Abstract
Huntington's disease (HD) is a fatal inherited neurodegenerative disorder. HD is caused by polyglutamine expansions in the huntingtin (htt) protein that result in neuronal loss and contribute to HD pathology. The mechanisms of neuronal loss in HD are elusive, and there is no therapy to alleviate HD. To find small molecules that slow neuronal loss in HD, we screened 1,040 biologically active molecules to identify suppressors of cell death in a neuronal cell culture model of HD. We found that inhibitors of mitochondrial function or glycolysis rescued cell death in this cell culture and in in vivo HD models. These inhibitors prevented cell death by activating prosurvival ERK and AKT signaling but without altering cellular ATP levels. ERK and AKT inhibition through the use of specific chemical inhibitors abrogated the rescue, whereas their activation through the use of growth factors rescued cell death, suggesting that this activation could explain the protective effect of metabolic inhibitors. Both ERK and AKT signaling are disrupted in HD, and activating these pathways is protective in several HD models. Our results reveal a mechanism for activating prosurvival signaling that could be exploited for treating HD and possibly other neurodegenerative disorders.
Keywords: caspase, ERK, survival signaling, drugs, neurodegeneration
Huntington's disease (HD) is an inherited, adult onset, progressive neurodegenerative disorder (1). HD is caused by a polyglutamine expansion (>36 glutamine repeats) in the huntingtin protein (htt) that leads to neuronal dysfunction and death (1, 2). The mechanism(s) by which the polyglutamine expansion in htt leads to HD pathology remain elusive. Numerous mechanisms including transcriptional dysregulation, altered intracellular trafficking, sequestration of critical cellular proteins in aggregates, aberrant caspase activation, and altered energy metabolism have been implicated in HD (2).
HD is a fatal disease with no therapy. To identify potential compounds for development as drugs and to use these compounds to gain mechanistic understanding of HD, we used a screening approach to identify small molecule suppressors of cell death in a cell culture model of HD. In this model, rat striatal neurons that were immortalized by expression of a temperature-sensitive large T antigen were engineered to express a mutant N-terminal, 548-aa fragment of human htt with 120 glutamine repeats to generate the N548 mutant cell line (3). Serum deprivation and a change to the nonpermissive temperature (39°C) causes T antigen degradation and N548 mutant cell death (3). Cell death can be used as an indicator of mutant htt toxicity because the cells expressing mutant htt die faster than parental cells (3). By using a previously described high-throughput assay (4), we discovered that metabolic inhibitors rescued cell death in this cell culture model and in two in vivo HD models. These compounds activated ERK and AKT prosurvival signaling. Furthermore, growth-factor-induced activation of ERK and AKT rescued cell death, thus elucidating a novel mechanism of rescue by these metabolic inhibitors.
Results
Mitochondrial Inhibitor Rotenone Rescues Cell Death in N548 Mutant Cells.
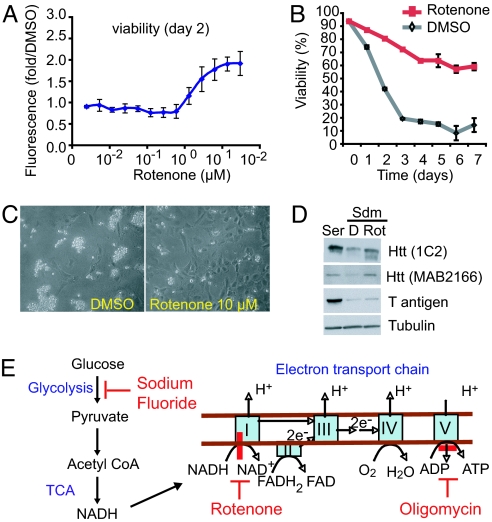
We screened a collection of 1,040 biologically active compounds (see Materials and Methods) by using a previously described high-throughput screening assay (4). We identified that rotenone, an inhibitor of complex I of the mitochondrial electron transport chain (ETC) (5), suppressed cell death in N548 mutant htt-expressing cells. Cell death rescue was confirmed by three independent cell viability assays and over a time course of 7 days (Fig. 1 A–C). Cell death rescue was verified in three independent clonal cell lines expressing N548 mutant htt (data not shown). We excluded the possibility that rotenone rescued cell death by decreasing the expression of htt transgene or altering T antigen levels (Fig. 1D). Because mutant htt aggregation is associated with HD (2), we tested the effects of rotenone on htt aggregation. In this model, occasional cells (<1%) showed visible aggregation by immunofluorescence [supporting information (SI) Fig. 6]. The low propensity of mutant htt to form microscopically visible aggregates may be due to the longer (N-terminal, 548-aa) htt fragment used in this model compared with other models because decreased aggregation of larger htt fragments has been reported in cell culture and in mouse models (6, 7). The percentage of cells with aggregates was not enhanced by rotenone, indicating that the effect on visible aggregation is unlikely to play a role in cell death rescue by rotenone.
Fig. 1.
Mitochondrial inhibitor rescues cell death in N548 mutant cells. (A) A dose–response experiment showing viability of N548 mutant cells treated with rotenone. Viability was determined by the calcein acetoxymethyl ester assay (see Materials and Methods) and is expressed as fold increase relative to DMSO-treated cells. The results are the averages ± SD of an experiment performed in triplicate. (B) Cells were incubated in serum-deprived media (Sdm) with DMSO or rotenone (10 μM), and cell viability was analyzed every day for 7 days by the trypan blue dye-exclusion assay. The data are averages ± SD from one of two independent experiments, each performed in duplicate. (C) Photomicrographs of cells in Sdm containing DMSO or rotenone for 2 days, as viewed by light microscopy. Dying cells detach, round up, and are brighter than attached cells. (D) Expression of mutant htt and T antigen assayed by Western blotting in cells treated with DMSO (D) or rotenone (Rot) for 12 h in Sdm at 39°C or in serum-containing media (Ser) at 33°C. 1C2 antibody detects expanded polyglutamine containing htt, and MAB2166 detects both WT and mutant htt. The temperature-sensitive T antigen is stable at 33°C and degrades at 39°C. Tubulin was the loading control. (E) Glucose metabolism pathway and sites of action of the diverse metabolic inhibitors used in this study (indicated by red bars). For details, see SI Fig. 7.
Diverse Metabolic Inhibitors Rescue Cell Death.
Because rotenone inhibits mitochondrial complex I of the ETC, we expanded our analysis and tested whether perturbing other aspects of metabolism would prevent cell death. We found that diverse inhibitors of metabolism prevented cell death. The compounds tested, their sites of action (Fig. 1E and SI Fig. 7), and their efficacy are shown in SI Table 1. We found that inhibitors of glycolysis (sodium fluoride), ATP synthetase (oligomycin), and mitochondrial coupling 2,4-dinitrophenol (8–10) all rescued cell death. These results indicated that mitochondrial ETC inhibition per se was not required for the rescue.
Rotenone Rescues Neuronal Loss in Multiple in Vivo HD Models.
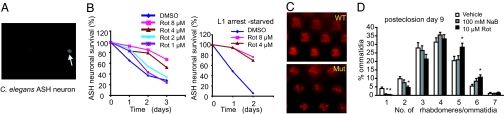
Next, we tested the ability of rotenone to alleviate neuronal loss and degeneration in two invertebrate models of HD. In a Caenorhabditis elegans model, an N-terminal, 171-aa fragment of human htt with 150 glutamines is expressed in ASH sensory neurons of polyglutamine enhancer-1 (pqe-1) genetic background animals. The pqe-1 background enhances mutant htt toxicity and causes age-dependent ASH neuronal death within 3 days (11). ASH neuronal death is monitored by the loss of GFP reporter expression (Fig. 2A) (11). Rotenone caused a dramatic and dose-dependent rescue of ASH neuronal cell death (Fig. 2B, left graph). Because mitochondrial inhibition can slow the aging process (12) and HD is linked to aging (13), we tested whether the effect of rotenone on aging could be dissociated from those on neuronal death by using an assay format in which the animals were growth arrested in the L1 larval stage by starvation (see Materials and Methods). Rotenone was effective in preventing ASH neuronal death of animals under starvation (Fig. 2B, right graph), indicating that it was effective independent of effects on growth or development.
Fig. 2.
Rotenone rescues neuronal loss in two in vivo HD models. (A) A GFP reporter was coexpressed with mutant htt in the ASH neurons of C. elegans and was used to assess neuronal cell viability in vivo. Live neurons express GFP (arrow), and loss of GFP expression indicates cell death. (B) Rescue of age-dependent ASH neuronal death by rotenone (Rot). Animals were treated with DMSO (vehicle) or rotenone in the presence of food (left graph) or under starvation conditions (right graph). ASH neuronal viability was assayed in live animals at the time indicated. One hundred neurons (50 animals) were counted per treatment per time point, and the data are representative of two independent experiments. (C) Mutant htt expression in the photoreceptors of Drosophila results in a time-dependent degeneration. WT animals have seven visible-light-collecting units (rhabdomeres) per ommatidium when viewed by light microscopy. Mutant htt-expressing animals (Mut) show a decrease in the number of rhabdomeres at day 1 after eclosion (emergence as adults); progressive degeneration was age dependent after eclosion (SI Fig. 8). (D) HD flies were treated with vehicle (0.1% DMSO), rotenone, or sodium butyrate (NaB). Sodium butyrate (positive control) increased the number of visible rhabdomeres. The average number of rhabdomeres per ommatidium was calculated at 9 days posteclosion and is shown as a distribution. (See SI Fig. 8 for a dose–response experiment for rotenone.) Error bars represent one SE. *, The significant differences in the number of rhabdomeres between vehicle- and compound-treated animals were based on Student's t test (P < 0.05).
Next, we tested rotenone in a Drosophila HD model in which an N-terminal, 170-aa fragment of human htt containing 120 glutamine repeats is expressed in the Drosophila eye (14). WT Drosophila have seven visible photoreceptor subunits (rhabdomeres) per light collecting unit (ommatidium). Expression of mutant htt transgene causes a progressive decrease in the number of rhabdomeres per ommatidium (Fig. 2C and SI Fig. 8). Treatment with rotenone rescued rhabdomere loss compared with vehicle-treated controls (Fig. 2D and SI Fig. 8). Rescue by rotenone was similar to that by sodium butyrate (Fig. 2D), a histone deacetylase inhibitor that is active in Drosophila and mouse HD models (15, 16). A few other mitochondrial inhibitors were also active in the C. elegans and Drosophila HD models (SI Table 1).
Mitochondrial Inhibitors Suppress Caspase Activation.
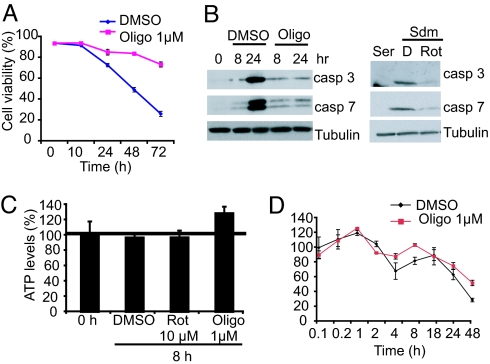
To gain insight into the mechanism of cell death rescue, we assessed the effects of mitochondrial inhibitors on the cell death machinery. Caspases are key regulators of cell death and are involved in HD toxicity, both as targets of mutant htt and as modulators of mutant htt toxicity, by inducing cleavage and generation of toxic htt fragments (17, 18). Cell death upon serum deprivation in N548 mutant cells is largely mediated by caspase activation because the broad-spectrum caspase inhibitor Boc-D-FMK rescues cell death (4). We found that both oligomycin and rotenone inhibited the generation of two downstream effectors in the apoptotic cascade, cleaved active caspase 3 and 7 fragments that correlated with suppression of cell death (Fig. 3 A and B), suggesting that this mechanism was relevant to rescue by these inhibitors.
Fig. 3.
Mitochondrial inhibitors suppress caspase 3 and 7 activation without decreasing ATP levels. (A) Time course of cell death rescue by oligomycin (Oligo). N548 mutant cells were treated with DMSO or oligomycin (1 μM) and analyzed by trypan blue viability assay. A total of 1,000 cells were counted per time point. The data are means ± SD of one experiment performed in triplicate. (B) Caspase (casp) 3 and 7 activation in serum-deprived cells treated with DMSO (D) or oligomycin (left gels) and rotenone (Rot) (right gels) was assayed by Western blotting. Tubulin was the loading control. (C) Cells were treated with different mitochondrial inhibitors or DMSO, and total ATP levels were assayed after 8 h in Sdm by using a luminescence-based assay (see Materials and Methods). ATP levels were normalized to cell number and expressed relative to untreated cells at 0 h, set as 100% in all experiments. All experiments were performed in triplicate, and the data are shown as means ± SD. The horizontal bar shows ATP levels in untreated controls. (D) Time course of ATP levels in DMSO and oligomycin-treated mutant cells. The data are means ± SD of an experiment that was performed in triplicate and is representative of three independent experiments. The data were normalized to ATP levels of DMSO-treated cells at 0.1 h that were arbitrarily set as a 100%.
Alterations in ATP, NADH, and Reactive Oxygen Species Levels Are Not Involved in Rescue.
Mitochondrial inhibitors can prevent cell death by altering cellular ATP levels (19, 20). We examined whether this mechanism contributed to suppression of cell death in our model. Mitochondrial inhibitors at concentrations that resulted in maximum rescue of cell death caused no decrease in ATP levels in N548 mutant cells (Fig. 3C). Furthermore, time course experiments failed to show a transient decline in ATP levels in oligomycin-treated cells relative to vehicle (DMSO)-treated cells (Fig. 3D). The decline in ATP levels at later times (48 h) is likely due to decreased viability because a larger decline in ATP levels was noted in DMSO-treated cells relative to oligomycin treated cells.
The result that cellular ATP levels did not decrease upon treating cells with mitochondrial inhibitors was unexpected. One explanation for this result could be that glycolysis was maintaining cellular ATP levels because both glycolysis and oxidative phosphorylation contribute to cellular ATP (SI Fig. 7). We found that oligomycin dramatically decreased cellular ATP levels when glycolysis was inhibited (cells grown in media without glucose) but not when glycolysis was functional (SI Fig. 9). This result suggests that both glycolysis and mitochondrial oxidative phosphorylation contribute to ATP production in N548 mutant cells and that glycolysis was likely maintaining cellular ATP levels upon mitochondrial inhibition.
Mitochondrial ETC inhibition also causes an accumulation of NADH (21), the principal product of the citric acid cycle, and can alter reactive oxygen species levels (21, 22). Increased NADH levels can prevent degeneration and cell death in certain models (21, 23). We found that neither exogenous NADH nor NAD+ nor a precursor of NADH (nicotinamide) rescued cell death at any of the concentrations tested (SI Fig. 10 and data not shown). Furthermore, mitochondrial uncouplers and glycolysis inhibitors rescued cell death (SI Table 1), despite the fact that they do not cause NADH accumulation (SI Fig. 7). Together, these data suggest that accumulation of NADH is not a relevant mechanism of rescue. Finally, we found no changes in reactive oxygen species levels upon treating cells with mitochondrial inhibitors, thus excluding this mechanism of rescue (SI Fig. 10).
Metabolic Inhibitors Activate Prosurvival ERK and AKT Signaling.
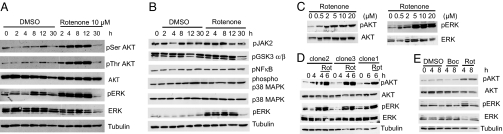
Altered cell survival signaling (ERK and AKT) is implicated in HD; components of these pathways are decreased or inhibited in HD (24–26), and their activation is protective in HD models (24, 27). We tested whether these pathways were activated by metabolic inhibitor treatment by measuring activating phosphorylation of specific amino acid residues on AKT and ERK proteins (24, 27). Rotenone treatment caused rapid and sustained activation of AKT and ERK (Fig. 4A). The activation of ERK/AKT was specific because several distinct kinase pathways: JAK2, NF-κB, p38 MAPK, and glycogen synthase kinase-3 α/β pathways were not activated by rotenone treatment (Fig. 4B). The activation of ERK/AKT was dose dependent and correlated with rescue of cell death (Figs. 4C and 1A), suggesting that this activation contributed to the rescue of cell death. We also confirmed activation of these pathways by rotenone in two additional clones of N548 mutant cells (Fig. 4D). This activation was not observed in cells treated with vehicle control (DMSO), confirming that it was not an indirect effect of serum starvation (Fig. 4 A–D). Furthermore, the pan-caspase inhibitor Boc-D-FMK (28) suppressed cell death of N548 mutant cells (4) but failed to activate these pathways (Fig. 4E), indicating that ERK and AKT activation was not an indirect effect of cell survival. Similar activation of ERK was observed by using the glycolysis inhibitor sodium fluoride (NaF) and the mitochondrial ATP synthetase inhibitor oligomycin, indicating that this activation was not an off-target effect of rotenone treatment (SI Fig. 11).
Fig. 4.
Rotenone specifically activates ERK and AKT. (A) N548 mutant cells were treated with DMSO or rotenone, and AKT and ERK activations were determined by Western blotting by using phospho-specific antibodies against serine 473 AKT, threonine 308 AKT, or threonine 202/tyrosine 204 ERK. Levels of total AKT, ERK, and tubulin served as controls. (B) The specificity of ERK activation was confirmed by testing for activation of distinct kinase pathways. Phospho-ERK served as a positive control. (C) Cells were treated with increasing concentrations of rotenone, and ERK/AKT activation was determined after 8 h. (D) Three independent clonal cell lines expressing the N548 amino acid N-terminal fragment of mutant htt were treated with 10 μM rotenone (Rot), and ERK/AKT activation was determined. (E) ERK/AKT activation was assayed in cells that were treated with DMSO, the Pan-caspase inhibitor Boc-D-fmk (50 μM) (Boc), or rotenone (Rot).
ERK/AKT Activation by Rotenone Is Independent of Mitochondrial Respiration.
Our results indicated that a decrease in total cellular ATP levels was not responsible for activating prosurvival signaling or the protection by rotenone. However, because cellular ATP levels are dependent on both glycolysis and oxidative phosphorylation, it was possible that inhibiting mitochondrial ATP production triggered ERK/AKT activation. To test this possibility, we generated two mitochondrial DNA-deficient (ρ0) N548 mutant cell lines by using established protocols (29). In ρ0 cells, mitochondrial DNA-encoded subunits of ETC complexes are not generated, making these cells deficient in ATP production by oxidative phosphorylation (30). Rotenone activated ERK and AKT and also rescued cell death in ρ0 cells to a similar extent as that in the parental N548 mutant cells (SI Fig. 12). These results indicate that decreased mitochondrial ATP production upon rotenone treatment is unlikely to cause rescue of cell death or to activate ERK and AKT.
ERK and AKT Activation Partially Explains Rescue by Metabolic Inhibitors.
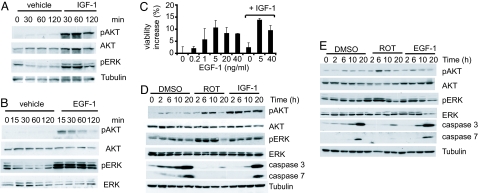
If activation of ERK and AKT by rotenone is important for rescue, then inhibiting their activation should abrogate the rescue by mitochondrial inhibitors, and, conversely, activating them should rescue cell death. We found that AKT and ERK inhibition by using specific chemical inhibitors of ERK and AKT abrogated the rescue by rotenone in a dose-dependent manner (SI Fig. 13). Moreover, the IC50 of abrogation of rescue by rotenone by these chemical inhibitors was similar to their reported cellular IC50 for the respective kinases (see Materials and Methods), suggesting that they were acting by their proposed mechanisms. Next, we tested whether activating ERK or AKT independently could rescue cell death. We used two growth factors, insulin-like growth factor 1 (IGF-1) and EGF-1 which are known activators of AKT and ERK, respectively (31, 32). As expected, IGF-1 caused a sustained activation of AKT but a transient activation of ERK (Fig. 5 A and D). In contrast, EGF-1 treatment caused rapid (15 min) and sustained (≈10 h) activation of ERK and a transient, but lesser, activation of AKT (Fig. 5 B and E). Cells treated with IGF-1 showed a slight rescue (≈5% increased viability), whereas EGF-1 increased viability by 10% (Fig. 5C). The rescue correlated with a small decrease in caspase 3 activity (Fig. 5 D and E). However, the rescue by growth factors was less than that by rotenone, which increased viability by 30% and substantially inhibited caspase 3 and 7 activation (Figs. 1B and 5 D and E). Because the rescue by these growth factors individually was less than that by rotenone, we tested whether a combination of IGF-1 and EGF-1 would recapitulate the rescue by rotenone. Together, IGF-1 and EGF-1 increased viability by 15% (Fig. 5C), suggesting an additive effect that was half as effective as rotenone (Fig. 1B). These results suggest that approximately half of the protective effect of rotenone can be explained by activation of ERK and AKT, with the ERK pathway being the major contributor to the rescue.
Fig. 5.
Growth factors activate ERK and AKT and rescue cell death. ERK and AKT activations after treatment with IGF-1 (90 ng/ml) (A) and EGF-1 (5 ng/ml) (B) in Sdm were assayed by Western blotting. (C) N548 mutant cells were treated with EGF-1 or IGF-1 alone or in combination, and cell viability was assayed after 2 days in Sdm by trypan blue dye exclusion assay. The results are the averages ± SD of one representative experiment. IGF-1 was used at one concentration (90 ng/ml); lower concentrations were ineffective, and higher concentrations did not further enhance viability. (D and E) N548 mutant cells were treated with DMSO, rotenone (Rot), or IGF-1 (90 ng/ml) (D) or EGF-1 (5 ng/ml) (E) in Sdm, and activation of AKT, ERK, and cleaved caspase 3 and 7 levels were monitored by Western blotting. Tubulin served as a loading control.
Discussion
By using a high-throughput screen, we discovered that small molecule inhibitors of metabolism (mitochondrial and glycolytic function) suppressed cell death in a striatal cell culture model of HD. Furthermore, several of these inhibitors were also efficacious in C. elegans and Drosophila HD models (SI Table 1 and Fig. 2), suggesting that this approach targets a conserved mechanism of HD toxicity. The role of energy metabolism in HD is controversial. Several studies implicate deficient energy production in HD pathophysiology; these data have been reviewed elsewhere (1, 33). However, data exist that are not readily explained by this hypothesis. First, several studies in HD animal models and patients show no changes or even higher metabolic rates and/or mitochondrial function compared with controls (34–38). Second, HD transgenic mice are more resistant to a mitochondrial complex II inhibitor (3-NP) than WT mice (39). Third, caloric restriction decreases energy metabolism (40, 41) and yet alleviates disease progression in HD mouse models (42). Finally, a recent study has identified multiple metabolic changes in HD patients that suggest a procatabolic phenotype (43). Together with our results, these data indicate that decreased energy metabolism may not be critical for the HD phenotype and that a more complex interplay likely exists among mutant htt, energy metabolism, and disease pathology.
The mechanism by which these metabolic inhibitors rescue cell death does not involve a decrease in cellular ATP levels (Fig. 3C) but involves caspase inhibition and activation of prosurvival signaling (Figs. 3B and 5 C–E). Caspase activation is implicated in HD pathology because caspase inhibition shows therapeutic value in mouse HD models (44). N548 mutant cell death is also caspase dependent because serum deprivation activates caspases and a broad-spectrum caspase inhibitor suppresses cell death (4). Metabolic inhibitors prevented “effector” caspase 3 and 7 activation (Fig. 3B), indicating that caspase inhibition by metabolic inhibitors is relevant to their protective effect. Furthermore, these inhibitors activated ERK and AKT; both of these enhance survival in diverse models (32). Activation of ERK and AKT by the growth factors IGF-1 and EGF-1 (32, 45) rescued cell death (Fig. 5C) and could partially explain the rescue of cell death by metabolic inhibitors in our cell culture model.
We investigated the mechanism by which the metabolic inhibitors activate ERK and AKT. Diverse inhibitors of metabolic function such as electron transport (rotenone), glycolysis (NaF), and ATP synthetase (oligomycin) activated ERK and AKT in our cell culture model. These results indicate that the prosurvival response is not limited to ETC inhibition or due to off-target effects of these compounds but is likely triggered by common metabolic effects of these compounds. We excluded potential mechanisms, such as changes in ATP, reactive oxygen species, NADH levels (21, 22), and mitochondrial respiration (Fig. 3 and SI Figs. 10 and 12), in triggering prosurvival signaling or rescuing cell death. Although mitochondria are involved primarily in energy metabolism and apoptosis (46), a role for mitochondria in sensing cellular environment and conveying signals to other cell compartments is emerging (47). Understanding the mode of prosurvival signal activation in this cell culture model could reveal conserved links between metabolism and cell survival signaling.
Altered growth factor signaling has been implicated in HD pathology; inhibition of EGF-1 and IGF-1 signaling is reported in HD models and patients (25, 26, 48). Conversely, activation of growth factor signaling protects cells from mutant htt toxicity (24, 27). Although extracellular administration of growth factors can activate these pathways, this approach is not feasible in brain tissue of whole animals. Our study reveals a novel means of activating ERK/AKT by using small molecules that could be exploited for therapeutic benefit in HD and possibly other neurodegenerative disorders. Of the active compounds, NaF and 2,4-dinitrophenol have a history of clinical use and can cross the blood–brain barrier (9, 49–51). These compounds are candidates for testing whether this approach would be efficacious in mouse HD models.
Materials and Methods
Cell Culture.
N548 mutant cells were maintained as described previously (4). Cell death was induced by a change to 0.5% FCS-containing medium (serum-deprived medium) and by incubating cells at 39°C. Glucose-free DMEM (catalog no. 90-113-PB; CellGro, Herndon, VA) was used for certain experiments.
Antibodies, Chemicals, and Growth Factors.
All chemicals were obtained from Sigma (St. Louis, MO), except IGF-1 (catalog no. RU020; Cell Sciences, Canton, MA) and EGF-1 (PMG0062; Invitrogen, Carlsbad, CA). Phosphoserine 473, phosphothreonine 308 AKT, phospho-ERK, phospho-JAK2, phospho-NF-κB (p65 subunit), phosphoglycogen synthase kinase α /β, phospho-p38 MAPK, AKT, and ERK antibodies were from Cell Signaling Technology (Beverly, MA). Mitochondrial complex II and IV antibodies were from Molecular Probes (Carlsbad, CA). Antibodies were used at the dilutions suggested by the suppliers. Other antibodies used have been previously described (4). The National Institute of Neurological Disorders and Stroke custom collection of 1,040 compounds was from Microsource Discovery, Inc. (Gaylordsville, CT); and U0126 and MEK1 inhibitor (52, 53), AKT inhibitor VIII, and phosphatidylinositol 3-kinase inhibitor LY294002 (54) were from Calbiochem (La Jolla, CA).
Western Blotting.
Cells were incubated with the compounds or vehicle (0.1% DMSO) in low serum or in 10% serum-containing media. Cells were harvested, and Western blotting was performed as described previously (4).
Cell Viability Assays.
Cell viability was assayed by using the calcein acetoxymethyl ester assay (Molecular Probes) as previously described (4). Trypan blue cell viability was performed on N548 mutant cells by using an automated trypan blue (0.4%) dye-exclusion assay (Vi-Cell 1.01; Beckman Coulter, Fullerton, CA) (4). At least 200 cells were counted per sample, and the percentage of trypan blue negative (viable) cells was calculated.
ATP Assay.
Cells were plated at 105 cells per 60-mm tissue culture dish and, after various treatments, were lysed in 200 μl of lysis buffer; ATP levels were measured by using a luminescence-based assay as suggested by the manufacturer (catalog no. K254-200; Biovision, Mountain View, CA). ATP controls were included to ensure linearity of assay. The light signal (integration, 12 s) was measured in a luminometer (Lumat LB9501; Berthold, Nashua, NH), and luminescence was normalized to cell number. The normalization based on protein content (protein assay reagent; Bio-Rad, Hercules, CA) was similar to that based on cell number.
Reactive Oxygen Species Assay.
The assay kit was used according to the instructions of the manufacturer (catalog no. D-399; Molecular Probes). In brief, 10 μM H2DCFDA dye (2′,7′-dichlorodihydrofluorescein diacetate) was added to 1,500 cells in a 384-well plate, and fluorescence (excitation, 490 nm; emission, 535 nm) was measured by using a plate reader (VICTOR3; PerkinElmer, Waltham, MA).
C. elegans Neuronal Survival Assay.
The C. elegans assay has been described previously (4, 55). In brief, L1 pqe-1;Htn-Q150 animals were incubated at 15°C in liquid S media with a food source (E. coli, OP50 strain) containing either a compound or DMSO (1.5%). GFP fluorescence in bilateral ASH sensory neurons was examined by using a fluorescence microscope (excitation, 485 nm; emission, 535 nm). One hundred neurons were scored in >50 animals. For the starvation assay, L1-arrested animals were obtained by continued absence of food because C. elegans larvae do not initiate growth under starvation (55). These experiments were conducted at 25°C.
Drug Testing in Drosophila.
The Drosophila HD strain (8534) was obtained from the Bloomington Drosophila stock center and has been described previously (14). Animals were maintained on standard fly food at room temperature. Drugs were mixed in the food every 2 days, and flies were fed on the food for the duration of the experiment at 25°C. The rhabdomere number was assessed by the pseudopupil technique (14). Equal numbers of males and females were included in the control (DMSO) and drug-treated samples. Eight to 10 animals were scored for each treatment, and 35 ommatidia were scored per animal. All scoring was performed in a blinded manner, and significance was calculated by Student's t test.
Supplementary Material
Acknowledgments
This work was supported by the High Q Foundation (H.V.), the Hereditary Disease Foundation (C.V. and B.R.S.), the Cure Huntington Disease Initiative (B.R.S.), and the National Institutes of Health (A.C.H.). B.R.S. also was supported by a Career Award at the Scientific Interface from the Burroughs Wellcome Fund and by the Arnold and Mabel Beckman Foundation.
Abbreviations
- ETC
electron transport chain
- HD
Huntington's disease
- htt
huntingtin protein
- IGF
insulin-like growth factor
- Sdm
serum-deprived media.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704482104/DC1.
References
- 1.Brouillet E, Condâe F, Beal MF, Hantraye P. Prog Neurobiol. 1999;59:427–468. doi: 10.1016/s0301-0082(99)00005-2. [DOI] [PubMed] [Google Scholar]
- 2.Ross CA. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 3.Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S, Sciorati C, Clementi E, Hackam A, Hayden MR, Li Y, et al. J Neurosci. 2000;20:3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varma H, Voisine C, DeMarco CT, Cattaneo E, Lo DC, Hart AC, Stockwell BR. Nat Chem Biol. 2007;3:99–100. doi: 10.1038/nchembio852. [DOI] [PubMed] [Google Scholar]
- 5.Scheffler IE. Adv Drug Deliv Rev. 2001;49:3–26. doi: 10.1016/s0169-409x(01)00123-5. [DOI] [PubMed] [Google Scholar]
- 6.Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodgson JG, Agopyan N, Gutekunst CA, Leavitt BR, LePiane F, Singaraja R, Smith DJ, Bissada N, McCutcheon K, Nasir J, et al. Neuron. 1999;23:181–192. doi: 10.1016/s0896-6273(00)80764-3. [DOI] [PubMed] [Google Scholar]
- 8.Penefsky HS. Proc Natl Acad Sci USA. 1985;82:1589–1593. doi: 10.1073/pnas.82.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harper JA, Dickinson K, Brand MD. Obes Rev. 2001;2:255–265. doi: 10.1046/j.1467-789x.2001.00043.x. [DOI] [PubMed] [Google Scholar]
- 10.Gumiânska M, Sterkowicz J. Acta Biochim Pol. 1976;23:285–291. [PubMed] [Google Scholar]
- 11.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Proc Natl Acad Sci USA. 2002;99:17131–17136. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 13.Troulinaki K, Tavernarakis N. Mech Aging Dev. 2005;126:23–33. doi: 10.1016/j.mad.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, Faber PW, MacDonald ME, Zipursky SL. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 15.Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL, Kazantsev A, Schmidt E, Zhu YZ, Greenwald M, et al. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 16.Ferrante RJ, Kubilus JK, Lee J, Ryu H, Beesen A, Zucker B, Smith K, Kowall NW, Ratan RR, Luthi-Carter R, et al. J Neurosci. 2003;23:9418–9427. doi: 10.1523/JNEUROSCI.23-28-09418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez Mejia RO, Friedlander RM. Neuroscientist. 2001;7:480–489. doi: 10.1177/107385840100700604. [DOI] [PubMed] [Google Scholar]
- 18.Graham RK, Deng Y, Slow EJ, Haigh B, Bissada N, Lu G, Pearson J, Shehadeh J, Bertram L, Murphy Z, et al. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Stefanelli C, Bonavita F, Stanic I, Farruggia G, Falcieri E, Robuffo I, Pignatti C, Muscari C, Rossoni C, Guarnieri C, et al. Biochem J. 1997;322:909–917. doi: 10.1042/bj3220909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicotera P, Leist M. Cell Death Differ. 1997;4:435–442. doi: 10.1038/sj.cdd.4400265. [DOI] [PubMed] [Google Scholar]
- 21.Sestili P, Brambilla L, Cantoni O. FEBS Lett. 1999;457:139–143. doi: 10.1016/s0014-5793(99)01027-3. [DOI] [PubMed] [Google Scholar]
- 22.Vrablic AS, Albright CD, Craciunescu CN, Salganik RI, Zeisel SH. FASEB J. 2001;15:1739–1744. doi: 10.1096/fj.00-0300com. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Zhai Q, Chen Y, Lin E, Gu W, McBurney MW, He Z. J Cell Biol. 2005;170:349–355. doi: 10.1083/jcb.200504028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apostol BL, Illes K, Pallos J, Bodai L, Wu J, Strand A, Schweitzer ES, Olson JM, Kazantsev A, Marsh JL, et al. Hum Mol Genet. 2006;15:273–285. doi: 10.1093/hmg/ddi443. [DOI] [PubMed] [Google Scholar]
- 25.Liâevens JC, Rival T, Ichâe M, Chneiweiss H, Birman S. Hum Mol Genet. 2005;14:713–724. doi: 10.1093/hmg/ddi067. [DOI] [PubMed] [Google Scholar]
- 26.Colin E, Râegulier E, Perrin V, Dèurr A, Brice A, Aebischer P, Dâeglon N, Humbert S, Saudou F. Eur J Neurosci. 2005;21:1478–1488. doi: 10.1111/j.1460-9568.2005.03985.x. [DOI] [PubMed] [Google Scholar]
- 27.Humbert S, Bryson EA, Cordeliáeres FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F. Dev Cell. 2002;2:831–837. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- 28.Dâeas O, Dumont C, MacFarlane M, Rouleau M, Hebib C, Harper F, Hirsch F, Charpentier B, Cohen GM, Senik A. J Immunol. 1998;161:3375–3383. [PubMed] [Google Scholar]
- 29.King MP, Attardi G. Methods Enzymol. 1996;264:304–313. doi: 10.1016/s0076-6879(96)64029-4. [DOI] [PubMed] [Google Scholar]
- 30.King MP, Attardi G. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 31.Hougardy BM, Maduro JH, van der Zee AG, Willemse PH, de Jong S, de Vries EG. Lancet Oncol. 2005;6:589–598. doi: 10.1016/S1470-2045(05)70281-3. [DOI] [PubMed] [Google Scholar]
- 32.Henson ES, Gibson SB. Cell Signal. 2006;18:2089–2097. doi: 10.1016/j.cellsig.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Grèunewald T, Beal MF. Ann NY Acad Sci. 1999;893:203–213. doi: 10.1111/j.1749-6632.1999.tb07827.x. [DOI] [PubMed] [Google Scholar]
- 34.Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, Hershey T, Perlmutter JS. Proc Natl Acad Sci USA. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guidetti P, Charles V, Chen EY, Reddy PH, Kordower JH, Whetsell WO, Jr, Schwarcz R, Tagle DA. Exp Neurol. 2001;169:340–350. doi: 10.1006/exnr.2000.7626. [DOI] [PubMed] [Google Scholar]
- 36.Pratley RE, Salbe AD, Ravussin E, Caviness JN. Ann Neurol. 2000;47:64–70. [PubMed] [Google Scholar]
- 37.Gaba AM, Zhang K, Marder K, Moskowitz CB, Werner P, Boozer CN. Am J Clin Nutr. 2005;81:1335–1341. doi: 10.1093/ajcn/81.6.1335. [DOI] [PubMed] [Google Scholar]
- 38.Panov AV, Lund S, Greenamyre JT. Mol Cell Biochem. 2005;269:143–152. doi: 10.1007/s11010-005-3454-9. [DOI] [PubMed] [Google Scholar]
- 39.Hickey MA, Morton AJ. J Neuro Chem. 2000;75:2163–2171. doi: 10.1046/j.1471-4159.2000.0752163.x. [DOI] [PubMed] [Google Scholar]
- 40.Madapallimattam AG, Law L, Jeejeebhoy KN. Am J Clin Nutr. 2002;76:1031–1039. doi: 10.1093/ajcn/76.5.1031. [DOI] [PubMed] [Google Scholar]
- 41.Feuers RJ. Ann NY Acad Sci. 1998;854:192–201. doi: 10.1111/j.1749-6632.1998.tb09902.x. [DOI] [PubMed] [Google Scholar]
- 42.Duan W, Guo Z, Jiang H, Ware M, Li XJ, Mattson MP. Proc Natl Acad Sci USA. 2003;100:2911–2916. doi: 10.1073/pnas.0536856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Underwood BR, Broadhurst D, Dunn WB, Ellis DI, Michell AW, Vacher C, Mosedale DE, Kell DB, Barker RA, Grainger DJ, et al. Brain. 2006;129:877–886. doi: 10.1093/brain/awl027. [DOI] [PubMed] [Google Scholar]
- 44.Pattison LR, Kotter MR, Fraga D, Bonelli RM. J Neurol. 2006;253:1137–1142. doi: 10.1007/s00415-006-0198-8. [DOI] [PubMed] [Google Scholar]
- 45.Kurmasheva RT, Houghton PJ. Biochim Biophys Acta. 2006;1766:1–22. doi: 10.1016/j.bbcan.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 46.Newmeyer DD, Ferguson-Miller S. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Butow RA. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 48.Song C, Perides G, Liu YF. J Biol Chem. 2002;277:6703–6707. doi: 10.1074/jbc.M110338200. [DOI] [PubMed] [Google Scholar]
- 49.Mattiasson G, Shamloo M, Gido G, Mathi K, Tomasevic G, Yi S, Warden CH, Castilho RF, Melcher T, Gonzalez-Zulueta M, et al. Nat Med. 2003;9:1062–1068. doi: 10.1038/nm903. [DOI] [PubMed] [Google Scholar]
- 50.Mullenix PJ, Denbesten PK, Schunior A, Kernan WJ. Neurotoxicol Teratol. 1995;17:169–177. doi: 10.1016/0892-0362(94)00070-t. [DOI] [PubMed] [Google Scholar]
- 51.Rubin CD, Pak CY, Adams-Huet B, Genant HK, Li J, Rao DS. Arch Intern Med. 2001;161:2325–2333. doi: 10.1001/archinte.161.19.2325. [DOI] [PubMed] [Google Scholar]
- 52.Favata MF, Horiuchi KY, Manos EJ, Daulerio AJ, Stradley DA, Feeser WS, Van Dyk DE, Pitts WJ, Earl RA, Hobbs F, et al. J Biol Chem. 1998;273:18623–18632. doi: 10.1074/jbc.273.29.18623. [DOI] [PubMed] [Google Scholar]
- 53.Wityak J, Hobbs FW, Gardner DS, Santella JB, III, Petraitis JJ, Sun JH, Favata MF, Daulerio AJ, Horiuchi KY, Copeland RA, et al. Bioorg Med Chem Lett. 2004;14:1483–1486. doi: 10.1016/j.bmcl.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Vlahos CJ, Matter WF, Hui KY, Brown RF. J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 55.Voisine C, Varma H, Walker N, Bates EA, Stockwell BR, Hart AC. PLoS One. 2007;2:e504. doi: 10.1371/journal.pone.0000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.