Abstract
Current understanding of the integration of cell division and expansion in the development of plant lateral organs such as leaves is limited. Cell number is established during a mitotic phase, and subsequent growth into a mature organ relies primarily on cell expansion accompanied by endocycles. Here we show that the three Arabidopsis cyclin D3 (CYCD3) genes are expressed in overlapping but distinct patterns in developing lateral organs and the shoot meristem. Triple loss-of-function mutants show that CYCD3 function is essential neither for the mitotic cell cycle nor for morphogenesis. Rather, analysis of mutant and reciprocal overexpression phenotypes shows that CYCD3 function contributes to the control of cell number in developing leaves by regulating the duration of the mitotic phase and timing of the transition to endocycles. Petals, which normally do not endoreduplicate, respond to loss of CYCD3 function with larger cells that initiate endocycles. The phytohormone cytokinin regulates cell division in the shoot meristem and developing leaves and induces CYCD3 expression. Loss of CYCD3 impairs shoot meristem function and leads to reduced cytokinin responses, including the inability to initiate shoots on callus, without affecting endogenous cytokinin levels. We conclude that CYCD3 activity is important for determining cell number in developing lateral organs and the relative contribution of the alternative processes of cell production and cell expansion to overall organ growth, as well as mediating cytokinin effects in apical growth and development.
Keywords: cell division, cyclin D, flowering time, plant development
The development of plants is strikingly different from that of animals, and their indeterminate growth pattern and repeated organogenesis provide plasticity to environmental signals and damage. This poses challenges in integrating cell division and cellular differentiation during the formation of lateral organs such as leaves, which grow from primordia initiated on the flanks of the shoot apical meristem (SAM), a dome-like structure containing stem cells at the shoot tip (1). Initial primordium growth is associated with mitotic cell cycles, but cell division ceases in a basipetal wave (from tip to base) once the leaf is a few millimeters in length, and subsequent growth depends primarily on the increasing size of the differentiating cells already present (2, 3). This cell expansion and differentiation are accompanied by endocycles in which nuclear DNA content is increased by rounds of full genome duplication without intervening mitoses, giving rise to cells with higher ploidy levels. There is a strong correlation between final cell size and nuclear ploidy level (4). Final organ size therefore depends on both contributing cell number and the ultimate size to which these cells expand (5), alternative processes that reflect, respectively, the number of mitotic cycles and endocycles.
Whereas the broad regulation of the cell cycle in plant cells is understood and has resulted in definition of plant-specific aspects, including a plant-specific mitotic cyclin-dependent kinase, there is little understanding of its overall integration with development (6). The unusually large number of genes encoding core cell cycle factors in plants (7, 8) complicates genetic analysis, and the focus of most previous work on ectopic expression of cell cycle regulators or their engineered mutant versions limits conclusions regarding their normal roles in development.
The CYCD family in Arabidopsis includes 10 genes in seven subgroups. CYCD proteins act in a pathway broadly conserved among plants, worms, flies, and higher animals (9, 10), in which CYCD-containing cyclin-dependent kinase complexes phosphorylate the retinoblastoma-related protein (RBR), allowing the activity of E2F regulated genes to be manifest and promoting S phase entry.
The Arabidopsis CYCD3 subgroup has three members (CYCD3;1, CYCD3;2, and CYCD3;3), and these are prime candidates for playing key roles integrating cell division in leaf and lateral organ development. CYCD3;1 activity rises as cells reenter the cell cycle (11, 12), and its ectopic expression in cell cultures results in the accumulation of cells in G2 after accelerated progression through the G1-to-S phase boundary (13). Plants overexpressing CYCD3;1 (CYCD3;1 OE) exhibit hyperplasia and ectopic divisions in leaves and other tissues (11, 14). Cellular expansion and its accompanying endoreduplication are also inhibited in CYCD3;1 OE plants, and in consequence leaf growth becomes largely dependent on cell proliferation (14). Trichomes are single-celled leaf hairs that normally contain an endoreduplicated nucleus, but overexpression of CYCD3;1 induces cell division in place of endocycles to produce a multicellular structure (15). These observations show the plasticity of overall leaf growth to the alternative contributing components of cell number and size and further suggest that CYCD3;1 promotes mitotic cycles. However, being based on high-level constitutive expression, they do not allow us to conclude that CYCD3;1 is a normal regulator of these processes.
Cytokinins are plant-specific hormones that influence numerous developmental programs, including shoot regeneration, leaf development, and greening, as well as promoting the cell cycle (16). Somatic plant tissues can dedifferentiate and proliferate in culture to form callus in response to the combined effects of the plant hormones auxin and cytokinin, with the resulting cellular identity dependent on their relative concentrations. Plants with reduced cytokinin levels as a result of ectopic expression of cytokinin-degrading enzymes or mutations in biosynthetic genes (17, 18) show that cytokinin has an important role for in promoting SAM function and cell number in developing leaves.
CYCD3;1 expression is induced by cytokinin, and its overexpression renders callus formation independent of exogenous cytokinin, suggesting that CYCD3;1 is a key target of cytokinin (11). Together with the action of CYCD3 at the point of commitment in the cell cycle, CYCD3 activity may integrate multiple input signals influencing the number and types of cell cycles (13).
Here we present a genetic loss-of-function analysis of the complete CYCD3 subgroup in shoot development. We show that these genes are not required for cell cycle activity or morphogenesis, but rather are necessary for determining the normal cell number in shoot lateral organs by promoting mitotic cycles and restraining endocycles. CYCD3 is shown to be essential for cytokinin-mediated functions, in particular the regeneration of shoots from callus.
Results
Expression of CYCD3 Genes in Arabidopsis Shoots.
The expression of the three Arabidopsis CYCD3 genes was analyzed in vegetative and flowering shoots using reporter fusions to GUS (encoding β-glucuronidase) and mRNA analysis by real-time PCR (RT-PCR). CYCD3;1 expression is restricted to the SAM, very young primordia, and young hydathodes (water-secreting glands on the leaf margin), whereas CYCD3;2 and CYCD3;3 reporters are also active in older leaf primordia, with CYCD3;2 expression persisting longest in young leaves (Figs. 1 and 2A). These overlapping but distinct patterns of spatial and temporal regulation are consistent with previous partial analysis of CYCD3;1 and CYCD3;2 expression by in situ hybridization (11, 14, 19) and confirm an association of CYCD3 expression with early stages of leaf and floral organ ontogeny.
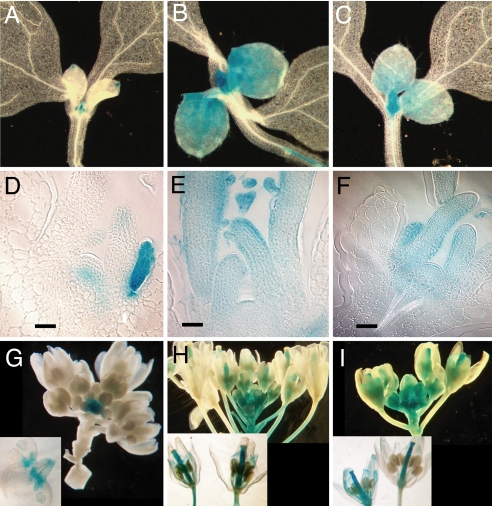
Fig. 1.
Arabidopsis CYCD3 genes display overlapping and distinct expression in proliferating tissues of the vegetative and floral shoot. CYCD3;1 expression is detected in young organs (A, D, and G), the periphery of the SAM, and the rib zone (D); CYCD3;2 and CYCD3;3 expression persists longer through lateral organ development (B, C, H, and I) and is detected throughout the vegetative SAM (E and F). In developing flowers CYCD3;2 (H) and CYCD3;3 (I) are maintained until opening. CYCD3;1 is detected only in very young flowers, most persistently in the petal (G).
Fig. 2.
Cellular development in cycd3;1-3 mutant and WT leaves and petals. (A) CYCD3 transcript levels in successive rosette leaves of a plant with seven visible leaves. L1 and L2 are the pair of juvenile leaves, and L3–L6 are sequential adult phase leaves (L6 being the youngest dissectible leaf). Relative levels are scaled to expression in the youngest leaf and show a faster decrease in the levels of CYCD3;1 mRNA in the older leaves. (B) Transcript levels of the mitotic gene CYCB1;1 in sequential leaves of cycd3;1-3 (d3;1-3) and WT, consistent with reduced cell division in young cycd3;1-3 leaves. (C–E) Adult phase leaves of cycd3;1-3 have fewer larger cells in both the adaxial epidermis and palisade mesophyll, whereas similar numbers were found in the juvenile leaves (L1+L2). (E) Traces of cell outlines for the adaxial epidermis (Upper) and photographs of palisade mesophyll (Lower) for WT (Left) and cycd3;1-3 (Right). (Scale bar: 50 μm.) (F) Cells in the adaxial epidermis of WT and cycd3 mutant petals. The number of cells in the adaxial epidermis is indicated for cycd3;1, cycd3;1-3, and WT. Loss of CYCD3;1 has the strongest effect. (Scale bar: 60 μm.) (G and H) Ploidy histogram for nuclei isolated from rosette leaves L3 (G) and petal (H) of WT and cycd3;1-3 at the time of flowering. The proportion of nuclei with 16C DNA content is increased in the cycd3;1-3 L3 (G), and a proportion of nuclei with 8C was detected in the mutant petals (H). (I) The growth rate of leaf 3 is similar for cycd3;1-3 and WT (Left), whereas the average ploidy in the mutant is higher (Right).
D3 Cyclins Regulate Cell Production and Size but Not Overall Organ Growth.
We noted that the basipetal cessation of the mitotic phase of leaf development when a leaf is a few millimeters long (2, 3) is mirrored by CYCD3 expression, being confined to the youngest leaves and persisting longest in the basal region [supporting information (SI) Fig. 4A]. This raised the possibility that CYCD3 activity directly influences the duration of the mitotic phase of leaf development, as suggested by its promotion of ectopic mitotic cycles in mesophyll and epidermal cells (13), trichomes (15), leaves (14), and seedlings (20).
To gain further insight into the role of CYCD3, we compared gene expression using Affymetrix (Santa Clara, CA) arrays in WT and plants overexpressing CYCD3;1 (CYCD3;1 OE). We have previously defined a set of 82 genes expressed at G2/M in synchronized cultured cells that provide a reliable expression fingerprint for mitosis (8). These include almost all mitotic cyclins and many other genes involved in exclusively mitotic processes. Ninety percent (74/82) of these mitotic genes were up-regulated in 7-day CYCD3;1 OE plants compared with WT (SI Fig. 4 and SI Table 1). When expression profiles were compared between 7- and 14-day plants, 22 of these mitotic genes are significantly reduced in WT reflecting the decreased proportion of proliferating tissues, whereas only a single mitotic gene was reduced in the CYCD3;1 OE (SI Table 2). This reflects the continued mitotic cycles in developing CYCD3;1 OE leaves (SI Fig. 4B), consistent with CYCD3;1 OE extending the developmental window during which leaf cells are maintained in the mitotic cycle.
To determine further the role of CYCD3 in lateral organs, we identified loss-of-function mutants in the three CYCD3 genes (SI Fig. 5). Remarkably, all single, double, and triple mutant combinations produced adult plants, demonstrating that CYCD3 function is not essential for cell cycles or morphogenesis.
The consequences of CYCD3;1 OE suggested that loss of CYCD3 function could affect the number of cells contributing to lateral organs. The Col-0 ecotype of Arabidopsis produces a basal rosette of ≈11 leaves followed by a flowering stalk. We quantified cell numbers in the third, fourth, and fifth leaves (L3–L5), of the triple homozygous cycd3;1 cycd3;2 cycd3;3 (cycd3;1-3) mutant and WT plants (Fig. 2C). The number of cells comprising both the adaxial (upper) epidermis and underlying mesophyll was reduced by 20–30% in cycd3;1-3, consistent with the earlier decline in expression of the mitotic B-type cyclin CYCB1;1 in young cycd3;1-3 leaves (Fig. 2B).
This reduction of cell number did not inhibit the organ growth rate, based on analysis of L3 (Fig. 2I Left) and L4 (data not shown) leaves from their emergence until mature rosette stage in midflowering, and no difference was observed in the size of equivalent adult leaves of the rosette (SI Fig. 6). However, measurement of cell areas in rosette leaves showed substantially larger mesophyll and adaxial epidermis cells (Fig. 2 D and E), accounting for the unaffected growth rate and leaf size.
Loss of CYCD3 therefore reduces cell number in developing leaves and hence leads to earlier cessation of mitotic phase. In combination with the reciprocal phenotype of CYCD3;1 OE plants (14), we conclude that CYCD3 activity plays a significant role in determining the cell number comprising leaves but does not affect overall organ size.
CYCD3 Restrains Endocycles.
Because cellular expansion upon exit from the mitotic cycle is normally accompanied by endoreduplication in Arabidopsis (2) and inhibition of endocycles correlates with reduced cell growth (21), we examined endoreduplication in cycd3;1-3. Endoreduplication in leaf 3, which has similar growth rates in the mutant and WT, was determined from its emergence until the rosette bore 10 leaves. The average ploidy level was found to be consistently higher in cycd3;1-3 (Fig. 2I Right). Furthermore, in a rosette with seven visible leaves, loss of even two CYCD3 genes results in the earlier onset of endoreduplication in young leaves (SI Fig. 7C, L5) and higher ploidy levels in older leaves (SI Fig. 7C, L4–L1), an effect more pronounced in the cycd3;1-3 mutant. The premature onset of endoreduplication in the mutant indicates that the normal function of CYCD3 is to delay the onset of endoreduplication by extending the “mitotic window” of leaf development during which cell division normally occurs.
Loss of CYCD3 Causes Initiation of Endoreduplication in Petals.
Unlike the leaf, which is multilayered and contains several cell types, the Arabidopsis petal is a lateral organ of two cell layers, each composed of a single cell type that does not endoreduplicate (4). Consistent with the leaf analysis, cycd3;1-3 mutant petals were of equivalent size to WT but contained 60% of the number of cells (Fig. 2F). Cell size was increased almost 100% in cycd3;1-3 petals (WT adaxial cells, 170 μm2; cycd3;1-3, 326 μm2) (Fig. 2F). Remarkably, this larger cell size was accompanied by the onset of endoreduplication, with increased 4C nuclei and the appearance of a distinct 8C peak (Fig. 2H). We note that the majority of the effect on petal cell number is contributed by loss of CYCD3;1 (Fig. 2F), whose expression persists relatively longer in petals than in other floral organs (Fig. 1G), suggesting that tissues may have differential requirements for specific CYCD3 activity.
Different Regulation of Cell Number and Organ Size Occurs in Juvenile Leaves.
The first two leaves of Arabidopsis that emerge after germination are a symmetric pair of juvenile leaves morphologically distinct from subsequent adult leaves (22). Analysis of this juvenile leaf pair showed they have the same number of cells in both the cycd3;1-3 and WT (Fig. 2C, L1+L2), indicating that, in contrast to later leaves and petals, CYCD3 does not influence cell number in these leaves. However, as in the adult leaves, CYCD3 function is required to restrain endoreduplication and cell size, which both increase in cycd3;1-3 L1 and L2 (Fig. 2D, L1+L2, and SI Fig. 7C), resulting in an increase in juvenile leaf area of 60% at the seven-leaf stage.
CYCD3 Activity Is Important for Normal SAM Function.
Longitudinal sections of the vegetative SAM in cycd3;1-3 revealed that it consists of fewer cells, demonstrating a further role for CYCD3 in controlling the size of the SAM cell pool (Fig. 3A). Impaired SAM function is further evidenced by the delayed initiation of the first pair of postembryonic leaves (SI Fig. 7B), reduced floral density (Fig. 3D), and an increased angle between sequential flowers from 135° to a more variable angle averaging 177° (data not shown). The cycd3;1-3 mutant also displays substantially delayed flowering, producing 14–15 rosette leaves compared with 11 WT leaves (Fig. 3E and SI Fig. 7B). These multiple effects on SAM function suggest that CYCD3 plays an important role in SAM function at different stages of development.
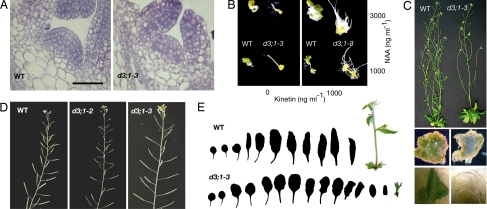
Fig. 3.
cycd3;1-3 mutants have SAM defects and reduced cytokinin responses. (A) Longitudinal sections of the SAM of WT (Left) and cycd3;1-3 (d3;1-3; Right); the mutant SAM contains fewer cells. (Scale bar: 50 μm.) (B) Callus induction on hypocotyl segments of triple cycd3 mutants. Unlike cycd3;1-3, WT explants form calli in the absence of cytokinin (kinetin) on both ends (Left). At 1,000 ng·ml−1 kinetin WT explants form green calli (Right), whereas cycd3;1-3 hypocotyls develop yellow root-like tissue [Upper, 3,000 ng·ml−1 NAA (auxin); Lower, 1,000 ng·ml−1 NAA]. (C) cycd3;1-3 floral shoots (Right) display reduced branching of axillary shoots. WT calli maintained for several weeks on 3,000 ng·ml−1 auxin and kinetin develop shoot-like structures of friable green cells (Left), whereas cycd3;1-3 develops yellow callus covered in roots (Right). (D) floral shoots of triple cycd3 mutants have a reduced density of siliques. (E) Dissected rosette leaves produced by WT and cycd3;1-3 before bolting. The floral stalk is presented at the time of analysis to illustrate late flowering.
Loss of CYCD3 Impedes Cytokinin Responses.
These SAM phenotypes, as well as the reduced branching and axillary shoots of cycd3;1-3 floral shoots (Fig. 3C), are strikingly similar to plants with reduced cytokinin levels (18–20). This could suggest either reduced cytokinin levels or deficient responses. We measured endogenous cytokinin levels in seedlings and found no significant changes in cycd3;1-3 seedlings (see SI Table 3). We therefore tested cytokinin responses using a callus assay (23), in which cytokinins promote shoot formation and greening, whereas higher relative concentrations of auxin promote root development.
Auxin alone was sufficient to induce callus on both ends of WT and single and double mutant hypocotyl stem segments in tissue culture, but, in contrast, cycd3;1-3 required exogenous cytokinin for calli to develop on both sides of the explants (Fig. 3B and SI Fig. 7D). In the presence of cytokinin WT, single and double cycd3 mutant calli were green and developed shoots, whereas cycd3;1-3 calli remained yellow and did not develop shoots. In response to higher concentrations of auxin and cytokinin (Fig. 3 B and C), cycd3;1-3 calli developed roots, but no case of shoot formation was observed, even at high cytokinin levels. We conclude that CYCD3 function is required for normal cytokinin responses and that the cycd3;1-3 mutant is strongly impaired in several aspects of cytokinin response, including normal meristem maintenance, branching, shoot formation, and greening of callus.
Discussion
Plant organ growth is determined by cell number and size. If either of these parameters is modulated within certain limits, a mechanism termed “compensation” generally ensures that the genetically determined size is achieved by compensatory changes in the other (24). Because cell proliferation and cell expansion are temporally separated during lateral organ development and require the mitotic cell cycle and endocycle, respectively (2, 3), a key aspect in leaf development is the transition between mitotic cycle and endocycle, which then determines the relative contribution of cell number and size to the organ.
In a mutant lacking the complete CYCD3 gene family in Arabidopsis, we find that most organ sizes are normal, but the extent and type of cell cycles during organ growth are strongly affected. In adult leaves and petals, a premature exit from cell proliferation occurs. This earlier exit from the mitotic cycle and consequential reduced cell number are compensated by an earlier onset and more extensive endoreduplication and a larger cell size, so that final organ sizes are unchanged. These data show conclusively that CYCD3 function is not required for initiation or progression of endocycles but that its loss in adult leaf development results in the premature onset of endocycles and increased cellular DNA content. Taken together with the data from ectopic expression, we conclude that CYCD3 acts on the mitotic population of cells both to maintain these cells “in cycle” and to restrain their transition to endocycling. Within the framework of the compensation mechanism, CYCD3 is therefore important to determine the type of cycle and mode of organ growth.
Previous analysis of the mitotic/endocycle transition during Arabidopsis organ development has largely depended on engineered phenotypes using ectopic expression strategies, which demonstrate in general that the transition to endocycles requires, and is promoted by, a decrease in mitotic kinase activity (6). This can be brought about by increased expression of the cyclin-dependent kinase inhibitory proteins KRP2 (2, 25) and SIM (26), up-regulation of the APC/C component CCS52A (27), or dominant negative forms of the mitotic kinase CDKB1;1 (28). Mutants of the A-type cyclin CYCA2;1 or its regulator ILP1, or of CYCA2;3, also result in increased endocycling without apparent effects on the population of proliferating cells (29, 30). Whereas increased expression of such mitosis-promoting genes enhances mitotic cycles, genes that stimulate DNA replication, such as CDC6, E2Fa, and CDT1, notably promote both the mitotic cycle and endocycle when ectopically expressed, rather than affecting the timing of the transition (6).
In contrast, ectopic expression of CYCD3;1, whose expression and activity are also normally activated in late G1 phase (8, 12, 31), specifically promotes the mitotic cell cycle, leading to increased cell number, reduced cell size, and reduced endocycling (14, 15).
The effect of CYCD3 on endocycles could be either due to direct inhibition or an indirect consequence of the organ size compensation mechanism, such that reduced cell number is compensated by increased cell size and consequentially increased endocycles. Analysis of juvenile leaves in cycd3;1-3 suggests that the CYCD3 indeed plays a direct role in restraining endocycles. In these leaves cell number is unaffected in cycd3;1-3, showing that CYCD3 activity is not rate limiting for mitotic activity, but endoreduplication and cell size are nevertheless increased. This also indicates that the cell number determination and the compensation mechanism operate differently in juvenile leaves. In petals, as in adult leaves, reduced cell number and increased cell size are observed, and endocycles are initiated in cells that do not normally endoreduplicate. We conclude that CYCD3 has the specific ability to inhibit endocycles, consistent with the effects of its ectopic expression (14, 15).
There are two explanations of how CYCD3 promotes mitotic cycles over endocycles, unlike other genes operating at the G1/S transition that promote S phase entry rather than a specific type of cycle. First, CYCD3 could also act as a mitotic cyclin, boosting G2/M kinase activity and hence directly enhancing progress from G2 into mitosis. Although no association between CYCD and mitotic cyclin-dependent kinase has been detected in plant cell extracts using immunoprecipitation or proteomic approaches (32, 33), cyclin-dependent kinase can bind Arabidopsis CYCD4;1 and tobacco CYCD3 in vitro (34, 35), and histone H1-directed CYCD3 kinase activity is detected in mitotic tobacco suspension cultured BY-2 cells (36). The evidence for CYCD promotion of the G2/M transition is equivocal: Antirrhinum CYCD1, when induced at G2/M in synchronized BY-2 cells, indeed increased the rate of progression through mitosis (37), and ectopic expression of CYCD3;1 in Arabidopsis trichomes leads to additional mitoses (15). On the other hand, expression of CYCD3 in cell cultures promotes the G1/S transition in tobacco (36) and Arabidopsis cells (13) but does not increase the rate of progression through mitosis. Indeed, in Arabidopsis cell cultures and CYCD3;1 OE plants (13, 14), cells accumulate in G2 phase, inconsistent with a direct role in stimulating mitosis.
The second possible explanation of CYCD3 effects on both mitotic cycle and endocycle is that CYCD3 could trigger a commitment to the entire mitotic cycle at the G1/S transition. Because altering the levels of downstream components of the CYCD–E2F-RB pathway by E2Fa overexpression or RBR inactivation does not promote mitotic cycles over endocycles, but rather promotes both types of cycle depending on the developmental stage of the leaf (38, 39); CYCD3 would in this case have to act through specific targets. Two possible downstream candidates are the E2F-related genes E2Fc and DEL1 (40, 41). However, although del1 mutants have increased endoreduplication, there is no effect on cell division, suggesting that DEL1 acts after the switch to endocycles (41). E2Fc RNAi plants phenocopy CYCD3;1 OE with smaller cells and reduced endocycles (40), but the E2Fc OE phenotype of severe dwarfing does not resemble cycd3;1-3 (40), and E2Fc expression is conversely up-regulated in CYCD3;1 OE plants (14). As the upstream component of the CYCD-RBR-E2F regulatory pathway, CYCD3 appears to be a primary determinant of mitotic over endoreduplication cycles and may act independent of E2Fc.
The cycd3;1-3 mutant also inhibit increased cell size, in contrast to the proposed role of CYCD in Drosophila in promoting cell growth (42). Our results demonstrate that in Arabidopsis CYCD3 function is not required for growth, and the increased cell size observed is probably due to the compensation mechanism.
Does CYCD3 play a part in normal development in determining cell number in organ growth? Previous work has proposed CYCD3;1 as a positively acting target of AINTEGUMENTA (ANT) affecting cell number in lateral organs, because ectopic ANT expression leads to extended CYCD3;1 expression, extra cell divisions, and increased leaf size (43). In this case, prolonged CYCD3;1 expression is associated with an increased number of cells of normal size. This apparent overriding of the compensation mechanism may be linked to ANT's proposed direct role in determining organ size (5). ANT is a downstream target of the auxin inducible gene ARGOS (44), providing a link between hormone control and cell cycle activity through ANT and CYCD3;1.
A further phenotype observed in cycd3;1-3 is a reduction in SAM size and reduced cytokinin responses, despite normal endogenous cytokinin levels. Callus derived from cycd3;1-3 hypocotyls was unable to produce shoots even in response to high concentrations of cytokinin, reciprocating the cytokinin-independent production of green callus from CYCD3;1 OE leaf tissue and the induction of CYCD3;1 expression by cytokinin (11, 45). Similar effects on meristem size were observed in ectopic expressers of cytokinin oxidase and mutants of biosynthetic genes (17, 18), suggesting that CYCD3 genes may be a major factor in responses of meristem cells to cytokinin. Indeed, CYCD3;1 expression is up-regulated in amp1 mutants that have an enlarged SAM in response to elevated zeatin levels due to an increased number of cells (11, 46).
A further cycd3;1-3 phenotype is the late onset of flowering with at least three additional leaves. The RBR-binding protein FVE is known to be involved in the vegetative to reproductive shift (47), and, because CYCD3-directed phosphorylation affects RBR function, CYCD3 may link cell cycle control with developmental switches.
We conclude that CYCD3 plays an important role as a regulator of the population of proliferating cells and in determining cell number and endoreduplication/cell size in developing shoot lateral organs. CYCD3 links these two aspects of organ development, impacting on the mode of growth that contributes to the organ. The proposed action of CYCD3 downstream of ARGOS and ANT in leaf development, and of AMP1 and cytokinin in the SAM, suggest that this gene family also plays key roles in integrating cell proliferation in development in response to hormonal signals.
Materials and Methods
Expression Analysis.
CYCD3;2 and CYCD3;3 reporters lines carry 2,541 and 2,398 bp, respectively, of 5′ upstream sequence (20). CYCD3;1::GUS and CYCD3;1 OE lines were described (11). CYCD3 transcripts were measured by quantitative RT-PCR; primers are available on request.
CYCD3 Mutants.
SET4061 contains an insertion in CYCD3;1 (48), 5580 in CYCD3;2 (19), and N174667 in CYCD3;3, in all cases within the first exon (SI Fig. 5). The absence of both full-length transcript and transcripts containing the cyclin box and LxCxE motif in these lines was confirmed, and they are therefore considered null alleles.
All lines were backcrossed twice to Col-0 before crossing for double and triple mutant combinations and grown under long days (18 h/6 h).
Silique angles were recorded on paper discs and analyzed by Student's t test (28 ≤ n ≤ 30).
Microscopic Examination and Cell Counting.
Shoots were embedded in Technovit 7100 resin, and 6-μm sections were cut on a Leica RM2145 microtome (14). Epidermal and palisade mesophyll cells were viewed in formaldehyde-fixed and ethanol-cleared leaves of seven-leaf rosettes, photographed by using Nikon Optiphot-2 and Nikon SMZ-U microscopes, and measured in ImageJ (49). Sizes and density were scored at 25% and 75% of the leaf length, and average cell density was used to estimate the total number of leaf cells. Cells were recorded at the distal portion of petals of stage 13 and 14 flowers (50) and analyzed as for leaves.
Ploidy Analysis.
Endoreduplication of six successive leaves of plants with seven visible leaves was measured by flow cytometry by pooling similar leaves from four rosettes (51). Reliable dissection of the seventh leaf was not possible because of size limitations.
Callus Induction Assay and Cytokinin Measurements.
The hormone response of WT and single, double, and cycd3;1-3 mutants was assayed by using callus induction on hypocotyl segments (23). Results were recorded 24 days after transfer of segments. Calli induced with 3,000 ng/ml kinetin and 3,000 ng/ml 1-napthaleneacetic acid were maintained on hormone-containing medium for a further 30 days. Cytokinin levels in 8-day-old in vitro grown seedlings were measured by liquid chromatography–tandem MS (52).
Analysis of M Phase Gene Expression.
Transcript level analysis of 7- and 14-day-old WT and CYCD3;1 OE shoot tissue using duplicate Affymetrix ATH1 arrays was as described (8). Average signals of genes defined as having cell-cycle-regulated expression with a peak at mitosis (M) (8) were plotted for WT and CYCD3;1 OE at 7 and 14 days (SI Fig. 4A).
Supplementary Material
Acknowledgments
We thank Susan Howroyd for technical help, the Swiss Centre for Functional Genomics (Zürich, Switzerland) for microarray analysis, and T. Jack (Dartmouth College, Hanover, NH) for line 5580. This work was funded by Biotechnology and Biological Sciences Research Council Grants BB/D011914/1 (to W.D.) and BBS/B/13268 (to M.M.), and a Cooperative Award in Science and Engineering studentship (to C.C.), the European Union Systems Biology of Stem Cell Function training network (A.A.A.), the Isaac Newton Trust (S.S.), the European Union Marie Curie fellowship program (S.C.M. and J.N.), and the European Union Auxin and the Cell Cycle training network (N.B. and J.N.).
Abbreviations
- SAM
shoot apical meristem
- RBR
retinoblastoma-related protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704166104/DC1.
References
- 1.Carraro N, Peaucelle A, Laufs P, Traas J. Plant Mol Biol. 2006;60:811–826. doi: 10.1007/s11103-005-2761-6. [DOI] [PubMed] [Google Scholar]
- 2.De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inze D. Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- 4.Sugimoto-Shirasu K, Roberts K. Curr Opin Plant Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Mizukami Y. Curr Opin Plant Biol. 2001;4:533–539. doi: 10.1016/s1369-5266(00)00212-0. [DOI] [PubMed] [Google Scholar]
- 6.Inze D, De Veylder L. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- 7.Vandepoele K, Raes J, De Veylder L, Rouze P, Rombauts S, Inze D. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menges M, de Jager SM, Gruissem W, Murray JA. Plant J. 2005;41:546–566. doi: 10.1111/j.1365-313X.2004.02319.x. [DOI] [PubMed] [Google Scholar]
- 9.Frei C. Cell Cycle. 2004;3:558–560. [PubMed] [Google Scholar]
- 10.Sherr CJ, Roberts JM. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 11.Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA. Science. 1999;283:1541–1544. doi: 10.1126/science.283.5407.1541. [DOI] [PubMed] [Google Scholar]
- 12.Riou-Khamlichi C, Menges M, Healy JM, Murray JA. Mol Cell Biol. 2000;20:4513–4521. doi: 10.1128/mcb.20.13.4513-4521.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menges M, Samland AK, Planchais S, Murray JA. Plant Cell. 2006;18:893–906. doi: 10.1105/tpc.105.039636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA. Plant Cell. 2003;15:79–92. doi: 10.1105/tpc.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnittger A, Schobinger U, Bouyer D, Weinl C, Stierhof YD, Hulskamp M. Proc Natl Acad Sci USA. 2002;99:6410–6415. doi: 10.1073/pnas.092657299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakakibara H. Annu Rev Plant Biol. 2006;57:431–449. doi: 10.1146/annurev.arplant.57.032905.105231. [DOI] [PubMed] [Google Scholar]
- 17.Miyawaki K, Tarkowski P, Matsumoto-Kitano M, Kato T, Sato S, Tarkowska D, Tabat S, Sandberg G, Kakimoto T. Proc Natl Acad Sci USA. 2006;103:16598–16603. doi: 10.1073/pnas.0603522103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaminathan K, Yang Y, Grotz N, Campisi L, Jack T. Plant Physiol. 2000;124:1658–1667. doi: 10.1104/pp.124.4.1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masubelele NH, Dewitte W, Menges M, Maughan S, Collins C, Huntley R, Nieuwland J, Scofield S, Murray JA. Proc Natl Acad Sci USA. 2005;102:15694–15699. doi: 10.1073/pnas.0507581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inze D, De Veylder L. Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerstetter RA, Poethig RS. Annu Rev Cell Dev Biol. 1998;14:373–398. doi: 10.1146/annurev.cellbio.14.1.373. [DOI] [PubMed] [Google Scholar]
- 23.Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Nature. 2001;109:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- 24.Horiguchi G, Ferjani A, Fujikura U, Tsukaya H. J Plant Res. 2006;119:37–42. doi: 10.1007/s10265-005-0232-4. [DOI] [PubMed] [Google Scholar]
- 25.Jasinski S, Riou-Khamlichi C, Roche O, Perennes C, Bergounioux C, Glab N. J Cell Sci. 2002;115:973–982. doi: 10.1242/jcs.115.5.973. [DOI] [PubMed] [Google Scholar]
- 26.Churchman ML, Brown ML, Kato N, Kirik V, Hulskamp M, Inze D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, et al. Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi E. EMBO J. 1999;18:4476–4484. doi: 10.1093/emboj/18.16.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inze D, De Veylder L. Plant Cell. 2004;16:2683–2692. doi: 10.1105/tpc.104.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshizumi T, Tsumoto Y, Takiguchi T, Nagata N, Yamamoto YY, Kawashima M, Ichikawa T, Nakazawa M, Yamamoto N, Matsui M. Plant Cell. 2006;18:2452–2468. doi: 10.1105/tpc.106.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imai KK, Ohashi Y, Tsuge T, Yoshizumi T, Matsui M, Oka A, Aoyama T. Plant Cell. 2006;18:382–396. doi: 10.1105/tpc.105.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menges M, Hennig L, Gruissem W, Murray JA. J Biol Chem. 2002;277:41987–42002. doi: 10.1074/jbc.M207570200. [DOI] [PubMed] [Google Scholar]
- 32.Healy JM, Menges M, Doonan JH, Murray JA. J Biol Chem. 2001;276:7041–7047. doi: 10.1074/jbc.M009074200. [DOI] [PubMed] [Google Scholar]
- 33.Van Leene JSH, Eeckhout D, Persiau G, Van Slijke E, Van Isterdael G, De Clerq A, Bonnet E, Laukens K, Remmerie N, Hendrickx K, et al. Mol Cell Proteomics. 2007;6:1226–1238. doi: 10.1074/mcp.M700078-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Kawamura K, Murray JA, Shinmyo A, Sekine M. Plant Mol Biol. 2006;61:311–327. doi: 10.1007/s11103-006-0014-y. [DOI] [PubMed] [Google Scholar]
- 35.Kono A, Umeda-Hara C, Lee J, Ito M, Uchimiya H, Umeda M. Plant Physiol. 2003;132:1315–1321. doi: 10.1104/pp.103.020644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A. Plant Cell. 2002;14:1847–1857. doi: 10.1105/tpc.002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koroleva OA, Tomlinson M, Parinyapong P, Sakvarelidze L, Leader D, Shaw P, Doonan JH. Plant Cell. 2004;16:2364–2379. doi: 10.1105/tpc.104.023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. Plant Physiol. 2006;140:67–80. doi: 10.1104/pp.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Veylder L, Beeckman T, Beemster GT, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inze D. EMBO J. 2002;21:1360–1368. doi: 10.1093/emboj/21.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.del Pozo JC, Diaz-Trivino S, Cisneros N, Gutierrez C. Plant Cell. 2006;18:2224–2235. doi: 10.1105/tpc.105.039651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inze D, De Veylder L. Curr Biol. 2005;15:59–63. doi: 10.1016/j.cub.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 42.Emmerich J, Meyer CA, de la Cruz AF, Edgar BA, Lehner CF. Genetics. 2004;168:867–875. doi: 10.1534/genetics.104.027417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizukami Y, Fischer RL. Proc Natl Acad Sci USA. 2000;97:942–947. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Y, Xie Q, Chua NH. Plant Cell. 2003;15:1951–1961. doi: 10.1105/tpc.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soni R, Carmichael JP, Shah ZH, Murray JA. Plant Cell. 1995;7:85–103. doi: 10.1105/tpc.7.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaudhury AM, Letham S, Craig S, Denis ES. Plant J. 1993;4:907–916. [Google Scholar]
- 47.Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Nat Genet. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 48.Parinov S, Sevugan M, Ye D, Yang WC, Kumaran M, Sundaresan V. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rasband WS. ImageJ. Bethesda: Natl Inst Health; 2006. [Google Scholar]
- 50.Smyth DR, Bowman JL, Meyerowitz EM. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menges M, Murray JA. Plant J. 2002;30:203–212. doi: 10.1046/j.1365-313x.2002.01274.x. [DOI] [PubMed] [Google Scholar]
- 52.Prinsen E, Redig P, Strnad M, Galis I, Van Dongen W, Van Onckelen H. Methods Mol Biol. 1995;44:245–262. doi: 10.1385/0-89603-302-3:245. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.