Abstract
The SaPIs are 14- to 17-kb mobile pathogenicity islands in staphylococci that carry genes for superantigen toxins and other virulence factors and are responsible for the toxic shock syndrome and other superantigen-related diseases. They reside at specific chromosomal sites and are induced by certain bacteriophages to initiate an excision-replication-packaging program, resulting in their incorporation into small infective phage-like particles. These are responsible for very high transfer frequencies that often equal and sometimes exceed the plaque-forming titer of the inducing phage. The ability of the SaPIs to replicate autonomously defines them as individual replicons and, like other prokaryotic replicons, they possess replicon-specific initiation functions. In this paper, we report identification of the SaPI replication origin (ori) and replication initiation protein (Rep), which has helicase as well as initiation activity. The SaPI oris are binding sites for the respective Rep proteins and consist of multiple oligonucleotide repeats in two sets, flanking an AT-rich region that may be the site of initial melting. Plasmids containing the rep-ori complex plus an additional gene, pri, can replicate autonomously in Staphylococcus aureus but are very unstable, probably because of defective segregation.
Keywords: helicase, initiation protein, segregation kinetics, superantigen, bacteriophage
The SaPIs are small pathogenicity islands in staphylococci and in other Gram-positive bacteria that carry superantigen and other virulence genes and are responsible for certain staphylococcal toxinoses, most notably toxic shock syndrome. In the now classical paradigm, the SaPI, integrated at its specific chromosomal att site, is induced to excise and replicate by certain coreplicating bacteriophages (1–3). After replication, SaPI DNA is efficiently encapsidated in small infectious phage-like particles, resulting in extremely high transfer frequencies (4). Because their virion DNA is circularly permuted, as is that of the typical inducing phage, it is assumed that their replication products are multimeric, possibly linear concatemers, and that they are packaged by the headful mechanism. The SaPI genomes, for which sequences are now available for 15 (5), have a conserved modular organization similar to that of typical temperate phages. In separate studies, we have characterized this organization in some detail (C.Ú., Elisa Maiques, P.B., Avery Matthews, María Ángeles Tormo, et al., unpublished results), as shown in Fig. 1A for a prototypical member of the family, SaPIbov1. There is always a site-specific integrase at or very near the left junction, which is required for replication and transfer as well as for integration and excision [P.B. et al., unpublished results (6)]. This is followed by a pair of divergent genes encoding putative transcriptional regulatory proteins that may be analogous to λ cI and cro, respectively. Near the right junction is a six-gene operon involved in packaging (7), including a highly conserved homolog of the terminase small subunit of phages from Gram-positive bacteria. Between these two regions is a replication module encoding DNA helicase- and primase-like proteins, of which the former is required for SaPI DNA replication (8); (C.Ú., Elisa Maiques, P.B., Avery Matthews, María Ángeles Tormo, et al., unpublished results). Note that the helicase gene was originally thought to consist of two separate ORFs, 13 and 14 (2), but is now known to be a single gene (C.Ú., Elisa Maiques, P.B., Avery Matthews, María Ángeles Tormo, et al., unpublished work). Although certain other pathogenicity islands have recently been found to be capable of excision, only the SaPIs are capable of autonomous replication, suggesting they may possess specific replicons. Because prokaryotic replicons are distinguished by the specificity of their replication initiation, we addressed the question of whether this is also true for the SaPIs. In particular, prokaryotic replicons possess a specific replication origin and a unique means of activating it, usually but not always a protein that recognizes the origin and binds to it in a sequence-specific manner, causing localized melting that enables the initiation of replication. With certain small theta replicons, the initiator protein is relatively large and has helicase and, sometimes primase activities in addition to ori recognition and binding. Here, we confirm that the SaPIs each possess a specific replicon which includes a replication initiator protein (Rep) with helicase activity and a replication origin, a ≈300-bp iteron-containing region 3′ to the rep gene. We show also that SaPIs encode a primase homolog, which is important for SaPI replication, although not required, that a ≈3-kb segment containing these three elements can drive the autonomous replication of a plasmid in the absence of any phage, and that the product of this replication is a linear concatemer. This autonomously replicating unit, however, is highly unstable, probably because of the multimeric form of its replication product, which is designed for encapsidation and is not readily adaptable to the plasmid lifestyle.
Fig. 1.
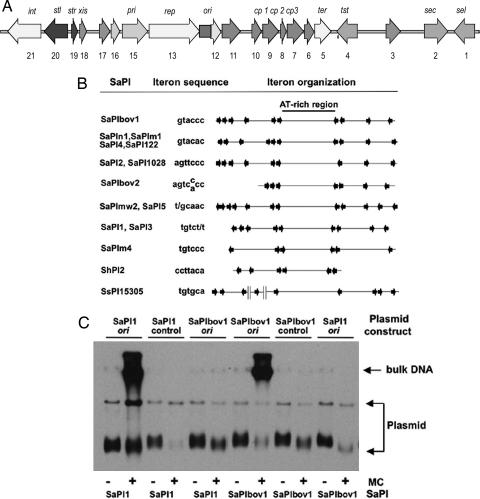
Identification of the SaPI replication origins. (A) Map of SaPIbov1. Arrows represent the localization and orientation of the different ORFs, with labels indicating known gene functions. No functions have been identified for ORFs 3, 6, 10, 11, 12, 16, and 17, which have no significant matches in the database. (B) Comparative map of the replication origins of several SaPIs. The iterons are represented by arrows, and their sequences are shown at left. Note that there are always two sets of iterons flanking an AT-rich region, which could be the melting site. (C) The sequences containing the iterons of SaPIbov1 or SaPI1 were cloned into the thermosensitive plasmid pBT2. Sequences not containing iterons of each island were cloned as controls. The resulting plasmids were introduced into 80α lysogens carrying SaPI1 or SaPIbov1. Bacterial cultures of the different strains were exposed to MC, then incubated in broth at 43°C (restrictive temperature for pBT2 replication). Samples were removed 90 min after MC induction. Standard minilysates were prepared, separated by agarose gel electrophoresis, and blotted by using a pBT2-specific probe.
Results
Identification of the SaPI Replication Origin.
Deletion of the SaPIbov1 region between ORFs 12 and 13 abolished SaPIbov1 replication (C.Ú., Elisa Maiques, P.B., Avery Matthews, María Ángeles Tormo, et al., unpublished work). This region contains 10 repeats of the hexanucleotide GTACCC, six in one direction and four in the other. Because the occurrence of multiple oligonucleotide repeats (iterons) in a short region is typical of many prokaryotic replication origins (9), it seemed likely that this 300-bp region corresponds to the SaPIbov1 replication origin. Indeed, all of 15 sequenced SaPIs contain sets of similar but SaPI-specific repeats at the same relative position, as shown in Fig. 1B. These repeats are always arranged in two sets, flanking an AT-rich region of ≈80 bp. Notably, SaPIn1, SaPI4, SaPI122, and SaPIm1 share a common set of repeats that have a very similar arrangement to those of SaPIbov1 and differ at only one position. Moreover, these four encode virtually identical helicase-like proteins, and the same is true of several other pairs of SaPIs.
To determine whether the iteron regions do, in fact correspond to the SaPI replication origin, we cloned these regions, from SaPI1 and SaPIbov1, into a thermosensitive plasmid vector, pBT2 (10) that replicates very poorly at 43°C, generating pRN9204 and pRN9205, respectively. We would expect to see significant replication of the plasmid at 43°C after induction of the cognate SaPI only if the cloned sequence contained the SaPI replication origin. This expectation was confirmed as shown in Fig. 1C: a high-Mr hybridization signal was seen in Southern blots of cellular lysates after mitomycin C (MC) induction of 80α lysogens containing a native SaPI plus a pBT2 derivative containing the putative cognate ori. No such signal was produced either with cloned segments containing other SaPI sequences (plasmids pRN9206 and pRN9207), or when the cloned segment and the endogenous SaPI did not match. These results together suggest that the cloned SaPI1 and SaPIbov1 iteron fragments contain the respective SaPI replication origins, that these are SaPI-specific, and therefore that the endogenous SaPIs encode protein(s) that induce replication of the cloned cognate iterons. It is suggested that the high-Mr signal represents a multimeric replication product, which is similar to that ordinarily seen after SaPI induction. It will be recalled that SaPI induction also results in a SaPI-specific DNA band that migrates ahead of the bulk DNA in agarose screening gels and represents SaPI monomers released from intracellular phage heads (1, 11). However, no such band was seen with the plasmids containing the cloned iterons, suggesting that this material could not be packaged into phage particles because the cloned segment does not contain the SaPI pac site. At the same time, high-Mr SaPI-specific material was seen in the same lysates. A transduction experiment showed clearly that this high-Mr material could not be packaged by a functional SaPI, nor was there significant recombination, because the two elements were transduced independently; in MC-induced lysates, the intact SaPI::tetM was transduced at its usual high frequency, with tetracycline-resistant transductants being produced at a frequency of ≈10−2, independently of the presence of the cloned ori, whereas chloramphenicol-resistant transductants representing the plasmid, with or without the cloned ori, appeared at ≈10−7, consistent with classical generalized transduction frequencies.
Identification of the SaPI Initiator (Rep) Protein.
The location of the helicase-like gene adjacent to the replication origin, its essentiality for replication, and the striking correspondence between its conservation and that of the replication origin suggested strongly that the helicase homolog would correspond to the SaPI Rep protein. We could not, however, obtain convincing results in attempts to drive replication of the ori plasmid with this protein cloned in trans and, because clones of both rep and ori to a single (suicide) plasmid inexplicably resulted in inactivating mutations in the rep gene, we constructed and tested clones containing an additional SaPIbov1 gene, ORF15, which is highly conserved, encodes a putative primase, and has been shown elsewhere to be important, although not essential, for SaPIbov1 replication (C.Ú., Elisa Maiques, P.B., Avery Matthews, María Ángeles Tormo, et al., unpublished results). The entire pri-rep-ori unit was cloned into a suicide plasmid as a PCR product under the control of a Cd-inducible promoter, generating pRN9211. This plasmid was successfully transferred to Staphylococcus aureus strain RN4220, where it generated high-Mr material similar to that seen with the cloned ori (Fig. 2). However, we were unable to transfer plasmids lacking the ori or rep (pRN9212 and pRN9213, respectively) to RN4220, indicating that the pri-rep-ori unit could drive plasmid replication in S. aureus. The ability to form colonies indicated also that the plasmid could segregate. Similar clones derived from SaPIs 1 and m4 (plasmids pRN9217 and pRN9218, respectively) could also be introduced into RN4220, also generating the typical high-Mr multimeric plasmid signal (not shown). On the basis of these results, we conclude that the SaPI pri-rep-ori complex can drive autonomous replication and segregation of a plasmid in S. aureus. We are unable to decide, at present, whether rep-ori alone is sufficient.
Fig. 2.
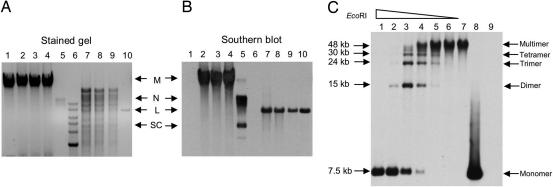
Replication products of SaPIbov1 pri-rep-ori subclone. (A) Total DNA from bacterial cultures was extracted and separated by agarose gel electrophoresis (lanes 1–4) or was initially digested with EcoRI, which cuts once in the plasmid (lanes 7–9). Lane 1, RN4220; lanes 2–4 and 7–9 correspond to three EmR transformants obtained in RN4220 with the SaPIbov1 pri-rep-ori subclone (pRN9211); lane 5 corresponds to the plasmid pRN9211 extracted from E. coli strain DH5α; lane 10, same plasmid DNA as lane 5 previously digested with EcoRI; lane 6, DNA ladder. (B) The same gel probed with a pRN9211-specific probe. The locations of multimers (M), nicked circular monomers (N), linear monomers (L), and supercoiled monomers (SC) are indicated. (C) Whole-cell DNA isolated from one pri-rep-ori transformant, digested with diminishing amounts of EcoRI (lanes 1–6) or undigested (lane 7), was separated by agarose gel electrophoresis, then Southern blotted as in B. Lane 8, plasmid pRN9211 extracted from E. coli and digested with EcoRI; lane 9, RN4220 DNA digested with EcoRI.
Binding Specificity.
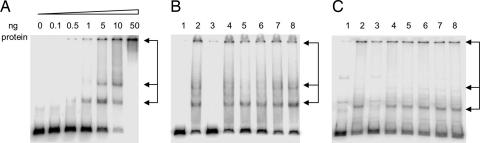
Because Rep is clearly the replicon-specific initiator for the SaPIs, it must bind specifically to its cognate origin. This was demonstrated by EMSA with purified 6xHis-tagged Rep protein from both SaPI1 and SaPIbov1. As shown in Fig. 3A, a clear gel shift was observed for SaPIbov1 Rep, demonstrating at least three different DNA–protein complexes as a function of increasing protein concentration, perhaps representing monomeric, dimeric, and higher multimeric forms; determination of the specific stoichiometry is in progress. As shown in Fig. 3B, the binding of SaPIbov1-Rep to its origin was specific. It was competed by an excess of unlabeled isologous DNA but not by DNAs representing the oris of other SaPIs, including SaPI2, SaPIn1, SaPI1, and SaPIm4. The same result was obtained for SaPI1 Rep. As shown in Fig. 3C, it bound specifically to the SaPI1-ori, and this binding was competed by an excess of unlabeled isologous DNA but not by DNAs representing the oris of other SaPIs, including SaPIbov1, SaPIbov2, SaPIn1, and SaPIm4 (Fig. 3C) or SaPI2 (not shown). These results fully confirm the role of the SaPI Rep proteins as replicon-specific initiators, a role that would necessitate the induction of ori-specific melting of the DNA, preparatory to the initiation of polymerization.
Fig. 3.
Specific binding of Rep protein to the cognate SaPI replication origin. (A) Gel mobility-shift assay by using SaPIbov1-ori as probe and increasing amounts of purified SaPIbov1 Rep protein. The different DNA–protein complexes are indicated by arrows. (B) Competition assays by using 5 ng of purified SaPIbov1-Rep protein, labeled SaPIbov1-ori DNA, and 10-fold excess of unlabeled DNA (lanes 3–8). SaPIbov1-ori DNA (lane 3), nonspecific DNA (lane 4), SaPI2-ori DNA (lane 5), SaPIn1-ori DNA (lane 6), SaPI1-ori DNA (lane 7), and SaPIm4-ori DNA (lane 8) were used as competitor unlabeled DNA. Lane 1 is a negative control without protein and unlabeled DNA. Lane 2 is a positive control without unlabeled DNA. (C) Competition assays by using 50 ng of purified SaPI1 Rep protein, labeled SaPI1-ori DNA, and 10-fold excess of unlabeled DNA (lanes 3–8). SaPI1-ori DNA (lane 3), nonspecific DNA (lane 4), SaPIm4-ori DNA (lane 5), SaPIbov1-ori DNA (lane 6), SaPIbov2-ori DNA (lane 7), and SaPIn1-ori DNA (lane 8) were used as competitor unlabeled DNA. Lane 1 is a negative control without protein and unlabeled DNA. Lane 2 is a positive control without unlabeled DNA.
Ori Recognition Specificity.
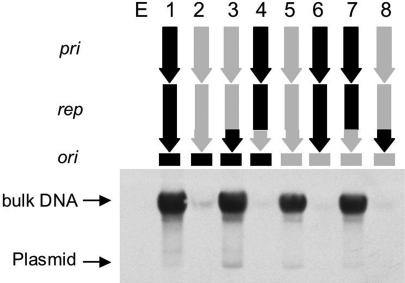
The results obtained with the EMSA strongly suggest that each SaPI Rep protein binds specifically to its cognate origin but not to others. Similarly, the results obtained with the thermosensitive plasmids containing the SaPIbov1- and SaPI1-ori also suggested that a SaPI Rep protein can initiate replication from its cognate origin but not from other SaPI oris (Fig. 1C). Comparison of the predicted Rep protein sequences revealed that certain pairs of these (e.g., SaPIbov1 and SaPI2) that were highly similar overall but had different specificities, had significant differences at their C termini [supporting information (SI) Fig. 7A]. Accordingly, we constructed two sets of replication module chimeras, as illustrated diagrammatically in Fig. 4. One set contained the pri and rep genes of SaPI2 and the SaPIbov1 ori (pRN9223), and vice versa (pRN9226) (lanes 2 and 6, respectively), and the other set contained the extreme C-terminal end of the rep gene plus either the matching or the nonmatching ori (lanes 3 and 4 and 7 and 8, respectively). The several chimeric constructs, along with the corresponding WT, were cloned into the thermosensitive plasmid, pJC1134, with pri and rep under the control of a Cd-inducible promoter. We used pJC1134, because it carries the exquisitely thermosensitive pT181 repC3 allele (12) and therefore has a much more stringent replication defect at 43°C than does pBT2. We introduced the chimera-containing plasmids to RN4220 and tested for the ability of the plasmids to replicate at the restrictive temperature (43°C). In each case, there was a strong signal in the bulk DNA only when the Rep C terminus matched the ori (lanes 3 and 7), similar to the signals seen with the intact pri-rep-ori clones (pRN9222 and pRN9225, lanes 1 and 5). No signal was seen in strains where these two elements did not match (lanes 2, 4, 6, and 8) or in the strain containing the empty vector (lane E). This result confirmed the predicted specificity of the rep-ori complex and the nonspecificity of the primase and showed that the ori recognition determinant is located at the extreme C-terminal end of the Rep protein, as predicted from the sequences.
Fig. 4.
Rep-ori replication specificity. Cultures of RN4220 strains containing the pri-rep-ori modules of SaPIbov1 and SaPI2 and chimeric constructs as shown were grown on CY broth at 43°C until OD540 = 0.6. Samples were removed and used to prepare minilysates. Lysates were separated by agarose gel electrophoresis and probed with a probe specific for the carrier plasmid, pRN9220. Above the blot is shown the composition of the pri-rep-ori module of each of the plasmids tested. Black indicates segments derived from SaPIbov1, gray, from SaPI2. E indicates empty vector.
Helicase Activity of Rep.
Although helicase activity is certainly necessary for the replication of any double-stranded DNA, this is not typically replicon specific and is often provided by the host cell. However, because the Rep protein of satellite phage P4 has helicase as well as initiation activity (13) and as the SaPI Rep proteins were initially annotated as helicases, we sought to determine whether the SaPI Reps also have helicase activity. Accordingly, the purified 6xhis-tagged proteins were initially tested for nonspecific helicase activity by a standard test using double-stranded oligonucleotides with nonhomologous ends (SI Fig. 8A), one strand of which was 32P-labeled. In SI Fig. 8B is shown the result of unwinding catalyzed by SaPI1 Rep; essentially the same result was obtained for SaPIbov1 Rep (not shown). As a further test of this helicase activity, we used a fully double-stranded oligonucleotide. This oligonucleotide served as a helicase substrate as well as the open-ended one (not shown), indicating that the SaPI Rep helicase can unwind a blunt-ended duplex. Because helicases use ATP, we looked for and found the classical Walker box ATPase motif (14) in the Rep protein sequences. We constructed a site-specific replacement (K186V) in the cloned SaPI1 enzyme to test for the role of this element in helicase activity. As predicted, helicase activity could no longer be demonstrated (not shown). Additionally, although the plasmid containing the WT helicase (pRN9003) could complement the defect in replication of a SaPI1 helicase knockout, the plasmid containing the K186V helicase mutant (pRN9201) could not (data not shown). Experiments to determine whether this mutation eliminates the initiation function of the protein, i.e., the ability to bind and melt the replication origin, as well as its helicase activity, are in progress.
Multimeric Nature of the SaPI Replicon.
To test for the predicted multimeric nature of the pri-rep-ori replication products, we analyzed the total DNA of three erythromycin-resistant (EmR) transformants obtained with pRN9211, the SaPIbov1 subclone, in comparison to the replication products of the same plasmid in Escherichia coli (Fig. 2). In S. aureus, none of the three EmR-transformants generated a DNA species matching any of the isomers generated by the same plasmid in E. coli, either in an ethidium bromide-stained gel (Fig. 2A) or in a Southern blot using a plasmid-specific probe (Fig. 2B). However, as previously observed, the bulk DNA strongly reacted with the probe, consistent with the production of multimers. To confirm this observation and to rule out the possibility of a chromosomal insertion, we digested the DNA with EcoRI, which cuts once in the plasmid, within the sequence complementary to the probe, so that an integrated plasmid would produce two bands in the Southern blot, whereas a multimeric form would generate linear plasmid monomers. As shown in Fig. 2B, the EcoRI digest generated a single band with the same mobility as the linear plasmid monomers generated by digesting the plasmid extracted from E. coli, ruling out a chromosomal insertion and confirming the multimeric nature of the replicating material in S. aureus. If this material is similar to that of temperate phage replication products, it would be a linear concatemer generated by late rolling circle replication. To test for a linear concatemer, we treated this same material with diminishing concentrations of EcoRI, separated these digests on agarose, and Southern blotted with the same probe. As shown in Fig. 2C, as the concentration of EcoRI decreased, multimeric forms began to appear, eventually generating a ladder of multimers, in which monomers, dimers, trimers, and tetramers could readily be distinguished, and higher multimers were very probably present also, but were not resolved.
Stability.
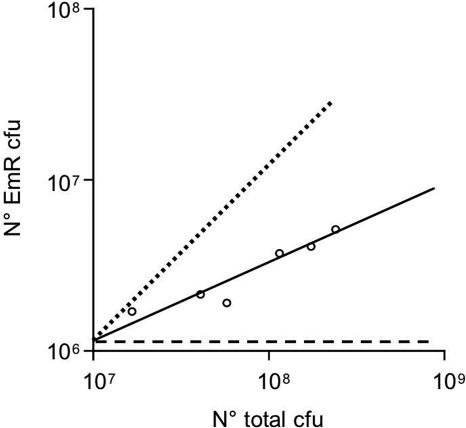
The ability to obtain colonies on an agar plate selective for the cloned SaPIbov1 replicon indicates not only that the pri-rep-ori plasmid can replicate in S. aureus, but also that it can segregate during cell division. Efficient segregation involves separation of the replicating complex into at least two physically separate units and distribution of at least one of these into each sister cell during cell division. Because the multimeric product of SaPI replication would not be designed for efficient segregation, we were not surprised to find that the pri-rep-ori plasmids were highly unstable. Even on selective medium, <10% of the cells in a colony were EmR, and a double-log plot of the relative growth of plasmid-positive vs. total cells (Fig. 5) had a slope, S, of 0.44. Because the probability, P, of replication/segregation per cell division is given by P = 2s−1, this means that a plasmid-carrying cell has only a 35% probability of producing two plasmid-containing progeny. Therefore, either fewer than two segregation units are produced during each cell cycle, or segregation is not random, or both.
Fig. 5.
SaPIbov1 pri-rep-ori subclone stability. Bacteria RN4220 containing the SaPIbov1 pri-rep-ori subclone grown on selective medium were grown on CY broth without antibiotic. Samples were taken every 30 min during 3 h and plated on GL plates with or without Em. The graph shows the number of EmR colonies obtained in relation to the total number of colonies obtained during the 3 h (solid line). The dashed line represents a plasmid that does not replicate. The dotted line represents a stable plasmid.
Discussion
In this report, we describe the identification of the replication origin and the replication initiation protein for a large series of related staphylococcal pathogenicity islands, the SaPIs. The rep gene, annotated as “helicase-like,” is conserved throughout the SaPIs, varying widely in sequence and is almost certainly functional in at least eight of the known SaPIs for which phage-induced replication has been demonstrated. The replication origin is immediately 3′ to the initiator protein coding sequence and consists of two sets of short oligonucleotide repeats, in a ≈300-bp region, with a variety of arrangements but always flanking an ≈80-bp AT-rich segment that could represent the site of initial melting. In the two cases analyzed, this region serves as a SaPI-specific binding site for the Rep protein. It remains to be determined, however whether the oligonucleotide repeats represent the actual Rep binding sites and/or correspond functionally to plasmid iterons.
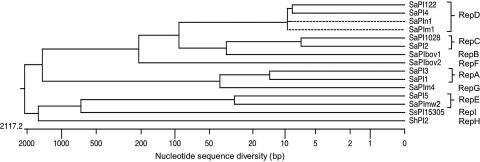
To rationalize SaPI nomenclature, we have adopted the convention used for plasmid genes, in which cognate genes carried by different plasmids are given arbitrary letters referring to the plasmid of origin. Thus, genes of SaPI1 are “A,” SaPIbov1 “B,” SaPI2 “C,” SaPIn1 “D”; SaPImw2 “E,” SaPIbov2 “F,” SaPIm4 “G,” ShPI2 “H,” and SsPI15305 “I” (Fig. 6). Additionally, with respect to Rep proteins and oris, any two Rep proteins that recognize the same ori are given the same letter. The predicted Rep proteins can be subdivided accordingly to their similarities (Fig. 6). One subtype contains the Rep proteins of SaPIbov1, SaPIn1, SaPIm1, SaPI4, SaPI122, SaPI1028, SaPI2, and SaPIbov2. Each of these is ≈569 amino acid residues in length and share 92–95% identities with the exception of SaPIbov2-Rep, which has 80% identity with the others. Identical or almost identical Rep proteins are associated with identical replication origins, e.g., SaPI122, SaPIn1, SaPIm1 and SaPI4, and also SaPI2 and SaPI1028, and are predicted to share ori recognition specificity. SaPIs with different oris have Rep proteins that differ correspondingly. For example, SaPIbov1 and SaPI2 iteron sequence are gtaccc and agttccc, respectively, and their Rep amino acid sequences, 96% identical, differ at the extreme C terminus, which determines recognition specificity.
Fig. 6.
Phylogenetic comparison of SaPI Rep proteins. The tree was constructed with Rep protein sequences of different SaPIs by using the DNASTAR MEGALIGN, JOTUN HEIN method. Images represent genetic distance as number of nucleotide substitutions. Rep proteins that recognize the same ori are given the same letter.
A second set of Rep proteins encoded by SaPI1, SaPI3 and SaPIm4 are ≈477 aa in length; SaPI1 and SaPI3 share identical oris, differing from SaPIm4-ori. This latter difference is, again, primarily at the extreme C terminus, which again represents the determinant of recognition specificity (SI Fig. 7B). Similarly, the predicted Rep protein of S. haemolyticus ShPI2 (15) is 455 aa in length, although its sequence is completely different.
A third set of Rep proteins, of 790 amino acids from SaPI5, SaPImw2, are associated with essentially identical oris, show 3% sequence divergence, but have identical C termini. These two SaPIs lack any primase-like ORF, suggesting that their Rep genes represent fusions between pri and rep. A blast search has confirmed that the N-terminal domain of these proteins is orthologous to primases. Similarly, the predicted Rep protein of SsPI15305 from S. saprophyticus (16) is 780 amino acids in length and may also represent a pri-rep fusion.
In tests for functionality, we first cloned the SaPIbov1 replication origin into a thermosensitive plasmid and demonstrated an ori-dependent multimeric plasmid replication product upon induction of a resident copy of SaPIbov1, which supplied the Rep protein. To construct a plasmid that could replicate autonomously on its own, we switched to a plasmid that could replicate in E. coli but not in S. aureus. We have not been able to test a derivative of this plasmid containing the SaPIbov1 rep and ori sequences for autonomous replication because all of three isolates of this complex, constructed in E. coli, contained inactivating mutations in the rep gene, suggesting that the rep–ori complex may be toxic. The smallest SaPIbov1 segment for which autonomous replication could be demonstrated contained the conserved primase gene in addition to SaPIbov1 rep and ori. Similar pri-rep-ori constructs from two other SaPIs could also support replication of a suicide plasmid as could a larger SaPIbov1 derivative, containing ORFs 11–20 (C.Ú., Elisa Maiques, P.B., Avery Matthews, María Ángeles Tormo, et al., unpublished work).
Because a prokaryotic minimal replicon is classically defined by its ability to drive replication of a plasmid and is necessarily evaluated by maintenance of the plasmid state, the interesting question arises of whether the multimeric nature of the replication product of elements such as SaPIs and pac phages could lend itself to maintenance in the plasmid state. Although replication must certainly be sufficient, there is no guarantee that replication would generate the two (or more) progeny replicas needed to enable segregation during cell division. In fact, the SaPI replicon transformants grew extremely slowly, requiring 2 days to produce colonies, because the plasmids were extremely unstable: even on selective medium, <10% of the cells in a colony had retained the plasmid, and a segregation curve revealed that any plasmid-positive cell had only a 35% probability of producing two plasmid-containing progeny. If the plasmid-positive and -negative cells were to grow at the same rate on nonselective medium, this would generate a colony in which <1 in 105 of the cells had retained the plasmid. It is suggested that segregation failure is responsible for this instability for the following reason: our evidence suggests that the replication product is a linear concatemer, which is typical of that produced during replication of a temperate phage genome. That is, initiation on circular monomeric DNA followed by conversion to the rolling circle, generating a linear concatemer. Encapsidation by the headful mechanism leads to the terminally redundant monomers that are found in phage and SaPI heads. In other words, this mode of replication, designed for encapsidation, is not at all suited to the plasmid lifestyle. Conceivably, the intact SaPI could function as a stable plasmid. Although this has not been ruled out, it is considered unlikely as it would require a monomerization mechanism (independent of encapsidation), and a mechanism to regulate and self-correct copy number. None of the unassigned ORFs appears likely to encode such functions.
Stable maintenance of any autonomous replicon involves regular segregation of replicas to progeny cells; low-copy plasmids encode specific partitioning systems, whereas high-copy plasmids, which are usually quite small, do not and have long been assumed to partition randomly, an assumption that is supported by mathematical predictions; however, recent microscopic analyses suggest that plasmid DNA forms a tight aggregate, so that random partitioning seems paradoxical. One suggested mechanism is that single copies of the plasmid “escape” from this aggregate, initiate replication, and are then randomly partitioned (17), implying that an unknown host function may be responsible, a function that must not be able to act on the multimeric SaPI replication product. It is interesting to consider the implications of the segregation requirement for replicons such as those of phages and related elements that are designed for encapsidation rather than segregation. Coliphage P1, whose prophage is a plasmid, has solved this problem by containing two independent replicons, one for vegetative phage replication, the other for maintenance as a low-copy plasmid (18). λdv, a high-copy plasmid derived from coliphage λ, does not form a multimeric replication product, probably because it lacks the mechanism for conversion to rolling circle replication but is maintained in the monomeric form (19); it is unstable despite a high-copy number, presumably because it possesses neither any explicit partitioning mechanism nor any self-correction mechanism; coliphage P4, a satellite phage similar in some ways to the SaPIs, can subvert the replication of its helper phage, P2, for the formation of specific small capsids and can replicate stably as a monomeric plasmid in the absence of its helper phage (20), because it is able to adapt itself to the plasmid lifestyle. Interestingly, the P4 replication initiation protein has helicase and primase as well as ori-binding activities (20). As noted above, some of the SaPIs have separate primase-like proteins, others have putative primase segments at the N-terminal ends of their Rep proteins. In either case, it seems clear that both activities are important for SaPI replication. As is the case with certain plasmids (21), the primase is not absolutely essential, suggesting that its function can be substituted by the host primase. The helicase activity of the Rep protein, however, seems to be absolutely essential for SaPI replication, which seems odd, because this activity does not appear to be sequence-specific, and because S. aureus encodes a helicase, PcrA that is required for replication of both plasmids and chromosome (22). Perhaps the helicase activity of SaPI Rep proteins is required for the actual initiation step as well as for unwinding during polymerization.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains used in these studies are listed in SI Table 1. Bacteria were grown at 32°C or 37°C overnight on glycerol-lactate agar medium, supplemented with antibiotics as appropriate. Broth cultures were grown at 32°C or 43°C in casamino acids–yeast extract broth with shaking (240 rpm). Procedures for transduction and transformation in S. aureus were performed essentially as described (23).
Induction of Prophages.
Bacteria were grown in CY broth to OD540 = 0.4 and induced by adding MC (2 μg/ml). Cultures were grown at 32°C or 43°C with slow shaking (80 rpm). Samples were removed after 90 min, and standard SDS minilysates were prepared and separated on 0.8% agarose gels, as described (1). Upon further incubation, the cultures underwent lysis, and the sterilized lysates were tested for SaPI transfer.
DNA Methods.
General DNA manipulations were performed by standard procedures (24, 25). Oligonucleotides used in this study are listed in SI Table 2. Oligonucleotides p1007/p1008 were used to generate the pBT2- and pRN9220-specific probes, and oligonucleotides p985/p628 were used to generate the pRN9211-specific probe. Labeling of the probes and DNA hybridization were performed according to the protocol supplied with the ECL Direct Nucleic Acid Labeling kit (Amersham, Piscataway, NJ).
Plasmid Constructs.
All plasmids used in this study are listed in SI Table 1. The primers used for each construction are also indicated in SI Table 1. Plasmids pRN9204-pRN9207 were constructed by cloning PCR products obtained with the appropriate primers into the thermosensitive plasmid pBT2 (10). The suicide plasmid pRN9210 was constructed by digesting pCN51 (26) with NarI and ApaI to remove the staphylococcal pT181 replicon. The digested plasmid was then blunt-ended and self-ligated. Plasmids pRN9211, pRN9212, pRN9213, pRN9217, and pRN9218 were constructed by cloning PCR products, obtained with the appropriate primers, into pRN9210. Strain JP225 was used as template to generate plasmid pRN9213. The replication module chimeras were constructed by cloning a 592-bp SphI-PstI fragment from pCN51, containing the Cd-resistance gene promoter from pI258, into the thermosensitive plasmid pJC1134 generating pRN9220. pJC1134 is a derivative of pCN50 (26) containing the thermosensitive (repC3) replicon from plasmid pSA0321 (12). PCR products containing SaPIbov1 or SaPI2 oris were cloned into pRN9220 generating pRN9221 and pRN9224, respectively. The SaPIbov1 pri-rep region was cloned as a PCR-amplified product into pRN9221 and pRN9224, generating pRN9222 and pRN9226, respectively. SaPI2 pri-rep was cloned as a PCR-amplified product into pRN9221 and pRN9224, generating pRN9223 and pRN9225, respectively. To switch the extreme C terminus of SaPIbov1 and SaPI2 Rep proteins and generate plasmids pRN9227-pRN9230, we performed two-step PCRs to obtain products containing pri and the N-terminal half of rep (amino acids 1–325) from the same SaPI and the C-terminal half (amino acids 326–569) from the other. Although the switching point is around the middle of the protein, the only differences are in the extreme C termini, as can be seen in SI Fig. 7A. The PCR products obtained were cloned into pRN9221 and pRN9224 containing SaPIbov1 and SaPI2 oris, respectively. pRN9003 was generated by cloning a PCR product containing SaPI1 rep into pCN51. pRN9201, a derivative of pRN9003 containing a SaPI1-rep with the specific amino acid substitution (K186V) was generated by using the Stratagene (La Jolla, CA) QuikChange mutagenesis kit and the primers p955/p956.
Rep Protein Expression and Purification.
SaPI1- and SaPIbov1-rep were cloned as PCR products into the E. coli expression vector pET15b (Novagen) generating pRN9202 and pRN9208, respectively. Plasmid pRN9203, derivative of pRN9202 containing a SaPI1-rep with the specific amino acid substitution (K186V) was generated by using the Stratagene QuikChange mutagenesis kit and the primers p955/p956. The resulting plasmids were transformed into E. coli BL21(DE3). Rep proteins, containing N-terminal histidine tags, were purified according to the manufacturer's (Qiagen, Valencia, CA) recommendations.
Mobility-Shift Assays.
Rep-DNA complexes were detected by EMSA by using purified Rep protein from SaPI1 and SaPIbov1 and PCR 32P end-labeled probes obtained with primers p982/p983 for SaPI1-ori probe and p984/p985 for SaPIbov1-ori probe. Typical 20-μl reactions containing 0.6 ng of labeled probe and different amounts of Rep protein were incubated in binding buffer (10 mM Hepes, pH 8/10 mM Tris·HCl, pH 8/5% glycerol/50 mM KCl/1 mM EDTA/1 mM DTT/1 μg of bulk carrier DNA/50 μg/ml BSA). After 20 min of incubation, the reaction mixtures were analyzed in a 4% native polyacrylamide gel. Gels were dried, and band shifts were analyzed with a Molecular Dynamics (Sunnyvale, CA) Phosphor Imager. For the binding-competition experiments, 10-fold molar excess of unlabeled DNA was added to the mixture. SaPI2-, SaPIn1-, SaPIm4-, SaPIbov2-ori, and the nonspecific DNA, used as DNA competitors, were amplified with the oligonucleotides p995/p996, p1003/p1004, p1005/p1006, p1011/p1012, and SaPIbov-46mE/36mX, respectively.
Helicase Assay.
To determine helicase activity, we adapted the method described in ref. 27. The substrate for helicase assays consisted of two 47-mer oligonucleotides, p946 and p947, which when annealed, create a partial duplex structure (SI Fig. 8A). The 3′ end of p946 was then filled in using exo− Klenow and 32P-labeled dATP. Helicase assay mixtures were assembled on ice, consisting of 1 μl of 10× helicase buffer (200 mM Tris·HCl, pH 8.5/100 mM MgCl2/2 mM DTT/100 μg/ml BSA), 1 μl of radiolabeled DNA substrate (≈100 fmol), 1 μl of ATP (50 mM, pH 7), 0.1 μg of Rep protein, and H2O to a final volume of 10 μl. Assay mixtures were incubated for 30 min at 37°C, at which point 3 μl of stop buffer was added (100 mM EDTA/1% SDS/50% glycerol/0.1% xylene cyanol/0.1% bromophenol blue) as well as 2 pmol of nonlabeled p946, and the assays were transferred to ice. Positive controls for substrate separation were heated to 100°C for 5 min before addition of stop buffer. Assay samples were then run on a 10% native polyacrylamide gel. The gels were dried under vacuum before being placed in PhophorImager cassettes for signal detection.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01-AI22159 (to R.P.N.) and by a grant from Ministerio de Educación y Ciencia from Spain (to C.Ú.). We thank John Chen (New York University School of Medicine) for providing plasmid pJC1134.
Abbreviations
- MC
mitomycin C
- Rep
replication initiator protein
- EmR
erythromycin-resistant.
Footnotes
The authors declare no conflict of interest.
See accompanying Profile on page 14179.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705994104/DC1.
References
- 1.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. Mol Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, Meaney WJ, Smyth CJ. J Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Úbeda C, Tormo MA, Cucarella C, Trotonda P, Foster TJ, Lasa I, Penades JR. Mol Microbiol. 2003;49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 4.Ruzin A, Lindsay J, Novick RP. Mol Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 5.Novick RP, Subedi A. Chem Immunol Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- 6.Maiques E, Úbeda C, Tormo MA, Ferrer MD, Lasa I, Novick RP, Penadés JR. J Bacteriol. 2007 doi: 10.1128/JB.00619-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Úbeda C, Maiques E, Tormo MA, Campoy S, Lasa I, Barbé J., Novick RP, Penades JR. Mol Microbiol. 2007;65:41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 8.Barry P. Microbiology. New York: New York University; 2006. [Google Scholar]
- 9.Filutowicz M, McEachern MJ, Mukhopadhyay P, Greener A, Yang SL, Helinski DR. J Cell Sci Suppl. 1987;7:15–31. doi: 10.1242/jcs.1987.supplement_7.2. [DOI] [PubMed] [Google Scholar]
- 10.Bruckner R. Gene. 1992;122:187–192. doi: 10.1016/0378-1119(92)90048-t. [DOI] [PubMed] [Google Scholar]
- 11.Úbeda C, Maiques E, Knecht E, Lasa I, Novick RP, Penades JR. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 12.Iordanescu S. Plasmid. 1979;2:207–215. doi: 10.1016/0147-619x(79)90039-8. [DOI] [PubMed] [Google Scholar]
- 13.Ziegelin G, Linderoth NA, Calendar R, Lanka E. J Bacteriol. 1995;177:4333–4341. doi: 10.1128/jb.177.15.4333-4341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haber LT, Walker GC. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi F, Watanabe S, Baba T, Yuzawa H, Ito T, Morimoto Y, Kuroda M, Cui L, Takahashi M, Ankai A, et al. J Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda M, Yamashita A, Hirakawa H, Kumano M, Morikawa K, Higashide M, Maruyama A, Inose Y, Matoba K, Toh H, et al. Proc Natl Acad Sci USA. 2005;102:13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordstrom K, Gerdes K. Plasmid. 2003;50:95–101. doi: 10.1016/s0147-619x(03)00056-8. [DOI] [PubMed] [Google Scholar]
- 18.Novick RP. Bacteriol Rev. 1969;33:210–235. doi: 10.1128/br.33.2.210-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Womble DD, Rownd RH. J Mol Biol. 1986;191:367–382. doi: 10.1016/0022-2836(86)90133-6. [DOI] [PubMed] [Google Scholar]
- 20.Lindqvist BH, Deho G, Calendar R. Microbiol Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohno K, Yasuzawa K, Hirose M, Kano Y, Goshima N, Tanaka H, Imamoto F. J Biochem (Tokyo) 1994;115:1113–1118. doi: 10.1093/oxfordjournals.jbchem.a124466. [DOI] [PubMed] [Google Scholar]
- 22.Iordanescu S. Mol Gen Genet. 1993;241:185–192. doi: 10.1007/BF00280216. [DOI] [PubMed] [Google Scholar]
- 23.Novick RP. In: Methods in Enzymology. Miller J, editor. Vol 204. Orlando, FL: Academic; 1991. pp. 587–636. [Google Scholar]
- 24.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: J Wiley; 1987. [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 26.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. Appl Environ Microbiol. 2004;70:6076–6085. doi: 10.1128/AEM.70.10.6076-6085.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soni RK, Mehra P, Choudhury NR, Mukhopadhyay G, Dhar SK. Nucleic Acids Res. 2003;31:6828–6840. doi: 10.1093/nar/gkg895. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.