Abstract
We describe the construction of a large-scale, orderly assembly of mutant ES cells, generated with retroviral insertions and having mutational coverage in >90% of mouse genes. We also describe a method for isolating ES cell clones with mutations in specific genes of interest from this library. This approach, which combines saturating random mutagenesis with targeted selection of mutations in the genes of interest, was successfully applied to the gene families of G protein-coupled receptors (GPCRs) and nuclear receptors. Mutant mouse strains in 60 different GPCRs were generated. Applicability of the technique for the GPCR genes, which on average represent fairly small targets for insertional mutagenesis, indicates the general utility of our approach for the rest of the genome. The method also allows for increased scale and automation for the large-scale production of mutant mice, which could substantially expedite the functional characterization of the mouse genome.
Keywords: G protein-coupled receptor, retroviral vector, ES cell, knockout mice
Knockout (KO) mice provide information on what biological functions are disrupted when individual genes are lost and thereby reveal insights on the roles played by those genes. Because there are an estimated 25,000 genes in the mouse genome, individually knocking out all of the genes in mice would involve a massive effort (1, 2). There are three types of technique in general use for the production of mouse mutants. One, gene targeting, involves preparation of a targeting construct for each individual gene to be knocked out and relies on homologous recombination of the construct with chromosomal DNA of ES cells (3). Recent modifications making this technique much more efficient use BAC modification in Escherichia coli to streamline construct production and provide very long regions of homology for recombination in ES cells (4–6). Gene trapping is a successful alternative for a genomewide mutagenesis (see, for example, refs. 7 and 8). Finally, direct genetics approaches using random chemical (9) and insertional (see ref. 10 as an example) mutagenesis remain useful options for revealing gene functions. Each of these techniques has its advantages and drawbacks (for recent discussions, see refs. 11 and 12).
Here we present an approach that combines saturating random insertional mutagenesis with the ability to identify specific insertions in genes of interest. It involves creating a large library of mutant ES cells with random insertions of a highly mutagenic retroviral vector and a method of isolating clones with inactivation of particular genes. We estimate that the library of 10 million independent mutant clones has >90% of genes mutated. As shown below, this approach allows for efficient isolation of KOs for a preset group of genes and may represent a valuable complement to the existing methods of mammalian gene inactivation. As a practical validation, we applied the technique to the family of G protein-coupled receptors (GPCRs), which constitutes a particularly hard target for insertional mutagenesis because approximately half of the GPCR-encoding genes are devoid of introns and are small (1–2 kb). We identified insertions in 90% of the genes we examined and produced mutant animals for 60 different GPCRs. We and others have used these mutants for functional studies and drug discovery (13–15). The mutagenic vector used to make the mutant library also contains features that allow production of inducible and reversible KO animals (H.Z., K.H., L.M., M.P., G.G., A.R., B. Shimpf, Y. Liang, E. Ojala, F. Kramer, P.R., O. Slobodskaya, I.D., E. Southon, L. Tessarollo, K. Bornfeldt, A.G., G.N.P., and G.A.G., unpublished work).
Results
Construction and Screening of the Library of Mutant ES Cells.
The retroviral vector we used has been described previously (16) and is depicted in Fig. 1, which shows important elements of the vector including (i) a splice acceptor, stop codons in all three reading frames, polyadenylation [poly(A)] signals, and a transcription terminator, all to assure target gene inactivation; (ii) a phosphoglycerate kinase (PGK) promoter-driven neo marker to select clones with insertions and LoxP sites to remove the marker, if necessary; (iii) an internal ribosome entry site (IRES) and a tetracycline (tet)-controlled transactivator, which are not needed for gene inactivation but serve as a plug-in to an inducible gene-inactivation system (H.Z., K.H., L.M., M.P., G.G., A.R., B. Shimpf, Y. Liang, E. Ojala, F. Kramer, P.R., O. Slobodskaya, I.D., E. Southon, L. Tessarollo, K. Bornfeldt, A.G., G.N.P., and G.A.G., unpublished work). Upon insertion, only one orientation of the vector relative to the direction of transcription of the target gene (as shown in Fig. 1, assuming transcription from left to right) is inactivating. Data shown below confirm the high mutagenicity of the vector.
Fig. 1.
Vector for insertional mutagenesis. The vector consists of the following components i–xi. (i) Packaging and integration sequences based on the Moloney murine leukemia virus. The vector lacks the viral enhancers and contains the bacterial supF gene in the 3′ LTR. Upon genome integration, the 5′ LTR enhancer is also deleted (Δen), and the supF sequence is copied to the 5′ LTR. (ii) The adenovirus major late transcript splice acceptor (SA) is included to facilitate the fusion of retroviral transcripts to the endogenous gene transcript in situations where retroviral integration occurs within an intron. (iii) Nonsense codons in all three reading frames ensure translational termination. (iv) The internal ribosome entry site (IRES) from the encephalomyocarditis virus provides translation initiation of the rtTA gene. (v) The reverse tetracycline transactivator (rtTA) stimulates the expression of genes placed under the control of the tetracycline operator in the presence of tetracycline derivatives (27). rtTA is expressed under the control of the endogenous gene, which has been mutated by the insertion. Although not essential for KO generation, rtTA is a key component of an inducible KO system (H.Z., K.H., L.M., M.P., G.G., A.R., B. Shimpf, Y. Liang, E. Ojala, F. Kramer, P.R., O. Slobodskaya, I.D., E. Southon, L. Tessarollo, K. Bornfeldt, A.G., G.N.P., and G.A.G., unpublished work). (vi) Polyadenylation signal (pA) from the bovine growth hormone gene provides for the expression of rtTA mRNA. (vii) The recognition sequences of bacteriophage P1 Cre recombinase LoxP (L) provide the option of removing the phosphoglycerate kinase (PGK) promoter (P) and neomycin phosphotransferase (neo), if desired. (viii) The PGK promoter (P) drives the expression of neo. (ix) The neo-selectable marker renders ES cells containing the provirus resistant to G418. (x) Synthetic poly(A) signal (spA) facilitates the expression of the neo mRNA. (xi) Also included is the transcription terminator (t) from the human complement gene (28) to terminate transcription from both the PGK and cellular promoters.
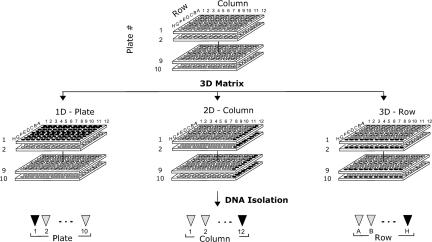
We have generated a library that contains 10 million independent mutant ES cell clones. To develop this library, 129S1/SvImJ ES cells were infected with the retroviral vector shown in Fig. 1. Cells were selected in G418 and distributed at ≈500 clones per well into 96-well plates. After a period of growth, a portion of the cells from each 96-well plate was frozen for later retrieval. The rest of the cells were pooled as shown in Fig. 2. In particular, the cells were split into three parts and used to prepare three types of pools: (i) plate pools, combining all of the wells from the entire plate; (ii) column pools, combining identical columns from a group of plates; and (iii) row pools, combining identical rows from the same group of plates. Thus, a group of 10 plates (960 wells) resulted in 10 plate pools, 12 column pools, and 8 row pools. The pooled cells were grown and used to isolate DNA.
Fig. 2.
Pooling scheme for a single library unit. Equal amounts of cell suspension from wells of a group of 10 96-well plates were transferred to the following: (i) deep-well tube racks with 2× freezing medium for storage and later retrieval (data not shown), (ii) 15-cm dish combining each well of each 96-well plate to make plate pools P1–P10, (iii) 15-cm dish combining each well from a given column from all of the plates in the group to make column pools C1–C12, and (iv) 15-cm dish combining each well from a given row from all of the plates in the group to make row pools RA–RH. Deep-well plates were stored in liquid nitrogen. Pooled cells were grown and treated in the following way. Half of each column and row pool was used to prepare three freezing vials for the future regrowth of the pools. The other half was used for genomic DNA isolation. Each plate pool was split at a ratio of 1:3 to 3 × 15-cm dishes (total of 30 dishes per library unit) and, after growth, each was processed the same way as the column and row pools. This step was needed to produce more DNA and more frozen copies of the pools for later regrowth, because plate pool DNAs were used the most during library screening for the inactivation of specific genes.
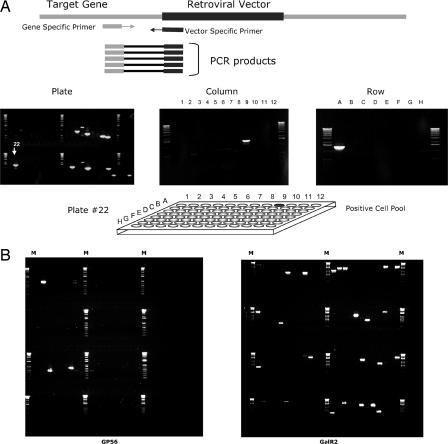
To identify insertions in genes of interest, we screened genomic DNA isolated from the library pools by nested PCRs. Primers specific to the target gene of interest and to the vector were used in sequential steps, and the resulting amplified fragments were analyzed on agarose gel (Fig. 3). Under our nested PCR conditions, the background was greatly reduced and the vast majority of “bands” represented integration events in genes of interest. The amplified fragments were subsequently sequenced to precisely determine the insertion site within the target gene. Detection of the identical, sequence-confirmed PCR fragment in a particular plate pool, column pool, and row pool yielded a 3D address for the well containing the specific ES cell clone of interest. The entire screening process was quite rapid, and the desired mutant ES cell could be located to a specific well in less than a week.
Fig. 3.
Library screening. (A) Nested PCR with gene- and vector-specific primers (Top) was used to screen plate pools from multiple library units (Middle Left). One lane corresponds to one plate pool. The example shows screening of four units, with 10 plate pools in each unit for a gene of interest. Multiple insertions (PCR bands) were detected in different plate pools. Then column and row pools (Middle Center and Right, respectively) of the unit(s) that demonstrated the presence of a sequencing-confirmed insertion in the gene were screened by using the same pairs of primers to find the 3D address of the positive well in the library (e.g., the positive well shown at Bottom gave rise to identical PCR fragments in plate pool no. 22, column pool no. 9, row pool A). (B) The results of screening eight library units (≈4 × 106 mutant ES cell clones) for insertions in the genes of two different GPCRs. M, marker.
Mutant ES cells were isolated from a mixture of ≈500 clones through a two-step procedure. First, cells from a positive well were sorted at ≈40 ES cells per well into 96-well plates by using a cell sorter. Two copies of each plate were then created. One copy was frozen while the other was used for DNA isolation and PCR analysis, with the same gene- and vector-specific primers that were used for the screening of the library. Second, cells from positive wells were sorted from a mixture of ≈40 clones into 96-well plates, this time at one cell per well. Positive wells in this step contained isolated mutant ES cell clones. This procedure enabled the isolation of many mutant ES cell clones to proceed in parallel, facilitating the large-scale production of KO mice.
Validation Through Isolation of Hypoxanthine-Guanine Phosphoribosyltransferase (HPRT) Mutants.
To evaluate the mutagenic capability of our retroviral vector, we analyzed several insertions into the HPRT locus. As many as 33 different integration sites were detected in our library and were confirmed by sequencing (Fig. 4A). Ten of these mutations, one in the 5′ UTR and nine in the first HPRT intron, were evaluated for their ability to confer resistance to 6-thioguanine (6-TG), a phenotype associated with the inactivation of the X-linked HPRT gene. The corresponding 10 ES cell lines, each harboring a different HPRT mutation, were isolated and plated on duplicate plates in the presence or absence of 6-TG. Nine of the 10 cell lines were 6-TG-resistant (Fig. 4B). Only one insertion (Fig. 4, clone 7) did not inactivate HPRT and later was found to harbor a rearranged provirus. (Examination of the retroviral vector integrity of most of the GPCR insertions described below showed that, in a low fraction of cases, portions of the vector, including those important for gene inactivation, were deleted). These results demonstrate the high mutagenicity of our retroviral vector for HPRT, which has long been considered a retroviral “cold spot” (17).
Fig. 4.
Inactivation of the HPRT gene by retroviral insertions. (A) Positions of vector insertions in HPRT, an X-linked gene, which allowed the direct assessment of gene inactivation in ES cells. Insertion locations were determined by sequencing vector–genome junctions for individual insertions. (B) HPRT activity of 10 ES cell clones marked by numbers in A. Activity was determined by the ability of clones to grow in the presence of 6-thioguanine (6-TG). All but one of the clones, no. 7, showed a lack of HPRT activity.
Genome Mutagenesis.
To examine the extent of mouse genome coverage of our mutant ES cell library in terms of retroviral integration events, we searched for insertions in the members of two large gene families, GPCRs and nuclear receptors (NRs).
Primers for 355 mouse nonchemosensory GPCR (18) and for 48 NR genes were designed and used to screen our library. We found that 319 of the 355 GPCR genes examined had one or more retroviral insertions. This represents almost 90% coverage, which is deemed significant because approximately half of GPCRs are encoded by small, intronless genes, making them difficult targets for insertional mutagenesis.
Fig. 3B demonstrates the results of screening 4 million clones from the 10 million clone library for insertions in two GPCR genes, GalR2 and GPR56. In each case, bands represent retroviral integration events. Three retroviral insertions can be seen in GPR56 and 22 in GalR2. Although GalR2 appears to be the preferred locus for retroviral integration as compared with GPR56, multiple retroviral insertions were found at both loci.
Retroviruses have been thought to preferentially integrate into active genes. We have been able to compare insertion frequencies for genes that are expressed in ES cells with those that are not, because we have profiled the expression of a number of GPCR genes in ES cells as well as in other tissues (18). Here, we analyzed retroviral integration events in 108 GPCR genes, 53 expressed and 55 not expressed in ES cells (no PCR product was found after 37 cycles with 20 ng of total reverse-transcribed RNA). Although we observed a slight preference for retroviral insertions into active genes, both active and inactive genes were successfully targeted (Fig. 5), and in most cases we were able to find one or more insertions per gene.
Fig. 5.
Ability of the retroviral vector to inactivate genes that are expressed in ES cells as well as silent genes. Frequencies of insertions into 53 GPCR genes expressed in ES cells (active genes) and 55 GPCR genes not expressed in ES cells (inactive genes) are compared and demonstrate similar potentials for these loci to be targets for the vector integration. Overall, 89% of active genes and 77% of inactive genes had one or more viral insertions.
Similarly, for another gene family, 44 of 48 (92%) NR genes screened had at least one retroviral insertion. These results strongly suggest that a large majority of mouse genes have been mutagenized in this library. Searches for genes that do not belong to the above two families were also successful.
Our PCR-based screening strategy detects only insertion(s) in the gene of interest, but the vector may be also present in other sites. For isolated ES cell clones with mutations in GPCR genes, we used Southern or quantitative slot-blot analysis to determine the copy number of the vector. As shown in Table 1, we found up to seven insertions per clone, but in most cases the clones had only one or two insertions. Considering the number of chromosomes in the mouse genome, the breeding necessary to produce KO animals would result in only one insertion left in the majority of cases.
Table 1.
Insertions per mutant ES cell clone
| No. of insertions per clone | ES clones, % |
|---|---|
| 1 | 38 |
| 2 | 32 |
| 3 | 12 |
| 4 | 8 |
| 5 | 6 |
| 6 | 1 |
| 7 | 3 |
Purified clones with insertions in GPCR genes (total of 105 characterized clones) were checked for the total number of the vector insertions by quantitative slot-blot hybridization. We found up to seven insertions per mutant clone.
GPCR KO Mice.
To validate our overall mouse gene KO approach, we created a series of mouse strains from ES cells containing retroviral insertions in a subset of the screened GPCR genes. After screening of the library, identification and isolation of 86 different mutant GPCR ES cell clones, blastocyst injection, and production of chimeric mice, we generated 60 mutant mouse strains of which the following 57 had the target gene expression eliminated or significantly reduced as explained below: 5HT1D*, 5HT2a, 5HT7, ADORA2b, ADRA2C*, ADRB2*, BLT2*, C3AR1*, CCR6*, CELSR2, CHRM4*, CTR1 (KD), CYSLT2*, DRD3, ECPN, EDG3 (KD)*, EDG8*, FKSG79*, GABABR, GALR2, GIPR, GLP2R, GPR19 (KD)*, GPR20*, GPR22*, GPR39, GPR43*, GPR44*, GPR54, GPR62, GPR63*, GPR68*, GPR77*, GPR85 (KD)*, GPR88 (KD)*, HistH3, HM74*, MC1R*, MC5R*, NMUR2, NPFF1, NTR2, P2Y5*, P2Y6 (KD)*, PG12*, PG208, PG5, PG57, PG63, PTGDR, RAIG1, RE2, SREB3*, SSTR5*, TRHR, VIaR, and VIPR1. In this list, KD stands for knockdown for strains with incomplete gene inactivation, and an asterisk indicates genes that have the entire coding region within a single exon (29 of 57).
We have examined, by either RT-PCR or Northern blotting, the RNA levels of the mutated genes in 23 of the GPCR mutant strains. Taking into account the results of the HPRT mutagenesis analysis, which demonstrated a high mutagenic rate for our retrovirus when integrated into introns, and the anticipated gene-inactivating effects of exonic insertions, we examined mostly retroviral insertions at the 5′ UTR of genes. In particular, we analyzed (i) all 11 lines where the retrovirus was integrated in the 5′ UTR exons or introns, (ii) 7 lines with insertions in coding exons, and (iii) 5 lines with retroviral insertions in introns within the coding region (i.e., downstream of the ATG start codon). As expected, all intronal and exonal insertions within coding regions produced null alleles. However, the mutagenic profile of the retrovirus when integrated into the 5′ UTR segment of a gene was rather different. Only 2 of these 11 events led to null mutations, whereas in 3 of 11 instances transcription of the target gene was not affected. The majority (6 of 11) of retroviral insertions in the 5′ UTR resulted in a significant decrease of the target transcript (knockdown), a reduction of ≥10-fold. We suspect that incomplete gene inactivation on vector insertion in the 5′ UTR is the consequence of activity of additional promoters that were recently reported to be prevalent in the mammalian genome (19). We did not explore this hypothesis further but noticed that it was clearly the case for the calcitonin receptor CTR1. The CTR gene contains 16 exons of which the last 14 encode the protein. In our mutant clone, the insertion was localized to the intron between 5′ UTR exons 1 and 2, and two minor promoters were observed downstream of the insertion site (20).
We suggest, based on these findings, that retroviral insertions upstream of the coding regions of genes should not be used to generate mutant animals. Instead, it is advisable that mutant animals be generated from ES cells containing the provirus in the coding region. This should not be a problem, considering the multitude of independent integration sites per gene within our library.
Discussion
The approach we describe is best suited for generating KOs for relatively large groups of genes. We were able to achieve a steady-state production rate of two mutant ES cell clones per month per person. The technique is scalable, and final productivity was proportional to the number of people involved. The isolated ES cell clones resulted in high germ-line transmission rates. Specifically, of 86 different mutant GPCR ES cells that were injected, we obtained 68 high-percentage chimeras (79%) and 60 heterozygous mutant mouse strains. This represents an overall germ-line transmission rate of 70% for a single mutant ES cell clone. Because we have detected two or more mutations for the majority (70%) of the genes in our library, the probability of making a KO animal for these genes would be 1 − (1 − 0.7)2 = 0.9. Taking into account these estimates (90% probability of creating a mutant animal for 70% of the genes and 70% probability for the remaining 30% of the genes), our overall success rate for producing a KO mouse for any gene for which we have a mutant ES cell would be (0.9 × 0.7) + (0.7 × 0.3) = 0.84 (or 84%).
Although similar to the gene-trapping insertional mutagenesis approaches, our technique is conceptually different because it allows the searching of the entire mutant library for inactivation of a particular gene before isolating the respective ES cell clone. Unlike gene trapping, our system does not depend on active transcription in ES cells or on splicing events for mutant ES cell selection, thus avoiding drug selection biases: In the promoter trap approach, vector insertions into genes silent in ES cells cannot be selected (21), and, in polyadenylation [poly(A)] traps, insertions into the upstream regions of genes are lost because of nonsense-mediated decay of the selection marker transcript (22). Although these problems were partially addressed by showing that very little ES cell expression is required for promoter trapping (23) and that nonsense-mediated mRNA decay can be suppressed (22), strong preference of vector insertion into certain “hot spots” still caused repeated reisolation of the same trapped clones. The high complexity of our mutant library circumvented many problems associated with the insertion hot spots and allowed us to identify the desired mutants with a probability of ≈90%. It is worth noting that, of the 57 genes listed in the GPCR KO Mice section, we were able to find, in the National Center for Biotechnology Information (NCBI) database of mouse gene trap sequences (www.ncbi.nlm.nih.gov/genome/seq/BlastGen/BlastGen.cgi?taxid=10090), only 12. This database also included clones from the Lexicon Pharmaceuticals (The Woodlands, TX) OmniBank (http://omnibank.lexgen.com/blast_form.jsp) [supporting information (SI) Table 2]. Moreover, a similar search for 139 more ES cell clones with GPCR insertions that we isolated from our library but did not use yet for KO production found only 24 trapped genes in the NCBI database; this confirmed our suggestion that gene trapping selects against small and poorly expressed genes in ES cells (SI Table 3).
Unlike other insertional mutagenic approaches, ours does not attempt to identify the vector integration site for each individual clone and then try to identify which gene has been mutated in the clone. Instead, we devised an efficient DNA-pooling, PCR-screening, clone-isolation strategy to directly screen for and isolate insertions into a specific gene of interest from a large number of ES cell clones. We chose to construct the 10 million clone library mainly for practical reasons. If necessary, the size of the library can be further increased to expand coverage to hard-to-catch genes.
Methods
Retroviral Vector-Producing Cell Line.
To make the viral producer cell line, the retroviral vector was transfected into an NIH 3T3-based packaging cell line, GP + E-86 that has gag-pol and env genes of the Moloney murine leukemia virus integrated in two different locations (24). G418-resistant clones were selected. To identify high-titer viral producer clones, each clone was treated with mitomycin C and cocultured with ES cells for 2 days, and the efficiency of viral transduction was determined by a titer of G418-resistant ES cell clones performed after replating ES cells infected by each individual viral producer clone with or without G418.
Construction of Mutant ES Cell Library.
Before library construction, 129S1/SvImJ ES cells [line CJ7 (25)] were rederived to obtain cells with high-probability germ-line transmission. It is known that the pluripotency of ES cells gradually deteriorates with culturing because of accumulated mutations and epigenetic changes. Some mutations and chromosomal abnormalities, notably trisomy 8, confer growth advantage, causing the mutant cells to gradually overtake the ES cell population upon passaging (26). Therefore, we first isolated a number of individual ES cell colonies and karyotyped them and used euploid clones (aneuploids did constitute a significant portion of the clones) for blastocyst injections. Clones yielding ≥90% chimeras were chosen for further testing of germ-line transmission. A few of the ES cell clones tested contributed to the germ line with probabilities of >90%.
Retroviral infection was conducted in 10-cm dishes by cocultivating ES cells and virus-producing fibroblasts for 48 h. For the selection of G418-resistant clones, infected cells were transferred to 96-well plates (one 96-well plate from one 10-cm dish to minimize the presence of multiple copies of the same insertion in the library). The efficiency of infection of ES cells with the retroviral vector was 40%, and their plating efficiency was 30%. To achieve a density of 500 neoR clones per well under these conditions, the plates were seeded with ≈4,000 ES cells per well.
The library was constructed in a stepwise manner, in units of 5 × 105 infected ES cells (10 × 96-well plates). A total of 20 units was generated, containing collectively 107 independent mutant ES cells. To facilitate the subsequent screening of the library, the content of each unit was pooled in a 3D orthogonal matrix, as shown in Fig. 2. Specifically, infected cells were split into three parts and used to prepare three types of cellular pools: (i) plate pools, combining all of the wells from the entire plate; (ii) column pools, combining identical columns from a group of plates; and (iii) row pools, combining identical rows from the same group of plates. A library unit generated 10 plate pools, 12 column pools, and 8 row pools, each representing a different number of wells: 96 wells in the plate pools, 120 wells in the row pools, and 80 wells in the column pools. Considering that each well contained ≈500 clones, the complexity of each pool was between 4 × 104 and 6 × 104 clones. After a period of growth, a part of the cellular pools was used for genomic DNA isolation for the subsequent screening of the library, while another part was frozen in aliquots for the regrowth of the pools, if necessary at a later time.
Library Screening.
The library was screened by using nested PCR with gene- and vector-specific primers. The vector-specific primers were common to all genes screened and were carefully selected to avoid the amplification of false positive fragments derived from endogenous retroviral sequences. Gene-specific primers were designed for each gene and were usually targeted to the 5′ end of the gene. All primers were designed by using Oligo Primer Analysis software, version 6 (Molecular Biology Insights, Cascade, CO). The annealing temperature of each primer pair was set to 65°C, and the parameters for intra- and intermolecular interactions between the primers were set to the highest stringency allowed by the program. The specificity of the primers used was evaluated by a BLAST search of the mouse genome. Qiagen (Valencia, CA) HotStarTaq DNA polymerase was used for all PCRs. The first designed screening primer worked for ≈75% of the genes. For the rest, primers were redesigned.
We first screened genomic DNA from plate pools and subsequently determined the 3D address of the mutation of interest by screening genomic DNA from column and row pools. We reliably detected insertions within 2–3 kb of the gene-specific primer. On average, we ran ≈100 PCRs per gene to identify mutations in a target gene and an additional ≈100 PCRs per gene to determine the 3D address of the mutation of interest in our library.
DNA pools being screened had an average complexity of ≈5 × 104 individual ES cell clones (≈100 wells × ≈ 500 clones per well). A minimum of ≈2 μg of pool DNA was used in each PCR. If we assume a molecular weight of the diploid mouse genome of ≈4 × 1012, then 2 μg of genomic DNA would contain ≈3 × 105 copies of diploid genome. Therefore, 2 μg of pooled genomic DNA would include, on average, approximately six copies of genome from each individual ES cell clone, which is close to the limit of PCR detection. The total amount of genomic DNA isolated from a plate pool in our library construction protocol was ≈2,500 μg, enough to screen 1,250 genes. After that, frozen copies of cell pools would have to be regrown to replenish the source of genomic DNA. In practice, it is convenient to screen 8 units of a 24-unit library at a time. In our experience, insertions in ≈50% of genes could be found by screening only one-third of a library; another third is required for ≈30% of genes, and the remaining 20% of genes have to be screened over the entire library (genes that have no insertions also fall into this category). The ability to screen many genes over a partial library increases the total number of genes that can be screened before it becomes necessary to regrow the pools. The number of genes that can be screened by using our current protocol is ≈2,000.
To use the library to isolate additional mutants, the cellular pools would have to be regrown and the DNA would have to be reisolated. Because of the differential growth rates of different individual clones, the complexities of the pools would be reduced with the repeated rounds of regrowth, resulting in an inability to use one constructed library indefinitely. In our experience, however, at least two rounds of regrowth are possible without a significant loss in the complexity of the pools. That would bring the estimate for the total number of genes that could be screened by using a single library to ≈5,000–6,000, which is a significant part of the entire genome. Furthermore, construction of new ES libraries could be accomplished fairly easily within several months by using the same retrovirus or modified versions of it.
Isolation of Mutant ES Cell Clones.
Once the 3D address of the mutant ES cell of interest was established, the desired clone was isolated from a mixture of 500 clones through two rounds of cell sorting and PCR screening.
In particular, the content of the appropriate well was sorted at 40 ES cells per well into four 96-well plates, with a cell sorter (the light-scattering profile allowed for the separation of ES cells from feeder mouse embryonic fibroblasts). The plating efficiency of sorted ES cells was ≈40%. The sorted cells were allowed to grow for 2 days; on day 3 they were trypsinized. At this stage, cells from each 96-well plate were evenly distributed into two 96-well plates. After an additional 2 days of growth, one copy of the plates was frozen while the other was used for genomic DNA isolation and PCR analysis with the same gene- and vector-specific primers that were used for the screening of the library.
For the second round of subcloning, the frozen content of positive wells was thawed and expanded for 2–3 days, and the cells were sorted into four 96-well plates, this time at one cell per well. The cells were treated similarly to those used in the first subcloning round, with the addition of another trypsinization step to prevent the differentiation associated with the longer growth of colonies derived from single cells.
To assure gene inactivation by vector insertion, we examined the integrity of the vector in each isolated clone. In each case, the precise site of vector insertion was known from the sequence of PCR products obtained during library screening. Based on this information, a pair of primers was designed upstream and downstream of the insertion site. These primers were used with vector-internal primers to confirm the integrity of the vector. An upstream primer with the vector primer in the rtTA gene ensured that the gene-inactivating sequences, including splice acceptor and stop codons, and the rtTA gene itself were still in place, and a downstream primer with the vector primer in the Neo gene further confirmed vector integrity. In our experience ≈5% of insertional mutants contain rearranged or deleted vector sequences. Although we observed that rearranged vector still sometimes causes gene inactivation, the clones that did not have intact vector were not cleared for blastocyst injections.
Mouse Production.
Standard techniques were used to produce GPCR KO mice from ES cells mutagenized with our retroviral vector. In brief, mutant ES cells were injected into blastocysts of the C57BL/6J strain, and these blastocysts were then transferred into the uteri of day 2.5 pseudopregnant CD1 females. Live-born pups were scored for fur color, and chimeric mice (black and agouti color) with a high contribution of agouti fur (≥50%) were bred directly with 129S1/SvImJ mice to maintain the mutants in the inbred background. The resulting progeny were genotyped by PCR to identify those with the retroviral insertion in the target gene. Inbred heterozygous animals were subsequently intercrossed to produce homozygous mice. All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee of Omeros in accordance with National Institutes of Health guidelines.
Supplementary Material
Acknowledgments
We thank Demetri Spyropoulos for help with ES cell rederivation. This work was supported in part by National Institutes of Health Grant MH070241 (to A.G.).
Abbreviations
- KO
knockout
- GPCR
G protein-coupled receptor
- HPRT
hypoxanthine-guanine phosphoribosyltransferase.
Footnotes
Conflict of interest statement: A.G., M.P., L.M., H.Z., G.G., A.R., I.D., A.E., S.M., C.N., E.F., J.B., D.K.V., and G.A.G. are or were employees of privately owned Omeros Corporation and declare competing financial interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700608104/DC1.
References
- 1.Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, et al. Nat Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas KR, Folger KR, Capecchi MR. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 4.Copeland NG, Jenkins NA, Court DL. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 5.Heintz N. Nat Rev Neurosci. 2001;2:861–870. doi: 10.1038/35104049. [DOI] [PubMed] [Google Scholar]
- 6.Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 7.Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 8.Hansen J, Floss T, Van Sloun P, Fuchtbauer EM, Vauti F, Arnold HH, Schnutgen F, Wurst W, von Melchner H, Ruiz P. Proc Natl Acad Sci USA. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A. Hum Mol Genet. 1999;8:1955–1963. doi: 10.1093/hmg/8.10.1955. [DOI] [PubMed] [Google Scholar]
- 10.Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, et al. Mol Cell Biol. 2003;23:9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson CM, Largaespada DA. Nat Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- 12.Glaser S, Anastassiadis K, Stewart AF. Nat Genet. 2005;37:1187–1193. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 13.Krasnow SM, Hohmann JG, Gragerov A, Clifton DK, Steiner RA. Neuroendocrinology. 2004;79:268–277. doi: 10.1159/000079632. [DOI] [PubMed] [Google Scholar]
- 14.Gottsch ML, Zeng H, Hohmann JG, Weinshenker D, Clifton DK, Steiner RA. Mol Cell Biol. 2005;25:4804–4811. doi: 10.1128/MCB.25.11.4804-4811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H, Gragerov A, Hohmann JG, Pavlova MN, Schimpf BA, Xu H, Wu LJ, Toyoda H, Zhao MG, Rohde AD, et al. Mol Cell Biol. 2006;26:9352–9363. doi: 10.1128/MCB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaitanaris GA. US Patent 6. 2001;228:639. [Google Scholar]
- 17.King W, Patel MD, Lobel LI, Goff SP, Nguyen-Huu MC. Science. 1985;228:554–558. doi: 10.1126/science.3838595. [DOI] [PubMed] [Google Scholar]
- 18.Vassilatis DK, Hohmann JG, Zeng H, Li F, Ranchalis JE, Mortrud MT, Brown A, Rodriguez SS, Weller JR, Wright AC, et al. Proc Natl Acad Sci USA. 2003;100:4903–4908. doi: 10.1073/pnas.0230374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Nat Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 20.Anusaksathien O, Laplace C, Li X, Ren Y, Peng L, Goldring SR, Galson DL. J Biol Chem. 2001;276:22663–22674. doi: 10.1074/jbc.M007104200. [DOI] [PubMed] [Google Scholar]
- 21.Friedrich G, Soriano P. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 22.Shigeoka T, Kawaichi M, Ishida Y. Nucleic Acids Res. 2005 Feb 1; doi: 10.1093/nar/gni022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedel RH, Plump A, Lu X, Spilker K, Jolicoeur C, Wong K, Venkatesh TR, Yaron A, Hynes M, Chen B, et al. Proc Natl Acad Sci USA. 2005;102:13188–13193. doi: 10.1073/pnas.0505474102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markowitz D, Goff S, Bank A. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swiatek PJ, Gridley T. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- 26.Liu X, Wu H, Loring J, Hormuzdi S, Disteche CM, Bornstein P, Jaenisch R. Dev Dyn. 1997;209:85–91. doi: 10.1002/(SICI)1097-0177(199705)209:1<85::AID-AJA8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 27.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 28.Ashfield R, Enriquez-Harris P, Proudfoot NJ. EMBO J. 1991;10:4197–4207. doi: 10.1002/j.1460-2075.1991.tb04998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.