged mononucleated cells blocked at the G2/M boundary and causes eventual cell death.
CDT was first reported in 1987 as a new type of protein toxin produced by E. coli O128 isolated from children with gastroenteritis (2, 3). Since then, the toxin has been detected in different categories of pathogenic E. coli and various other Gram-negative bacteria (2). At least five different CDTs (CDT-I through CDT-V) have been reported in E. coli strains associated with different syndromes (4–8). Whereas CDT-I, CDT-II, and CDT-IV have been detected in E. coli strains mainly causing human gastrointestinal and urinary infections (7–9), CDT-III was detected in E. coli strains carrying genes for cytotoxic necrotizing factors (cnf1 or cnf2) and/or Shiga toxins (stx1 and/or stx2), isolated from diarrhea patients or animals with diarrhea or septicemia (7, 9, 10). CDT-V was detected in E. coli O157 isolates from humans with diarrhea or hemolytic-uremic syndrome (7, 10, 11). CDTs of all strains are composed of three polypeptides (CdtA, CdtB, and CdtC), which form a ternary complex required for the toxin activity (2). Whereas the holotoxin binds to an unknown receptor on the target cell by using CdtA and CdtC subunits, the CdtB subunit enters into the cell to reach the nucleus (2) and causes DNA damage through its DNase I activity (12, 13). Therefore, CDT has recently been recognized as a genotoxin (14) and a member of the growing family of bacterial toxins and effectors designated as “cyclomodulins,” which interfere with the eukaryotic cell cycle (1).
Although CDTs are produced by diverse pathogenic bacterial species, the mechanism associated with possible horizontal transfer of the various cdt gene clusters resulting in wide distribution of the toxin genes among pathogenic bacteria is not adequately known. Here, we show that Cdt-I produced by enteropathogenic E. coli (EPEC) strains isolated from diarrheal patients is encoded on a lysogenic bacteriophage, Cdt-I converting phage (CDT-1Φ). Furthermore, the virulence region of the phage genome contains not only the cdtI gene cluster but also possible other virulence-associated genes, including those encoding a truncated cycle inhibiting factor (Cif) and a type 3 effector protein. Infectious CDT-1Φ particles produced from these strains infected a recipient strain, resulting in horizontal transfer of these virulence gene clusters.
Results
Cdt-I Is Encoded by a Lysogenic Bacteriophage.
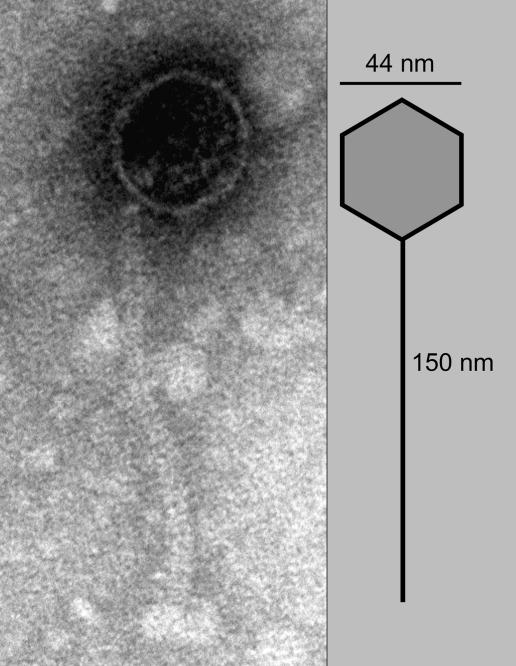
The CDT-positive E. coli strains were initially identified by screening isolates from patients with acute diarrhea by using DNA probes or PCR assays and later confirmed by using the HeLa cell assay for CDT production (15). The CDT-positive isolates belonged to the enteropathogenic category of diarrheagenic E. coli (15) and were of the O86a, O127, and O142 serogroups [see supporting information (SI) Table 3]. The CDT-positive strains were analyzed for possible production of extracellular phage particles by induction with mitomycin C. Filter-sterilized supernatants of mitomycin C-induced cultures were tested for the presence of phages by plating on a potential recipient strain E. coli C600. The supernatant fluid appeared to have infectious phage particles as indicated by formation of clear or translucent plaques on the bacterial lawn (Table 1). Plaque hybridization was used to investigate whether these plaques were formed by a bacteriophage carrying genes for Cdt-I. All plaques formed on C600 reacted with a cdtIB gene probe, but the probe did not hybridize with colonies of native C600 or plaques formed by Stx2 phage used as a control. Thus Cdt-I is encoded on a phage genome, which is viable and infectious. This finding was further confirmed by analyzing DNA prepared from isolated phage particles, which were pretreated with DNase I to remove any exogenous DNA, while retaining the phage DNA packaged in particles, and hence protected from DNase I activity. Furthermore, strain E. coli C600 when lysogenized with CDT-1Φ produced biologically active Cdt-I as was determined in the HeLa cell assay. Finally, electron microscopic examination of CDT-1Φ revealed that the phage particles had hexagonal heads (≈44 nm) and long tails (≈150 nm), characteristic of members of the family Siphoviridae (Fig. 1).
Table 1.
Induction of CDT-1Φ in Cdt-I-positive EPEC strains using mitomycin C
| Strain | Description | Titer of CDT-1Φ particles |
|---|---|---|
| NT3363 | Cdt-I-positive O127:H7 EPEC strain | 1.0 × 103 |
| GB469 | Cdt-I-positive O127:H7 EPEC strain | 1.0 × 103 |
| GB1807 | Cdt-I-positive O127:H7 EPEC strain | 1.0 × 101 |
| VTE1456 | Cdt-I-positive O142:H6 EPEC strain | 4.5 × 105 |
| GB1371 | Cdt-I-positive O86a:H34 EPEC strain | Not detectable |
| VTE1488 | Cdt-I-positive O86a:H34 EPEC strain | Not detectable |
| D05491 | Cdt-I-positive O86a:H34 EPEC strain | Not detectable |
| C600 (cdtI-phage) | E. coli C600 lysogenized with CDT-1Φ prepared from E. coli strain NT3363 | 2.5 × 106 |
Shown are median values of five observations.
Fig. 1.
Electron micrograph showing the morphology of a CDT-1Φ virion. The hexagonal head (≈44 nm) and the long tail (≈150 nm) are characteristic of the family Siphoviridae.
CDT-1Φ Carries Other Virulence Genes In Addition to cdtI Genes.
Determination of the nucleotide sequence of CDT-1Φ genome revealed that the phage genome comprises 47,021 nucleotide pairs and contains 60 predicted ORFs (Fig. 2 and SI Fig. 4). CDT-1Φ appeared to be a lambdoid phage, considering the sequence homology with other lambdoid phages (SI Table 4) and its genome organization. Genome structure of CDT-1Φ could be functionally grouped into six regions, including genes for head and tail, virulence, integrase, unknown function, regulation, and lysis (Fig. 2). The cdtI gene cluster is located adjacent to and upstream of the integrase gene and contains genes for all three subunits (CdtA, CdtB, and CdtC) of the Cdt-I holotoxin. Interestingly, we found that the genomic region of CDT-1Φ corresponding to virulence contains sequences homologous to a number of other putative virulence-associated genes in addition to the complete cdtI gene cluster (SI Table 4).
Fig. 2.
Comparison of the functional structure of the CDT-1Φ genome with those of prophage UTI89, prophage-like element B171, and bacteriophage SfV. The numbers in the bars indicate the homology between CDT-1Φ and each phage (UTI89 prophage, B171-prophage-like element, or bacteriophage SfV). The numbers below the bars indicate the size of each region, and the vertical arrow at the top indicates the attP site.
EPEC strains are known to use a type 3 secretion system (TTSS) encoded by the “locus of enterocyte effacement” (LEE) to subvert and attach to epithelial cells through the injection of effector molecules (16). Remarkably, the virulence region of the CDT-1Φ genome was found to contain sequence homologous to the gene for a TTSS-translocated effector molecule, Cif, which blocks cell cycle at the G2/M transition (1). The cif-like gene in the CDT-1Φ was however, apparently truncated and presumably nonfunctional. It was reported previously that Cif is not encoded by the LEE but by a lambdoid prophage present in EPEC and enterohemorrhagic E. coli. Besides the truncated cif gene, we also found the gene for a non-LEE encoded type 3 effector protein belonging to the NleH family of effector proteins (17), located adjacent and upstream of the cif gene. In addition, a gene homologous to DNA adenine methylase (dam) gene in SfV, a serotype-converting bacteriophage of Shigella flexneri (18), was also found in the putative regulatory region of the CDT-1Φ genome (SI Fig. 4 and SI Table 4). Dam has been demonstrated to be a virulence factor in Salmonella typhimurium (19). Thus, CDT-1Φ may have a role in clustering and interstrain transfer of diverse virulence-associated genes.
The genomic organization of CDT-1Φ was found to be more similar to those of phage SfV of S. flexneri (18) and prophage UTI89 (20) of an uropathogenic E. coli strain than Stx2Φ-I (21) (Fig. 2 and SI Fig. 5). Although the overall sequence homology of CDT-1Φ with UTI89 prophage was high, UTI89 lacked the virulence region containing the cdt gene cluster. The location of toxin genes in CDT-1Φ was also considerably different from that of the Stx phages. In the CDT-1Φ, the cdt gene cluster is located upstream of the attP site (Fig. 2 and SI Fig. 5), whereas in the Stx phage, stx genes are located in the late regulatory region close to the 3′ end of antiterminator gene Q (22). Thus despite similarity in the overall genomic organization, the region related to virulence is quite unique in each phage. The sequence homology of the different genomic regions of CDT-1Φ genome and other prophages are summarized in Fig. 2 and SI Table 4.
Outer Membrane Protein OmpC Is a Putative Receptor for CDT-1Φ.
Because bacterial outer membrane proteins such as LamB, FadL, OmpC, and OmpF are known to be the receptors for lambda phage, Stx2Φ-I, and phages T2 and T4 (23), we examined the effect of defined mutations of lamB, fadL, ompC, and ompF in E. coli K-12 strains (23) on the susceptibility of the bacterium to CDT-1Φ infection. The plaque-forming ability of CDT-1Φ on the wild type and corresponding mutants are summarized in Table 2. When a lamB, ompF, or fadL mutated E. coli strain was infected by CDT-1Φ, adequate number of plaques (2 × 103 to 1 × 104) were formed on the bacterial lawns. However, an ompC-mutated strain was completely resistant to plaque formation by CDT-1Φ. Additionally, when we introduced a plasmid containing a cloned ompC gene (pMA1) in the ompC-mutated strain, the susceptibility was restored to normal level (Table 2). This observation suggested that the OmpC protein might play a crucial role in the infection of E. coli by CDT-1Φ. Given that OmpC was identified as a receptor for phage T4 (23), and that certain uropathogenic E. coli strains including a strain UPI89 harboring a prophage UTI89 hyperexpressed OmpC protein (20), it is likely that OmpC also acts as a receptor for CDT-1Φ. However, further studies will be required to confirm this assumption.
Table 2.
Average plating efficiency of CDT-1Φ on different E. coli strains
| Strain | Description | Titer of CDT-1Φ particles |
|---|---|---|
| C600 | Reference E. coli strain | 2.0 × 103 |
| C600 (stx2Φ-I) | C600 lysogenized with stx2Φ-I | 2.5 × 103 |
| C600 (stx2Φ-II) | C600 lysogenized with stx2Φ-II | 2.6 × 103 |
| MW47 | ΔlamB | 1.0 × 104 |
| RAM191 | ΔompC | Not detectable |
| RAM191 (pMA1) | RAM191 transformed with a cloned ompC gene | 2.5 × 103 |
| RK4786 | ΔompF | 9.6 × 103 |
| MW72 | ΔfadL | 8.0 × 102 |
CDT-1Φ used in the assay was isolated from E. coli strain NT3363.
CDT-1Φ Genome Integrates Into the Gene for Peptide Chain Release Factor RF-3.
Although CDT-1Φ was produced in the supernatants of mitomycin C-induced cultures of Cdt-I-positive EPEC strains, no detectable phage was produced in the absence of mitomycin C. This observation suggested that the CDT-1Φ existed in a lysogenic form in these strains. Subsequently, we were also able to isolate CDT-1Φ lysogens of strain C600 under laboratory conditions, by analyzing bacteria from translucent plaques produced by CDT-1Φ on a lawn of C600. We therefore conducted further analysis to define the region where CDT-1Φ genome integrates into the bacterial chromosome.
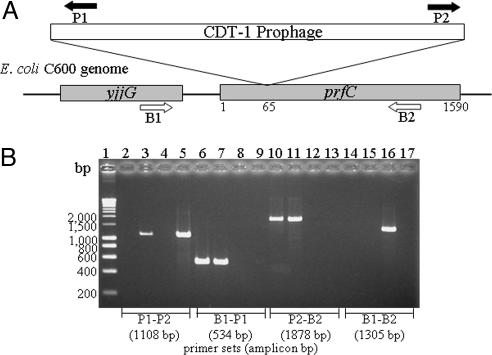
The integrase gene of CDT-1Φ showed a 98% homology with that in prophage UTI89 of uropathogenic E. coli strain (Fig. 2 and SI Table 4). Because prophage UTI89 has been reported to reside in the gene coding for peptide chain release factor RF-3 (20), we examined whether CDT-1 prophage also integrated into a similar region of the E. coli chromosome. Accordingly, PCR assays were conducted by using a combination of primers, which can either amplify the junctions of attP and attB sites in the lysogen or the bacterial attB site and phage attP site of the unintegrated phage genome (Fig. 3 and SI Table 5). As expected, when amplified with B1 and P1 primers or with P2 and B2 primers, amplicons of 530 or 1,900 bp, respectively, were produced from E. coli strains NT3363 and C600 (CDT-1Φ), which had integrated phage genome. However, these primer sets did not produce a PCR product when isolated phage genome or the native bacteria was used as templates (Fig. 3). On the other hand, amplification was observed from the isolated phage or bacterial genomes when primer sets corresponding to the attP site (P1 and P2) and attB site (B1 and B2) were used, and the size of the amplicons agreed with the expected sizes. These data indicated that CDT-1 prophage integrates into the gene coding for peptide chain release factor RF-3 in the E. coli chromosome. This integration site is unlike the lambdoid phages encoding Stxs, which are known to integrate into the wrbA gene downstream of the phage PR′ promoter and upstream of the lysis region (21).
Fig. 3.
Determination of the region of insertion of CDT-1Φ into the E. coli genome. (A) Schematic diagram showing the region of integration of CDT-1Φ genome in the chromosome of strain C600. The yjjG gene encodes nucleoside 5′-monophosphate phosphohydrolase, and prfC encodes peptide chain release factor RF-3. Arrows indicate the regions where PCR primers P1 and P2, as well as B1 and B2, anneal to the integrated phage and bacterial genome (see Results for details). (B) Ethidium bromide-stained agarose gel showing the amplicons from different combinations of PCR primers and template DNA. Lane 1, HyperLadderI; lanes 2–5, P1 and P2; lanes 6–9, B1 and P1; lanes 10–13, P2 and B2); lanes 14–17, B1 and B2; lanes 2, 6, 10, and 14, E. coli strain NT3363; lanes 3, 7, 11, and 15, E. coli strain C600 with CDT-1Φ; lanes 4, 8, 12, and 16, E. coli strain C600; lanes 5, 9, 13, and 17, CDT-1Φ genome. The combination of primers used and expected sizes of the respective amplicons are indicated below the gel.
CDT-1Φ Induction Enhances Cdt-I Activity.
To determine whether induction of the prophage in CDT-1Φ lysogens increased toxigenicity, Cdt activity of E. coli strain NT3363 and E. coli C600 lysogenized with CDT-1Φ was examined by using the HeLa cell assay. Cultures were grown in the presence and absence of mitomycin C, and different dilutions of crude toxin samples from cell-associated fraction (CF) and supernatant fraction (SF) were tested on HeLa cells (SI Table 6). When cultures were grown in the presence of mitomycin C, the mean titer of Cdt activity in CF was between 8- and 32-fold higher, and that in SF was 8- to 64-fold higher compared with the corresponding cultures grown in the absence of mitomycin C (SI Table 6). These results suggested that mitomycin C-stimulated phage induction might have enhanced Cdt activity in the lysogens. Notably, it has been proposed previously that expression of Stx also requires phage induction (22).
Genome Diversity of CDT-1 Prophage.
Although all strains analyzed in our study were positive for the presence of cdt genes and produced biologically active toxin, all Cdt-I-positive strains did not produce infectious CDT-1Φ particles. We presumed that there might be diversity in CDT-1 prophages carried by different strains. Southern hybridization analysis was conducted to determine possible genetic diversity among the integrated CDT-1Φ genome in different Cdt-I-positive strains. We used six different DNA probes specific for the genomic regions encoding head and tail, integrase, virulence, unknown functions, regulation and lysis. Hybridization of HindIII-digested genomic DNA of different Cdt-I-positive strains revealed restriction fragment length polymorphisms indicating genomic diversity in the CDT-1 prophages carried by different Cdt-I-positive strains (SI Fig. 6). Although the regions encoding integration and virulence functions of the CDT-1Φ genomes were found to be mostly conserved, there were possible variations in the other genomic regions, because additional bands were detected corresponding to these probes. However, these additional bands could also have generated from other phages having homology to CDT-1 prophage and that may exist in these strains together with the CDT-1 prophage. Notably, the genomic sequence of CDT-1Φ has homology with UTI89 prophage (20) and the prophage-like element of EPEC strain B171 (Fig. 2 and SI Table 4), although these latter phages do not have the virulence regions containing the cdt gene cluster.
Discussion
Bacterial protein toxins such as diphtheria toxin, botulinum neurotoxin, Stx, and cholera toxin are known to be encoded on bacteriophages (24). CDT constitutes a novel class of bacterial toxin termed genotoxin (14) or cyclomodulin (1) because of its unique mode of action. In a previous study, sequence flanking cdtV genes in E. coli was suggested to have homology with bacteriophages P2 and lambda (8), although infectious phage production by the bacterial strain was not reported. The present study provides direct evidence for production of CDT-1Φ by EPEC strains isolated from acute diarrhea patients (15). We further showed that the cdtI gene cluster is transferred by CDT-1Φ to a recipient strain, which then produce biologically active Cdt-I toxin.
The genome of CDT-1Φ reveals a highly mosaic structure with homology to sequences derived from several bacteriophages and bacterial genomes of different categories of pathogenic E. coli, as well as Shigella sonnei, S. flexneri, and S. typhimurium (SI Table 4), suggesting that the evolution of these phages and bacterial strains involved extensive horizontal exchange of genetic material. Interestingly, the genes responsible for head and tail, integrase, regulation, and lysis are highly conserved between CDT-1Φ and a previously described putative prophage UTI89 of uropathogenic E. coli (20). However, the genomic regions corresponding to virulence and “unknown functions” were not conserved between CDT-1Φ and prophage UTI89. Although the UTI89 prophage is not known to encode all functions required for production of infectious phage, we propose that the CDT-1Φ might have evolved from a precursor phage similar to prophage UTI89 by acquisition of the cdtI gene cluster.
Although the overall GC content in CDT-1Φ genome is ≈49.1%, which is similar to that of E. coli (50%) (25), the GC content in the virulence region and two ORFs located in the late region close to the 3′ end of the antiterminator gene Q is ≈30–40%. These results further indicated that the latter genes might have been recently acquired by a previously existing E. coli phage through multiple gene transfer events. This observation constitutes another example of how phage evolution contributes to increasing the virulence of its host bacterium. Although the donor for these recently acquired genes remains to be identified, the family of lambdoid phages encoding bacterial virulence factors appears to be expanding.
In diarrheagenic E. coli strains, the most well studied phage-encoded toxins are known to be encoded by Stx phages of Stx-producing E. coli (16, 22). However, genetic organization of the CDT-1Φ was found to be more closely related to those of SfV and prophage UTI89 than the Stx-converting phages. Although the regions encoding head and tail proteins and the lysis region in CDT-1Φ have homology to that in lambda and Stx phages, the locations of virulence regions in CDT-1Φ and Stx phages were found to be significantly different (SI Fig. 5). Whereas the cdtI gene cluster is located upstream of the attP site on the CDT-1Φ genome, the stx genes are located in the late regulatory region close to the 3′ end of antiterminator gene Q (22) of Stx phages (SI Fig. 5). The mechanism of Stx expression and its release by Stx phage lysogens is known to involve the activation of RecA by an SOS response-inducer such as mitomycin C, leading to autocleavage of the repressor CI. Subsequently, a series of transcriptions including that of the stx genes are initiated, and a regulatory cascade leads to the lytic phase. Thus, expression and release of Stx are associated with the lytic phase of the Stx phage (22). On the other hand, although expression and release of Cdt-I were also induced by mitomycin C (SI Table 6), the location of cdtI gene is not associated with the late regulatory region. Further studies are thus required to determine the mechanism of CDT-1Φ induction.
When cultured in the presence of mitomycin C, Cdt-I and CDT-1Φ particles were efficiently released from most wild-type CDT-1Φ lysogenic E. coli strains, e.g., NT3363. However, although expression of Cdt was enhanced, the Cdt protein was not efficiently released from E. coli C600 lysogenized with CDT-1Φ and a few other naturally occurring strains studied (SI Table 6). As a result, these strains showed high Cdt activity in the CF but low activity in the SF of the cultures (SI Table 6). We presume that some of the wild-type strains may have a unique system to enhance Cdt-I secretion, which is absent in the laboratory strain C600. Together these observations suggest that the biology of lambdoid phages encoding bacterial virulence factors may have considerable diversity. However, the relationship of phage induction with virulence may be common for other lambdoid phages, including Stx phages, and the phage induction probably occurs more efficiently under in vivo conditions. It has been suggested that Stx production in the human intestine could be enhanced through the infection of susceptible nontoxigenic E. coli with phage released by toxigenic E. coli (26).
The coevolution of bacterial pathogens and genetic elements, including pathogenicity islands and phages encoding virulence factors, has been observed in several species. E. coli is a member of the normal intestinal microflora of human and animals. However, certain E. coli strains have acquired virulence potential by acquisition of virulence-associated genetic loci through a variety of mobile elements, including transposons or phages. Often these elements encode multiple factors enabling the bacteria to colonize its host and cause disease. The genomic region related to virulence in CDT-1Φ contains genes for other virulence-associated factors in addition to the cdt genes (Fig. 2, SI Fig. 4, and SI Table 4). These genes include those coding for a truncated Cif and an NleH-like type 3 effector protein. Cif was identified as the first type 3 effector molecule to be encoded outside the LEE in EPEC (1). NleH was identified as non-LEE-encoded type 3 effector protein in Citrobacter rodentium and enterohemorrhagic E. coli O157 Sakai strain (17). Tobe et al. (17) identified 39 proteins as non-LEE-encoded type 3 effector proteins, which occur in >20 exchangeable effector loci in enterohemorrhagic E. coli O157 Sakai strain. These loci are always located just downstream of the tail fiber genes, and each locus contains more than one effector gene that stands out from their host phage backbone in possessing an extreme bias toward low GC content. The location and GC contents for the truncated Cif and NleH-like protein in CDT-1Φ genome was thus similar to the previously described prophages. Moreover, two ORFs (53 and 54), which were found to be located immediately downstream of the gene for Q protein in CDT-1Φ, resemble the location of stx genes in Stx phages (22, 23). These two ORFs have low GC content (32% and 35%), indicating that they might have also been acquired recently from an exogenous donor other than E. coli. Further studies are required for identifying the function of these two additional putative virulence-associated genes in CDT-1Φ. Thus despite some diversity the lambdoid prophages encoding virulence in E. coli share an underlying general theme, which allows their host strains to acquire additional genes. Thus the assembling of virulence genes by phages and the enhanced virulence of their host E. coli strains are evolutionary linked.
The OmpC protein was identified as a putative receptor for CDT-1Φ in this study. Interestingly, OmpC was identified as a receptor molecule for phage T4 (23), and some uropathogenic E. coli strains including a strain harboring prophage UTI89 highly expressed OmpC protein (20). It is possible that CDT-1Φ preferably infects diverse E. coli strains, which highly express OmpC. We presume that the phage conversion occurs more efficiently in human or animal guts than in laboratory experiments. The genetic mosaicisms observed among different lambdoid phages could also be a result of extensive genetic exchanges among different phages, which likely occur in the mammalian intestine. It may be mentioned that CDT-1Φ genome has considerable homology with a wide variety of prophages detected in diverse E. coli and Shigella strains (Fig. 2 and SI Table 4).
Pathogenic E. coli can cause three clinically different syndromes, which include enteric/diarrheal disease, urinary tract infections, and sepsis/meningitis (16). Thus pathogenic E. coli strains have been classified into specific pathotypes on the basis of virulence factors. Although a number of CDT-producing E. coli (CTEC) were isolated from patients with diarrhea, urinary tract infection, and septicemia, the etiologic significance of CTEC in diseases including diarrhea has not yet been established. CTEC isolated from patients with diarrhea in developing countries mainly belong to the EPEC serogroup (2), although all EPEC do not produce Cdt (16). On the contrary, CTEC reported from developed countries belong to a variety of serogroups, mostly other than EPEC serogroups (2). Okuda et al. (27) demonstrated that Cdt produced by Shigella dysenteriae could cause diarrhea and damage in the descending colon in suckling mice. Recent reports (15, 28) also suggested that excretion of high titer Cdt-I-producing E. coli were associated with bloody diarrhea in infants. Therefore, CTEC strains could constitute another category of diarrheagenic E. coli associated with bloody diarrhea.
Bacterial toxins, which interfere with the eukaryotic cell cycle and are known as cyclomodulins, are of two categories (1). While one category inhibits cell cycle, the other promotes cellular proliferation (e.g., cytotoxic necrotizing factor, dermonecrotic toxin, and Pasteurella multocida toxin). Cdt or Cif are inhibitory cyclomodulins. Recently, cyclomodulins have also been suggested to be associated with an increased risk of developing cancer (1). Therefore, the discovery of Cdt-I-converting phage in the present study is a significant step toward understanding the possible mode of spread of genes encoding bacterial cyclomodulins, which potentially have greater public health implications.
Materials and Methods
Bacterial Strains.
Description of the E. coli strains used in this study is listed in SI Table 3. E. coli C600 was used as a recipient strain for CDT-1Φ. E. coli strain lysogenized with Stx2 phage-I (23) was used as a positive control for phage induction studies.
Isolation of CDT-1Φ from the E. coli Strains.
CDT-1Φ was induced and isolated from lysogenic strains using previously described methods with minor modification (23). Briefly, an aliquot of overnight culture of the bacterial strain was inoculated into fresh LB supplemented with 2.5 mM CaCl2 and 2.5 mM MgCl2 (Ca2+, Mg2+) and grown for 1 h. Mitomycin C was added to the culture at a final concentration of 0.5 μg/ml, and the culture was incubated with aeration for 4 h or overnight at 37°C. The culture supernatant was sterilized by filtration through 0.22-μm filters (Millipore, Bedford, MA). Aliquots of the filtrate were titrated for infectious phage particles by mixing with a fresh culture of E. coli C600, added to Luria soft agar, and poured onto Luria agar (Ca2+, Mg2+). After overnight incubation at 37°C, plates were observed for plaque formation, and the number of plaques was expressed as pfu/ml of supernatant. Whereas some plaques were clear showing lysis, some had a hazy appearance suggesting possible lysogenic plaques. Plaque hybridization assay was conducted with Hybond N+ membrane (Amersham Biosciences, Upsala, Sweden) and PCR-amplified cdtIB gene probe by using the digoxigenin DNA labeling and detection kit (Boehringer, Mannheim, Germany). When hazy plaques were observed, the bacterial cells in the plaques were isolated and tested for possible lysogeny.
Electron Microscopy of Phage Particles.
CDT-1Φ prepared from mitomycin C-induced culture supernatant of NT3363 was concentrated by ultracentrifugation (100,000 × g for 1 h) and resuspension of the precipitate. The crude preparation was used to determine the morphology of the phage as described (29). The phage particles were negatively stained with 2% uranyl acetate and examined under a transmission electron microscope (model 420T; Philips, Amsterdam, The Netherlands).
Assay for Cdt Activity.
The HeLa cell assay for Cdt was conducted as described with minor modifications (15). Briefly, HeLa cells were grown in minimal essential media with 10% FCS and 0.01% gentamycin. Aliquots of ≈104 cells in 0.2 ml of culture medium were seeded in a 96-well plate. To prepare toxin samples, overnight culture of each E. coli strain was inoculated into 3 ml of fresh tryptic soy broth and grown at 37°C to an OD600 of 0.1. The culture was continued for another 6 h in the absence and presence of mitomycin C (0.5 μg/ml). The culture was centrifuged at 12,000 × g for 10 min, and supernatant was filtrated through a 0.22-μm pore-sized Millipore filter to prepare the SF of the toxin sample. The CF was prepared by sonicating the bacterial pellet (held on ice) in PBS and collecting the supernatant after centrifugation at 12,000 × g for 10 min. The supernatant was filtered as described above and used as the CF toxin sample. Aliquots (20 μl) of the toxin samples (SF and CF) were incubated with the HeLa cells under 5% CO2 in air at 37°C. After cultivation for 72 h, the HeLa cells were treated with Giemsa stain and observed by microscopy. The toxin titer was expressed as the reciprocal of the highest dilution of the toxin sample that caused 50% of the HeLa cells in a well to be distended.
Phage DNA Extraction.
Concentrated phage suspension was prepared by pooling a number of plate lysates of strain C600 infected with CDT-1Φ. The lysates suspended in a solution containing 20 mM Tris·HCl (pH 7.5), 60 mM KCl, 10 mM MgCl2, and 10 mM NaCl was centrifuged twice at 10,000 × g, and the supernatant was filtered through a 0.22-μm pore-sized filter. The filtrate was treated with pancreatic DNase I (1 μg/ml) and RNase A (50 μg/ml) at 37°C for 2 h and extracted with phenol-chloroform to disrupt phage particles, and the DNA was precipitated with ethanol. The DNA was used in enzymatic digestion, Southern blot hybridization, and sequencing.
Recombinant DNA Techniques, PCR, and Sequencing.
DNA fragments were cloned into appropriate vectors by using standard protocols (30). For sequencing, DNA fragments were initially cloned into pBluescript II SK (+). The recombinant plasmids were purified with a plasmid extraction kit (Qiagen, Hilden, Germany), and the appropriate inserts were amplified by PCR. The PCR amplicons were purified with a QIAquick PCR Products purification kit (Qiagen) and used as a template for sequencing with an ABI PRISM 377 automated sequencer (Applied Biosystems, Tokyo, Japan). Sequencing was performed by the chain termination method with the Big dye terminator kit (Applied Biosystems) using T3, T7, or synthetic primers. The sequence was analyzed with the DNA Lasergene software package (DNAstar, Madison, WI). Homology searches were performed with BLAST and FASTA programs, made available by the DNA Data Bank of Japan.
Supplementary Material
Acknowledgments
We thank Alejandro Cravioto for helpful suggestions. This work was supported in part by Ministry of Education, Culture, Sports, Science, and Technology of Japan Grant-in-Aid 15406019. M.S.A. was a recipient of a postdoctoral fellowship from the Japan Society for the Promotion of Science. Research at the International Center for Diarrhoeal Disease Research was funded by the center and its donors who provide unrestricted support to the center for its operations and research.
Abbreviations
- CDT
cytolethal distending toxin
- CDT-1Φ
Cdt-I converting phage
- Stx
Shiga toxin
- Cif
cycle inhibiting factor
- EPEC
enteropathogenic Escherichia coli
- LEE
locus of enterocyte effacement
- CF
cell-associated fraction
- SF
supernatant fraction
- CTEC
CDT-producing Escherichia coli.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the DNA Data Base of Japan (accession no. AB285204).
This article contains supporting information online at www.pnas.org/cgi/content/full/0706695104/DC1.
References
- 1.Nougayrede J-P, Taibe F, Rycke JD, Oswald E. Trends Microbiol. 2005;13:103–110. doi: 10.1016/j.tim.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Yamasaki S, Asakura M, Tsukamoto T, Deb R, Faruque SM, Ramamurthy T. Toxin Rev. 2006;25:61–88. [Google Scholar]
- 3.Johnson WM, Lior H. FEMS Microbiol Lett. 1987;43:19–23. doi: 10.1016/0378-1097(91)90131-s. [DOI] [PubMed] [Google Scholar]
- 4.Scott DA, Kaper JB. Infect Immun. 1994;62:244–251. doi: 10.1128/iai.62.1.244-251.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickett CL, Cottle DL, Pesci EC, Bikah G. Infect Immun. 1994;62:1046–1051. doi: 10.1128/iai.62.3.1046-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peres SY, Marches O, Daigle F, Nougayrede J-P, Herault F, Tasca C, Rycke JD, Oswald E. Mol Microbiol. 1997;24:1095–1107. doi: 10.1046/j.1365-2958.1997.4181785.x. [DOI] [PubMed] [Google Scholar]
- 7.Toth I, Herault F, Beutin L, Oswald E. J Clin Microbiol. 2003;41:4285–4291. doi: 10.1128/JCM.41.9.4285-4291.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janka A, Bielaszewska M, Dobrindt U, Greune L, Schmidt MA, Karch H. Infect Immun. 2003;71:3634–3638. doi: 10.1128/IAI.71.6.3634-3638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickett CL, Lee RB, Eyigor A, Elitzur B, Fox EM, Strockbine NA. Infect Immun. 2004;72:684–690. doi: 10.1128/IAI.72.2.684-690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielaszewska M, Fell M, Greune L, Prager R, Fruth A, Tschape H, Schmidt MA, Karch H. Infect Immun. 2004;72:1812–1816. doi: 10.1128/IAI.72.3.1812-1816.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedrich AW, Lu S, Bielaszewska M, Prager R, Bruns P, Xu JG, Tschape H, Karch H. J Clin Microbiol. 2006;44:1844–1846. doi: 10.1128/JCM.44.5.1844-1846.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara-Tejero M, Galan JE. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 13.Elwell C, Chao K, Patel K, Dreyfus L. Infect Immun. 2001;69:3418–3422. doi: 10.1128/IAI.69.5.3418-3422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nesic D, Hsu Y, Stebbins CE. Nature. 2004;429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 15.Pandey M, Khan A, Das SC, Sarkar B, Kahali S, Chakraborty S, Chattopadhyay S, Yamasaki S, Takeda Y, Nair GB, Ramamurthy T. J Clin Microbiol. 2003;41:5277–5281. doi: 10.1128/JCM.41.11.5277-5281.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaper JB, Nataro JP, Mobley HL. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 17.Tobe T, Beatson SA, Taniguchi H, Abe H, Bauley CM, Fivian A, Younis R, Matthews S, Marches O, Frankel G, et al. Proc Natl Acad Sci USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison GE, Angeles D, Tran-Dinh N, Verma NK. J Bacteriol. 2002;184:1974–1987. doi: 10.1128/JB.184.7.1974-1987.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heithoff DM, Sinsheimer RL, Low DA, Mahan MJ. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 20.Chen SL, Hung C, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, et al. Proc Natl Acad Sci USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato T, Shimizu T, Watarai M, Kobayashi M, Kano S, Hamabata T, Takeda Y, Yamasaki S. Gene. 2003;309:35–48. doi: 10.1016/s0378-1119(03)00487-6. [DOI] [PubMed] [Google Scholar]
- 22.Waldor MK, Friedman DI. Curr Opin Microbiol. 2005;8:459–465. doi: 10.1016/j.mib.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Watarai M, Sato T, Kobayashi M, Shimizu T, Yamasaki S, Tobe T, Sasakawa C, Takeda Y. Infect Immun. 1998;66:4100–4107. doi: 10.1128/iai.66.9.4100-4107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brussow H, Canchaya C, Hardt WD. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, et al. DNA Res. 2001;8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 26.Gamage SD, Patton AK, Hanspn JF, Weiss AA. Infect Immun. 2004;72:7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuda J, Fukumoto M, Takeda Y, Nishibuchi M. Infect Immun. 1997;65:428–433. doi: 10.1128/iai.65.2.428-433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinenoya A, Nagita A, Asakura M, Tsukamoto T, Ramamurthy T, Nair GB, Takeda Y, Yamasaki S. Microbiol Immunol. 2007;51:435–438. doi: 10.1111/j.1348-0421.2007.tb03917.x. [DOI] [PubMed] [Google Scholar]
- 29.Faruque SM, Naser IB, Fujihara K, Diraphat P, Chowdhury N, Kamruzzaman M, Qadri F, Yamasaki S, Ghosh AN, Mekalanos JJ. J Bacteriol. 2005;187:4095–4103. doi: 10.1128/JB.187.12.4095-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Fritsch EF, Sambrook. J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.