Abstract
The Par-1/MARK protein kinases play a pivotal role in establishing cellular polarity. This family of kinases contains a unique domain architecture, in which a ubiquitin-associated (UBA) domain is located C-terminal to the kinase domain. We have used a combination of x-ray crystallography and NMR dynamics experiments to understand the interaction of the human (h) MARK3 UBA domain with the adjacent kinase domain as compared with ubiquitin. The x-ray crystal structure of the linked hMARK3 kinase and UBA domains establishes that the UBA domain forms a stable intramolecular interaction with the N-terminal lobe of the kinase domain. However, solution-state NMR studies of the isolated UBA domain indicate that it is highly dynamic, undergoing conformational transitions that can be explained by a folding–unfolding equilibrium. NMR titration experiments indicated that the hMARK3 UBA domain has a detectable but extremely weak affinity for mono ubiquitin, which suggests that conformational instability of the isolated hMARK3 UBA domain attenuates binding to ubiquitin despite the presence of residues typically involved in ubiquitin recognition. Our data identify a molecular mechanism through which the hMARK3 UBA domain has evolved to bind the kinase domain, in a fashion that stabilizes an open conformation of the N- and C-terminal lobes, at the expense of its capacity to engage ubiquitin. These results may be relevant more generally to the 30% of UBA domains that lack significant ubiquitin-binding activity, and they suggest a unique mechanism by which interaction domains may evolve new binding properties.
Keywords: catalytic domain, dynamics, Par-1, relaxation, ubiquitin-associated
Protein kinases in the Par-1/MARK family are the subject of intense interest because of the crucial roles they play in establishing cell polarity (1–4), regulating the cell cycle progression (5–7), and controlling microtubule dynamics (8–11). Functional orthologs of Par-1 have been described from yeast to mammals (1–3, 8, 12). In the human (h) kinome, four Par-1 paralogs (hMARK1–4) are grouped within the AMP-activated protein kinase (AMPK)/Snf1 family of kinases (13). These mammalian kinases originally were shown to phosphorylate microtubule-associated proteins (MAP2, MAP4, and Tau), resulting in their detachment from microtubules (8, 10), and hence were named MAP/microtubule affinity-regulating kinases, or MARKs. They have been implicated in the etiology of Alzheimer's disease because of their involvement in Tau phosphorylation on S262 (reviewed in ref. 14).
The Par-1/MARK family kinases have a conserved domain architecture, comprising an N-terminal serine/threonine kinase domain, a short linker sequence of 20 aa, a ubiquitin (Ub)-associated (UBA) domain, a spacer region of ≈300 residues, and a C-terminal kinase-associated (KA) domain. This arrangement of kinase and UBA domains is unique to the AMPK/Snf1 family. Predominantly, UBA domains have been characterized as binding monoUb and K48-linked and K63-linked polyUb (15, 16), although a recent survey of 30 yeast and mammalian UBA domains revealed that ≈30% of UBA domains do not detectably interact with monoUb or polyUb chains in vitro (16). Further studies indicated that the MARK UBA domains belong to this latter class, as they were unable to detectably bind Ub or Ub-like species in vitro (17). However, comparison of the amino acid sequences of Ub-binding and nonbinding UBA domains did not show any obvious differences that would explain at a molecular level why a significant fraction of UBA domains fail to detectably bind Ub in vitro (16). Consequently, the basis for the two groups of UBA domains (Ub-binding and nonbinding), and the role of the MARK UBA domain in kinase regulation rather than Ub-binding, has remained enigmatic.
Here, we describe a structural characterization of the hMARK3 UBA domain when fused to the kinase domain and in isolation by using x-ray crystallography and NMR spectroscopy, respectively. In the hMARK3 kinase:UBA domain crystal structure, the UBA domain binds to the N-terminal lobe (N-lobe) of the kinase domain via a collection of conserved hydrophobic interactions. These interactions are facilitated by a noncanonical UBA domain topology, where the α3 helix is inverted relative to the canonical UBA domain fold, which sequence alignments suggest is likely to be conserved within the UBA domains of other AMPK/Snf1 family kinases. Analysis of multidimensional NMR spectra and characterization of the isolated hMARK3 UBA domain by relaxation-dispersion NMR experiments, which probe millisecond time-scale conformational exchange processes in proteins, demonstrates that the hMARK3 UBA domain is highly dynamic in solution and subject to an unfolding equilibrium that most severely affects the α3 helix. By using the capacity of NMR spectroscopy to detect extremely weak intermolecular interactions through binding-induced changes in NMR chemical shifts, we detected monoUb-binding with an isolated hMARK3 UBA, with a Kd in excess of 2 mM. The conformational instability and very weak Ub affinity of the isolated UBA domain support the notion that the AMPK/Snf1 family UBA domains have evolved a function as stabilizing interactors of their adjacent kinase domains at the expense of their capacity to bind Ub species.
Results and Discussion
The Structure of the Kinase and UBA Domains of Human MARK3.
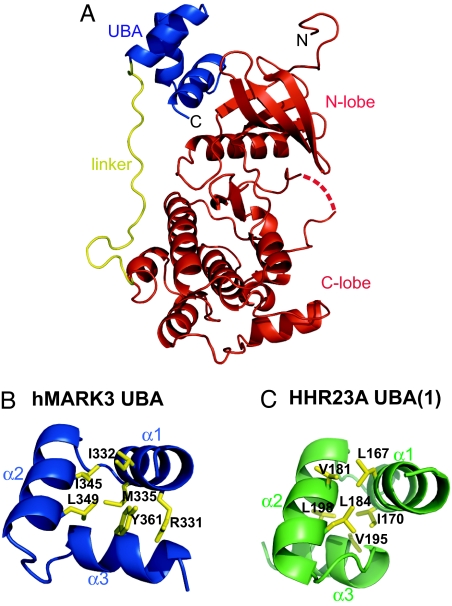
To understand how the kinase and UBA domains are organized within hMARK3, we crystallized and solved the structure of hMARK3 (residues 48–370) by x-ray crystallography to 2.7-Å resolution [Fig. 1, supporting information (SI) Fig. 5, and SI Table 1]. This fragment of hMARK3 incorporates the kinase domain (D48–N308), a short linker sequence (A309–D328), and the UBA domain (Q329–R366). The kinase domain adopts a characteristic bilobal fold with the subsequent short linker sequence running up the face of the catalytic domain opposing the active site toward the N-lobe of the catalytic domain, to which the UBA domain binds via hydrophobic contacts (SI Fig. 5). The N-lobe–UBA domain interaction is possible because the UBA domain adopts an atypical fold in which the α3 helix is inverted compared with the canonical UBA domain topology (Fig. 1 B and C). An analogous UBA domain fold was observed within the kinase:UBA domain crystal structures of hMARK1 (19) and rat MARK2 (20) reported during the course of this work. From the MARK crystal structures, it is apparent that a tyrosine residue within the UBA domain α3 helix (Y361 in hMARK3), which is conserved throughout the AMPK/Snf1 kinase family, plays a key role in stabilizing the topology because of its contributions to the hydrophobic core and a π–π stacking interaction with an arginine guanidinium side chain at the N terminus of the UBA α1 helix (R331 in hMARK3). Additionally, the UBA domain α3 helix features prominently in mediating hydrophobic contacts with the catalytic domain N-lobe in all three MARK crystal structures, suggesting that this UBA domain topology has evolved to optimize interactions with the catalytic domain (SI Fig. 5). It is noteworthy that with the exception of the MGY motif and hydrophobic core residues (SI Fig. 6) (16, 17, 20), UBA domains show little sequence conservation, and a tyrosine residue at the position homologous to Y361 of hMARK3 is seldom observed outside of the AMPK/Snf1 kinase family. Because of this poor sequence conservation, it therefore is difficult to predict whether a UBA domain is likely to adopt the topology identified in MARK kinases or a canonical fold.
Fig. 1.
Crystal structure of hMARK3 catalytic and UBA domains. (A) Cartoon of the hMARK3 (residues 48–370) crystal structure. The N- and C- termini are labeled in black text. The kinase domain (AMGS from vector and D48–N308) is colored red; the linker (A309–D328) is yellow; and the UBA domain (Q329–R366) is blue. The dashed red line corresponds to the activation segment residues (205–208) for which no electron density was observed. (B) Comparison of the hMARK3 UBA domain and the canonical UBA domain fold. Structure of hMARK3 UBA domain from hMARK3 (residues 48–370) crystal structure. Residues involved in the hydrophobic core of the UBA domain are depicted by yellow sticks. (C) Structure of HHR23A (1) UBA domain. Residues involved in the hydrophobic core of the UBA domain are depicted by yellow sticks. The hMARK3 UBA domain contains an inversion of the α3 helix compared with this canonical UBA domain topology. Figure was drawn from PDB ID code 1IFY (18).
The Isolated hMARK3 UBA Domain Is in Equilibrium with a Populated Unfolded State in Solution.
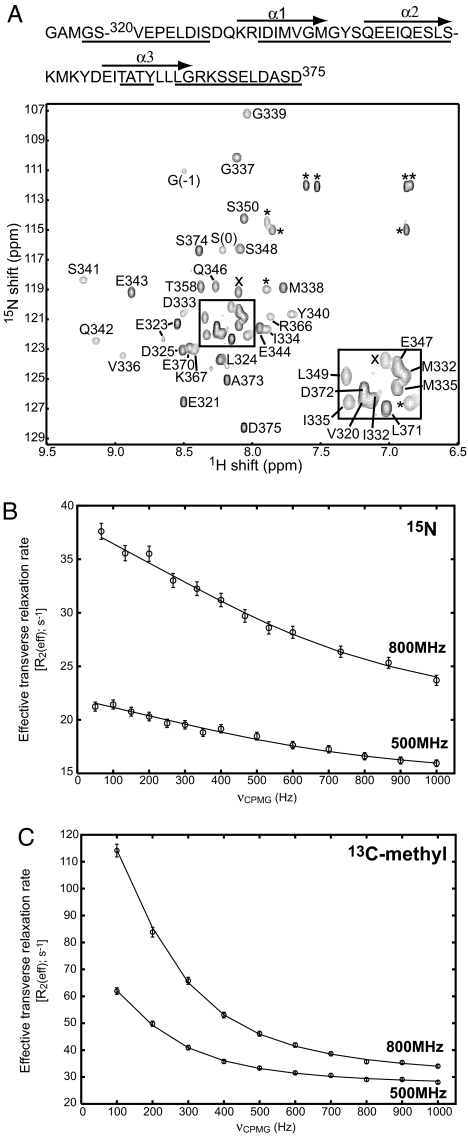
Because the topology of the hMARK3 UBA domain observed in the kinase:UBA domain crystal structure differs from the canonical UBA domain fold, we investigated the solution properties of the hMARK3 UBA domain by using NMR spectroscopy. We considered the possibilities that the isolated UBA domain might retain the folded conformation seen in the kinase:UBA polypeptide or convert to a more conventional UBA domain fold. UBA domains characterized by NMR spectroscopy to date, such as the two UBA domains of hHR23A (21), show well resolved, sharp peaks for which complete NMR assignment and structure determination were possible. In contrast, 1H-15N heteronuclear single-quantum correlation (HSQC) spectra of the 15N-labeled hMARK3 UBA domain (residues 320–375) are quite different (Fig. 2A) with extensive line-broadening apparent. For example, of the 60 non-proline residues in the construct (including the vector-encoded N-terminal GAMGS), only 40 backbone amide correlations are present in the 1H-15N HSQC spectrum (25°C), with the rest broadened beyond the level of detection. The absence of NMR peaks (or the presence of extremely low-intensity correlations) is indicative of millisecond to microsecond time-scale conformational exchange. Of interest, we noted that broadening is most pronounced for correlations with 1H chemical shifts that lie far from those typical of random coil values (≈8.1 ppm) (22), suggesting that the conformational exchange events responsible for the observed broadening are derived from a folding–unfolding equilibrium.
Fig. 2.
NMR spectroscopy characterization of millisecond to microsecond exchange processes within the hMARK3 UBA domain. (A) 1H-15N HSQC spectrum of the hMARK3 UBA domain. HSQC spectrum recorded at 25°C, 500 MHz. The protein sequence is drawn above the spectrum, with assigned residues underlined and helices indicated by arrows above the corresponding sequence. Assignments are annotated next to the corresponding peaks in the spectrum in black text. The boxed region is enlarged in Right Inset. Peaks marked with × were not assigned, and * marks a side chain resonance. G(−1) and S(0) arise from the cloning vector. (B) Representative 15N relaxation-dispersion profile. 15N relaxation-dispersion profile for the backbone amide resonance of E343 derived from the CPMG experiment at 25°C. The upper and lower curves are derived from data collected at 800 and 500 MHz, respectively. (C) Representative 13C relaxation-dispersion profile. 13C-methyl relaxation-dispersion profile for a Leu δ1 or δ2 methyl resonance (assignment unavailable) derived from the CPMG experiment at 5°C. The upper and lower curves are derived from data collected at 800 and 500 MHz, respectively.
To obtain site-specific information about the exchange process, we attempted to sequentially assign backbone amide resonances by using a 13C-15N-labeled hMARK3 UBA domain and 3D heteronuclear NMR spectroscopy. Although many of the backbone assignments could be obtained, correlations derived from the α3 helix were among the weakest. A significant fraction of the cross-peaks for the residues in this element were not observed in the spectra, and several of those that were (A359, T360, Y361, and G365) were only just visible above noise levels at high sample concentrations (>1.5 mM). This finding suggests that the α3 helix is less stable than the other two helices in the hMARK3 UBA domain. This pattern of broadening, which most significantly affects the α3 helix, and the decrease in peak intensity as a function of chemical shift offset from random coil values (see above) are consistent with a folding reaction: α1 − α2 − α3 ⇆ α1 − α2 − U3 ⇆ U1 − U2 − U3, where αI (I ∈ {1,2,3}) denotes helix I and UI is the unfolded state of this helix. Such a mechanism also is in keeping with circular dichroism spectroscopy studies in which denaturation by heat or guanidine hydrochloride treatment occurred over a broad transition without a steady, folded baseline (data not shown). These data are consistent with the UBA domain existing in a partially folded state under native conditions where, according to the scheme above, the unfolded state of the α3 helix would be highly populated but where the α1 and α2 helices retain secondary structure that requires extensive heat or chemical treatment for complete denaturation (see below).
NMR Characterization of the Putative Folding/Unfolding Equilibrium in the hMARK3 UBA Domain.
We have examined the conformational exchange processes within the UBA domain in more detail by using 15N and 13C Carr–Purcell–Meiboom–Gill (CPMG) NMR relaxation-dispersion techniques, which provide a powerful means to study motions occurring on the microsecond time scale at a large number of sites within the protein (23–26). In this method, the exchange contribution to transverse relaxation, Rex, resulting from differential chemical shifts of the probe spin in each of the exchanging states, is modulated by the application of a variable number of refocusing pulses within a fixed time period (27). The effective magnetization decay rate constant, R2eff = R2,int + Rex (where R2,int is an intrinsic transverse relaxation rate that is independent of exchange), measured as a function of the frequency of application of refocusing pulses, νCPMG, provides information about the rates of interconversion between states (kinetics), their populations (thermodynamics), and the differences in chemical shifts between exchanging sites (related to structure) (23, 24). Examples of relaxation-dispersion profiles, R2eff versus νCPMG, for selected backbone 15N and side-chain methyl 13C nuclei of hMARK3 UBA are shown in Fig. 2 B and C.
Initially, exchange dynamics were studied by using a 15N-labeled sample of the hMARK3 UBA domain. Of the 55 non-proline residues in this construct (excluding the sequence GAMGS at the N terminus that results from cloning), we were able to measure relaxation-dispersion profiles for 32 amide groups at 25°C and 28 amide groups at 15°C. A global fit of relaxation-dispersion data for these residues to a two-site exchange model
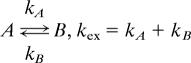 |
gave kex = 4,910 ± 225 s−1 at 25°C and 2,390 ± 65 s−1 at 15°C. These kex values indicate that exchange occurs in the fast limit [i.e., kex ≫ δω, where δω = ω̄N*δω, δω, and δω̄ are frequency (rad/s) and chemical shift differences (ppm) between states, respectively, and ωN is Larmor frequency of the 15N nucleus] so that neither the populations of the exchanging states nor the chemical shifts for each probe in each state could be reliably estimated. In this limit, it is only possible to extract the exchange rate constant kex = kA + kB and the product pApB(δω̄)2, where pA and pB are the fractional populations of interconverting sites A and B, respectively. In what follows, we have assumed that the populations of each state are equal (pA = pB = 0.5) so that the minimum chemical shift differences between each state, δω̄, could be obtained for each probe. The extracted minimum δω̄ values are reasonably large (1–2.2 ppm), which is consistent with an unfolding event (that would necessarily lead to large structural changes and hence significant concomitant changes in shifts).
Although the exchange process may well be more complex than that described by a two-state model (see discussion above), the quality of the data did not permit analysis with more complex models. In this regard, it is worth noting, however, that the majority of the correlations expected for the α3 helix are missing in the spectra and hence are not available for analysis. It is not expected that the unfolding of helix 3 in-and-of-itself would be the cause of the large changes in shifts between states for residues in the remaining two helices (minimum δω̄ values of between 1 and 2 ppm) that have been quantified. Thus, the measured dispersion profiles that were analyzed are consistent with the second exchange process in the folding reaction above (α 1 − α2 − U3 ⇆ U1 − U2 − U3) that would have a large effect on measured shift changes in helices 1 and 2; a two-state analysis thus is quite reasonable.
The conformational exchange process characterized by 15N relaxation-dispersion spectroscopy at 25°C and 15°C is expected to slow down at the lower temperature, potentially allowing a more quantitative analysis. However, the decrease in kex with temperature leads to increased broadening so that most of the 15N correlations could not be quantified at 5°C and 15N-based experiments were not possible. Because methyl spectra of proteins are in general much more sensitive than corresponding 1H-15N correlation maps, methyl 13C-CPMG relaxation-dispersion profiles were collected for the hMARK3 UBA domain at 5°C. A two-site exchange model was fitted to these data (as shown for a representative resonance in Fig. 2C), and a global fit of all methyl resonances yielded an exchange rate constant (kex) of 1,300 ± 30 s−1 along with a population of the minor state of 20%, with the minor state in the exchange model proposed above corresponding to the fully unfolded protein (see below).
To provide further evidence that the millisecond conformational exchange process we have characterized corresponds to an unfolding event, we have performed an amide-solvent hydrogen-exchange experiment (28) by using a 15N-labeled hMARK3 UBA domain sample at 25°C. From these data protection factors, Rintr/Rexp were calculated from the experimental (Rexp) and predicted intrinsic random coil (Rintr) hydrogen-exchange rates for resonances observed within the 2D 1H-15N HSQC spectrum to estimate the proportion of backbone amide resonances that are solvent-exposed (in an “open” conformation) at any time. In the absence of protection from solvent exchange, ratios of 1 would be predicted, with ratios increasing (from 1) as a function of increasing protection. The disordered regions, comprising residues from the expression vector, V320–D325, and K367–S374, all exhibit protection factors Rintr/Rexp in the range of 1.5–9. By comparison, Rintr/Rexp values determined for the α1 helix are between 8–50 and 14–20 for the α2 helix, i.e., about a factor of 5 more than those in the disordered regions. These small levels of protection are consistent with estimates of 20% unfolded protein in equilibrium with the folded state, based on the analysis of 13C methyl dispersion data. Because of the absence of most α3 helix resonances in 1H-15N HSQC spectra, it is only possible to quantify protection for the backbone amides of T358 within this element (Rintr/Rexp of 3.7) and R366 immediately after the α3 helix (Rintr/Rexp of 8.3), both of which are lower than those obtained for residues in the other two helices. In sum, these protection data demonstrate that the hMARK3 UBA domain is highly dynamic and are strongly consistent with an exchange process involving an unfolded state that is highly populated at ambient temperatures.
The hMARK3 UBA Domain Has a Weak Affinity for MonoUb.
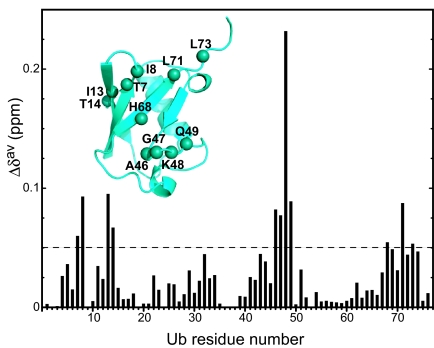
The NMR relaxation-dispersion and hydrogen-exchange studies presented above provide strong evidence for an exchange process that involves a highly populated unfolded state. Such an unfolding equilibrium would be expected to lower affinity for targets. Moreover, although we were unable to determine whether, in solution, the isolated UBA domain adopts the topology observed in the hMARK3/Par-1 kinase:UBA domain structure, it is plausible that this atypical fold would lower the affinity of this UBA domain for Ub. With these factors in mind, we next examined whether the hMARK3 UBA domain possesses an intrinsic ability to bind Ub. Previous biochemical experiments suggest that the MARK UBA domains do not measurably interact with Ub and Ub-like molecules in vitro (17, 20). Consequently, the MARK UBA domains can be categorized as one of the ≈30% of UBA domains that do not detectably bind Ub (16). Like the MARK UBA domains, most of these ≈30% of UBA domains contain sequence motifs characteristic of Ub-interacting UBA domains (SI Fig. 6) and exhibit no apparent sequence differences that would preclude the binding of Ub species. To definitively address this issue, we used the sensitivity of NMR spectroscopy for detecting weak intermolecular interactions and for mapping intermolecular surfaces involving weakly binding partners by monitoring binding-induced NMR chemical shift changes. Initially, we collected a series of 1H-15N HSQC spectra for 15N-labeled monoUb in the presence of up to 5.3 molar equivalents of unlabeled hMARK3 UBA domain. We observed chemical shift perturbations for a subset of resonances within the 15N-Ub 1H-15N HSQC spectrum upon titration with the hMARK3 UBA domain, thus providing evidence for the Ub·UBA interaction in solution (Fig. 3). Upon addition of successive aliquots of ligand, correlations in the Ub spectrum titrated in a linear manner, consistent with binding in the fast-exchange (two-state) regime and suggesting therefore that monoUb binds the UBA domain with low affinity. We calculated weighted-average chemical shift perturbations (Δδav) for each Ub amide backbone resonance (Fig. 3) as described in Materials and Methods, revealing that Ub engages the hMARK3 UBA domain via its canonical interaction interface (Fig. 3 Inset). The Ub residues exhibiting the largest chemical shift perturbations in the presence of the 5.3 equivalents of the hMARK3 UBA domain spatially cluster around K48 in the Ub structure and include L8, I13, L43, A46, G47, Q49, and L71 in addition to K48 (Fig. 3). The residues located in this patch of Ub previously have been shown to mediate binding to each of the two HHR23A UBA domains (21).
Fig. 3.
Ub binds the hMARK3 UBA domain via a classical interaction interface. Weighted-average chemical shift perturbation (Δδav) versus residue number. Δδav, calculated as described in Materials and Methods, for each residue in 15N-Ub 1H-15N HSQC spectra in the absence versus presence of 5.3 molar equivalents of unlabeled hMARK3 (residues 320–375). (Inset) The hMARK3 UBA domain binding surface of Ub is spatially clustered around K48. Backbone nitrogen atoms of Ub residues exhibiting Δδav > 0.05 ppm are drawn as spheres on the structure of Ub (PDB ID code 1UBQ). This UBA domain binding surface is analogous to that implicated in the canonical Ub interaction with the hHR23A UBA domains (21).
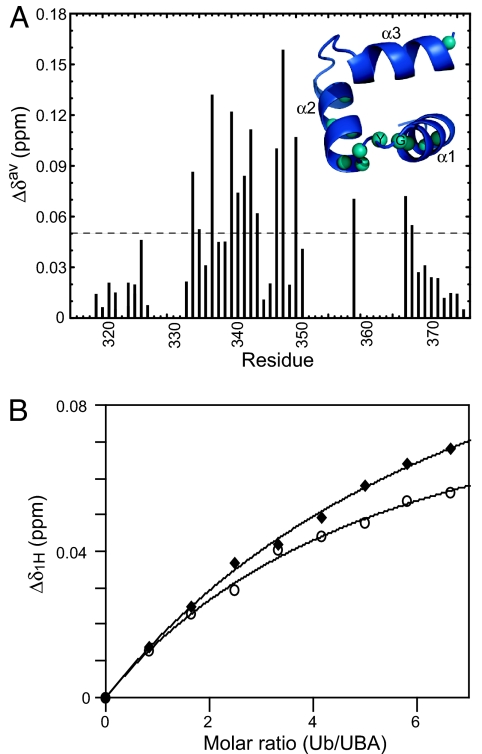
To determine the site of Ub-binding by the hMARK3 UBA domain, we performed the reciprocal titration by collecting 1H-15N HSQC spectra for the 15N-labeled UBA domain in the absence or presence of up to 6.65 molar equivalents of unlabeled monoUb. The biggest perturbations (Δδav) were observed for the α1 helix residues (D333 and V336), G339 and Y340 of the MGY motif, the neighboring residues S341 and Q342, and the α2 helix residues (Q346, E347, and L349) (Fig. 4A). Ub-binding by a canonical UBA domain typically is mediated by the MGY motif (α1–α2 loop), and residues located in the cleft between the α1 and α3 helices (underlined in SI Fig. 6 Lower), as exemplified by the Dsk2p UBA domain:Ub complex structure (29) (SI Fig. 7). The fact that residues outside this region also showed shifts may indicate that the hMARK3 UBA domain possesses an additional, secondary monoUb binding surface analogous to that located between the α2 and α3 helices of the Schizosaccharomyces pombe Mud1 UBA domain (30) (SI Fig. 6) and hHR23A UBA2 (31). However, because of unfolding and consequent absence of nearly all α3 helix resonances of the hMARK3 UBA domain from 2D spectra, it is difficult to speculate on the mode of the Ub interaction. Furthermore, on the basis of our characterization of conformational exchange dynamics within the isolated hMARK3 UBA domain in solution, we are unable to deduce whether the folded state of this UBA domain corresponds to a canonical UBA domain topology (characteristic of Ub-binding UBA domains described to date) or the fold present within the hMARK3 kinase:UBA domain crystal structure. Even though the mode of Ub-binding is unclear, we were able to estimate the dissociation constant (Kd) for the Ub–UBA domain interaction by performing nonlinear curve-fitting for the binding isotherms of V336, G339, and Q342, three well resolved residues in proximity of the canonical Ub-interaction interface (Fig. 4B). An average apparent Kd of 2.35 mM ± 0.50 mM has been obtained from the V336, G339, and Q342 binding isotherms. It should be noted that the binding isotherms do not reach saturation (Fig. 4B), even at 6.65 molar equivalents of Ub, which may affect the accuracy of the Kd estimation. However, the fact that saturation does not occur even at 6.65 equivalents of Ub is consistent with a Kd in excess of 2 mM.
Fig. 4.
A reciprocal NMR titration confirms that the hMARK3 UBA domain binds monoUb. (A) Δδav versus residue number for hMARK3 UBA domain in the presence of monoUb. Δδav was calculated for each residue in 1H-15N HSQC spectra in the absence versus presence of 5.8 molar equivalents of monoUb. No data, and thus no bars in this chart, were available for peaks absent from 2D spectra. (Inset) Ub-interacting residues of the hMARK3 UBA domain. Backbone nitrogen atoms corresponding to residues exhibiting Δδav > 0.05 ppm are indicated as spheres on the structure of the hMARK3 UBA domain from the kinase:UBA domain crystal structure (Fig. 1). The UBA domain is rotated 180° about the x axis compared with the orientation in Fig. 1B. The backbone N spheres of G339 and Y340 of the MGY motif are labeled in black text. (B) Characteristic plots of Δδ1H versus Ub/UBA molar ratio. Plot of absolute 1H chemical shift change as a function of addition of ligand for V336 (filled diamonds) and G339 (open circles). The binding isotherm for Q342, also used in the calculation of Kd, was omitted for clarity because this curve overlays with that of V336. Nonlinear curve-fitting was used to determine the Kd for the hMARK3 UBA domain interaction with monoUb, as described in Materials and Methods.
We were unable to perform a similar analysis of hMARK3/Par-1 kinase:UBA conformational dynamics and Ub-binding by using NMR spectroscopy because of a tendency for longer hMARK3 constructs to precipitate above 4°C. Thus, to assess whether the stabilization of the hMARK3 UBA domain fold in the kinase:UBA construct favors Ub binding, we compared the ability of the isolated UBA domain and kinase:UBA from hMARK3 to bind immobilized GST-Ub by using in vitro pull-down assays (SI Fig. 8 A and B). Although a control UBA domain from hHR23A, known to bind monoUb (16), detectably interacted with GST-Ub, no such interaction was observed for the isolated UBA domain or the kinase:UBA from hMARK3 (SI Fig. 8 A and B). These data indicate that stabilization of the atypical hMARK3 UBA domain fold by the kinase domain N-lobe interaction does not measurably increase the affinity of the hMARK3 UBA domain for Ub. Our findings are concordant with those of Jaleel et al. (17), who used pull-down assays to demonstrate that the AMPK/Snf1 family UBA domains have no detectable affinity for different Ub linkages and Ub-like molecules in the context of isolated UBA domains and full-length kinases. These findings can be rationalized structurally by superimposing the canonical Dsk2p UBA domain:Ub complex structure on our hMARK3 kinase:UBA domain crystal structure (SI Fig. 8C). This analysis illustrates that the hMARK3 UBA domain within the kinase:UBA domain construct could not mediate canonical Ub binding because of a steric clash that would occur between the kinase N-lobe and Ub.
Conformational Instability Modifies the Ligand-Binding Properties of the Par-1/MARK UBA Domain.
Although UBA domains are found in many proteins within eukaryotic proteomes, the arrangement of linked kinase and UBA domains only is observed in the MARKs and their homologs within the AMPK/Snf1 family of protein kinases. Many functions have been attributed to UBA domains in other proteins, including dimerization (32, 33), the binding of Ub-like (UBL) domains (34, 35), monoUb (15, 16), and K48-linked and K63-linked polyUb (16). In the present work, we have demonstrated that the hMARK3 UBA domain stably interacts with the N-lobe of the kinase domain in the hMARK3 (residues 48–370) crystal structure (average B factors for atoms in the kinase versus UBA domains were 33.05 versus 33.72, respectively) via a series of hydrophobic interactions. These are contributed primarily by the UBA domain α3 helix and are optimized by the formation of an unconventional UBA domain fold. In contrast to the conventional Ub-binding UBA domains, we found that the isolated hMARK3 UBA domain in solution is highly dynamic and in equilibrium with a highly populated unfolded state when no longer bound to the kinase domain. Our findings illustrate that interactions between the N-lobe of the kinase domain and the UBA domain α3 helix, the most unstable element in the isolated UBA domain, are essential for maintaining the helical character of the α3 helix. Characterization of the conformational exchange dynamics within this isolated UBA domain provides a plausible explanation for a lack of detectable Ub binding in simple in vitro assays (16), an observation that may be of more general relevance for the ≈30% of UBA domains that lack obvious Ub-binding activity. By using NMR spectroscopy, it was in fact possible to detect residual Ub-binding by the hMARK3 UBA domain, demonstrating that this class of UBA domains indeed may possess a previously unrecognized ultraweak Ub affinity beyond the dynamic range of pull-down experiments. It will be of interest to know whether, like the hMARK3 UBA domain, the absence of significant Ub-binding exhibited by many other UBA domains arises from millisecond to microsecond time-scale conformational exchange processes and whether these UBA domains have evolved functions and topologies that distinguish them from canonical Ub-binding UBA domains.
Previous studies have demonstrated the importance of the UBA domains within the Par1/MARKs and other AMPK/Snf1 family members for phosphorylation of the adjacent kinase domain's activation segment by the upstream activator complex, LKB1/Par4:Mo25:STRAD (17). These data are consistent with a functional role for the UBA domains in stabilizing an open conformation of the N- and C-terminal lobes of the kinase domain through interactions with the N-lobe (as observed in the kinase:UBA domain crystal structures), which in turn exposes the kinase domain activation segment for phosphorylation by LKB1/Par4. LKB1/Par4-mediated phosphorylation of the Par1/MARK activation segment threonine increases kinase activity >50-fold in vitro (36) and activates Drosophila Par1 in vivo (37).
In summary, our observations are consistent with the emerging view that different members of a family of related interaction domains may have quite different binding properties. In the case of the Par-1/MARK UBA domains, the conformational instability of the isolated domain appears to have evolved as a mechanism that contributes to destabilization of canonical Ub recognition and thus promotes an alternative intramolecular binding interaction with the kinase domain. This finding represents an elegant mechanism by which the binding potential of an interaction domain can be expanded.
Materials and Methods
hMARK3 (Residues 48–370) Crystallization and Structure Determination.
Crystals of protein expressed and purified as describing in SI Text were grown at 4°C in 1.5 M LiSO4·H2O and 0.1 M Hepes, pH 7.5. For data collection, crystals were washed in cryobuffer [1.5 M LiSO4·H2O, 0.1 M Hepes (pH 7.5), and 20% (vol/vol) glycerol] and flash frozen, and data were collected at the Advanced Photon Source (Argonne, IL) on SBC-CAT beamline BM-19. Phases were obtained by molecular replacement (PDB ID code 1ZMU). All structure figures were drawn with PyMOL (www.pymol.org).
NMR Spectroscopy.
NMR data for resonance assignments were collected for 1.6 mM 13C-15N-labeled hMARK3 (residues 320–375), and backbone resonances were assigned by using standard triple-resonance experiments (38, 39). Spectra were collected at 25°C. Samples were prepared as described in SI Text.
Conformational exchange processes in the isolated hMARK3 domain were studied via backbone 15N (27, 40) and side-chain methyl 13C (41) SQ CPMG relaxation-dispersion measurements, which are described in SI Text. Dispersion data were supplemented with backbone amide–solvent hydrogen exchange rates measured by using an 15N-labeled sample at 25°C, pH = 7.0, with the CLEANEX-PM experiment, as described by Hwang et al. (28). Additional experimental procedures are described in detail in SI Text.
Supplementary Material
Acknowledgments
We thank SBC-CAT, Advanced Photon Source, Argonne, IL, for access to the BM-19 beamline; S. Orlicky, R. Muhandiram, and T. Farquharson for technical assistance; and Drs. M. Plevin and S. Wiesner for helpful discussions. This work was supported by grants from the Canadian Institutes for Health Research (CIHR), the National Cancer Institute of Canada, and Genome Canada through the Ontario Genomics Institute (to T.P.) and grants from CIHR (to L.E.K. and F.S.). J.M.M. is the recipient of a C. J. Martin Fellowship from the National Health and Medical Research Council of Australia. D.M.K. and D.F.C. are the recipients of a CIHR training grant fellowship in protein folding and disease and a CIHR–Strategic Training Initiative in Health Research fellowship, respectively.
Abbreviations
- h
human
- AMPK
AMP-activated protein kinase
- MARK
microtubule-associated protein/microtubule affinity-regulating kinase
- Ub
ubiquitin
- UBA
Ub-associated
- N-lobe
N-terminal lobe
- HSQC
heteronuclear single-quantum correlation
- CPMG
Carr–Purcell–Meiboom–Gill.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Atomic coordinates and structure factors for hMARK3 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2QNJ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703012104/DC1.
References
- 1.Guo S, Kemphues KJ. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 2.Bohm H, Brinkmann V, Drab M, Henske A, Kurzchalia TV. Curr Biol. 1997;7:603–606. doi: 10.1016/s0960-9822(06)00260-0. [DOI] [PubMed] [Google Scholar]
- 3.Shulman JM, Benton R, St Johnston D. Cell. 2000;101:377–388. doi: 10.1016/s0092-8674(00)80848-x. [DOI] [PubMed] [Google Scholar]
- 4.Chen YM, Wang QJ, Hu HS, Yu PC, Zhu J, Drewes G, Piwnica-Worms H, Luo ZG. Proc Natl Acad Sci USA. 2006;103:8534–8539. doi: 10.1073/pnas.0509955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ, III, Wu Z, Stephenson MT, Piwnica-Worms H. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 6.Muller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. Mol Cell. 2001;8:983–993. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 7.Muller J, Ritt DA, Copeland TD, Morrison DK. EMBO J. 2003;22:4431–4442. doi: 10.1093/emboj/cdg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drewes G, Trinczek B, Illenberger S, Biernat J, Schmitt-Ulms G, Meyer HE, Mandelkow EM, Mandelkov E. J Biol Chem. 1995;270:7679–7688. doi: 10.1074/jbc.270.13.7679. [DOI] [PubMed] [Google Scholar]
- 9.Illenberger S, Drewes G, Trinczek B, Biernat J, Meyer HE, Olmsted JB, Mandelkow EM, Mandelkow E. J Biol Chem. 1996;271:10834–10843. doi: 10.1074/jbc.271.18.10834. [DOI] [PubMed] [Google Scholar]
- 10.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. Cell. 1997;89:297–308. doi: 10.1016/s0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 11.Nishimura I, Yang Y, Lu B. Cell. 2004;116:671–682. doi: 10.1016/s0092-8674(04)00170-9. [DOI] [PubMed] [Google Scholar]
- 12.Levin DE, Bishop JM. Proc Natl Acad Sci USA. 1990;87:8272–8276. doi: 10.1073/pnas.87.21.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning G, Whyte GB, Martinez R, Hunter T, Sudsarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 14.Drewes G. Trends Biochem Sci. 2004;29:548–555. doi: 10.1016/j.tibs.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. Nat Struct Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 16.Raasi S, Varadan R, Fushman D, Pickart CM. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 17.Jaleel M, Villa F, Deak M, Toth R, Prescott AR, Van Aalton DM, Alessi DR. Biochem J. 2006;394:545–555. doi: 10.1042/BJ20051844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller TD, Feigon J. J Mol Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 19.Marx A, Nugoor C, Müller J, Panneerselvam S, Timm T, Bilang M, Mylonas E, Svergun D, Mandelkow EM, Mandelkow E. J Biol Chem. 2006;281:27586–27599. doi: 10.1074/jbc.M604865200. [DOI] [PubMed] [Google Scholar]
- 20.Panneerselvam S, Marx A, Mandelkow EM, Mandelkow E. Structure (London) 2006;14:173–183. doi: 10.1016/j.str.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 21.Mueller TD, Kamionka M, Feigon J. J Biol Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, Sykes BD, Richards FM. J Mol Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 23.Palmer AG, III, Kroenke CD, Loria JP. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 24.Mulder FAA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Nat Struct Biol. 2001;8:932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 25.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AR, Dobson CM, Kay LE. Nature. 2004;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 26.Mittermaier A, Kay LE. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 27.Tollinger M, Skrynnikov NR, Mulder FAA, Forman-Kay JD, Kay LE. J Am Chem Soc. 2001;123:11341–11352. doi: 10.1021/ja011300z. [DOI] [PubMed] [Google Scholar]
- 28.Hwang T-L, van Zijl PCM, Mori S. J Biomol NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- 29.Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure (London) 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Trempe J-F, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble MEM, Endicott JA. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varadan R, Assfalg M, Raasi S, Pickert C, Fushman D. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Sasaki T, Funakoshi M, Endicott JA, Kobayashi H. Biochem Biophys Res Commun. 2005;336:530–535. doi: 10.1016/j.bbrc.2005.08.126. [DOI] [PubMed] [Google Scholar]
- 33.Bayrer JR, Zhang W, Weiss MA. J Biol Chem. 2005;280:32989–32996. doi: 10.1074/jbc.M507990200. [DOI] [PubMed] [Google Scholar]
- 34.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. J Mol Biol. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Lowe ED, Hasan N, Trempe J-F, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Acta Crystallogr D. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 36.Lizcano JM, Goransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Makela TP, Hardie DP, Alessi DR. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J-W, Imai Y, Lu B. J Neurosci. 2007;27:574–581. doi: 10.1523/JNEUROSCI.5094-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kay LE. Prog Biophys Mol Biol. 1995;63:277–299. doi: 10.1016/0079-6107(95)00007-0. [DOI] [PubMed] [Google Scholar]
- 39.Sattler M, Schleucher J, Griesinger C. Prog NMR Spectrosc. 1999;34:93–158. [Google Scholar]
- 40.Loria JP, Rance M, Palmer AG., III J Am Chem Soc. 1999;121:2331–2332. [Google Scholar]
- 41.Skrynnikov NR, Mulder FAA, Hon B, Dahlquist FW, Kay LE. J Am Chem Soc. 2001;123:4556–4566. doi: 10.1021/ja004179p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.