Abstract
Proteasomes can be markedly activated by associating with 19S regulatory complexes to form the 26S protease or by binding 11S protein complexes known as REG or PA28. Three REG subunits, α, β, and γ, have been expressed in Escherichia coli, and each recombinant protein can activate human proteasomes. Combining PCR mutagenesis with an in vitro activity assay, we have isolated and characterized 36 inactive, single-site mutants of recombinant REGα. Most are monomers that produce functional proteasome activators when mixed with REGβ subunits. Five REGα mutants that remain inactive in the mixing assay contain amino acid substitutions clustered between Arg-141 and Gly-149. The crystal structure of the REGα heptamer shows that this region forms a loop at the base of each REGα subunit. One mutation in this loop (N146Y) yields a REGα heptamer that binds the proteasome as tightly as wild-type REGα but does not activate peptide hydrolysis. Corresponding amino acid substitutions in REGβ (N135Y) and REGγ (N151Y) produce inactive proteins that also bind the proteasome and inhibit proteasome activation by their normal counterparts. Our studies clearly demonstrate that REG binding to the proteasome can be separated from activation of the enzyme. Moreover, the dominant negative REGs identified here should prove valuable for elucidating the role(s) of these proteins in antigen presentation.
A major mechanism for controlling viral infections involves cytotoxic T lymphocytes that recognize viral peptides presented on the cell surface by major histocompatability complex class I molecules and lyse the infected cells (1, 2). There is considerable evidence that some presented peptides, at least, are produced by the proteasome (3, 4). Crystal structures of Thermoplasma and yeast proteasomes reveal that they are cylindrical protein complexes, composed of four stacked rings (5, 6). The two end rings consist of catalytically inactive α-type subunits, whereas the two inner rings are composed of β-type subunits, some of which are catalytically active (7). The protease active sites are located within an inner chamber that is virtually sealed from the particles surface (5, 6). Thus, it seems clear that substrate entry to the sites of peptide bond hydrolysis must be tightly regulated.
Two proteasome activators have been identified so far. The proteasome can either associate with a 19S regulatory complex to form the 26S protease, which is capable of degrading intact proteins (8–11), or the proteasome can bind an 11S activator called REG or PA28. This association greatly enhances fluorogenic peptide hydrolysis by the proteasome (12, 13). As isolated from human red blood cells, REG is a hexameric or heptameric ring formed from two homologous subunits, REGα and REGβ; these two subunits are, in turn, homologous to KI antigen or REGγ. cDNAs for all three proteins have been expressed in Escherichia coli, and each recombinant protein is capable of activating the proteasome in vitro (14).
A variety of evidence suggests that REG is involved in antigen processing. For example, synthesis of REGα is up-regulated by interferon-γ (15), a cytokine that induces synthesis of several proteins involved in antigen presentation, including major histocompatability complex class I molecules and transporters associated with antigen presentation (1, 16). Overexpression of REGα in mammalian cells has been reported to enhance presentation of class I epitopes (17). Kloetzel and his colleagues have also reported that REG induces the proteasome to catalyze dual cleavages in precursor peptides containing naturally occurring epitopes (18).
To understand how REG activates the proteasome, we have isolated 45 single-site REGα mutants with altered activities by screening random-mutagenized REGα cDNA libraries. Here we describe the properties of 36 completely inactive, single-site REGα mutants. These findings demonstrate that a short stretch of amino acids highly conserved among REG homologs is critical for proteasome activation, that proteasome activation by REGα is separable from its binding to the proteasome, and that single-site mutations in REGα, β, and γ produce inactive proteins able to prevent proteasome activation by their wild-type counterparts.
MATERIALS AND METHODS
Generation of the REGα-Random-Mutation Expression Libraries.
Random mutations were introduced into REGα cDNA in pAED4 by using PCR under error-prone conditions (19, 20). The PCR products were digested with NdeI and BamHI and ligated into the pET11a expression plasmid. Aliquots of ligation mixtures were used to transform E. coli [BL-21(DE3)] by electroporation. Most mutants described here were isolated from one expression library that gave rise to about 60% single-site mutants on initial screening. The PCR conditions for construction of this library were: 0.2 mM each of dGTP and dATP; 1 mM each of dCTP and dTTP; 10 mM Tris⋅HCl, pH 8.3, 50 mM KCl, 7 mM MgCl2, 0.01% gelatin, and 2.5 units of Taq DNA polymerase (Perkin–Elmer) for 100-μl reactions. After an initial melting at 94°C for 4 min, 30 cycles of denaturation (1 min at 94°C), annealing (1 min at 50°C), and extension (3 min at 72°C) were performed.
Isolation of REGα Mutants with Altered Activities.
Transformants were picked and grown overnight in 50 μl H medium (10 tryptone, 8g NaCl per liter water) in each well of a 96-well microtiter plate. Aliquots of each culture were induced with 0.4 mM IPTG at 30°C for 2 hr and lysed with 30 μl of 20 mM Tris⋅HCl, pH 7.5, 1% Triton X-100, and 0.6 mg/ml polymixin B sulfate. Then 10 μl of 17 ng/μl proteasome and 50 μl of 200 μM Suc-LLVY-aminomethylcoumerin (MCA) was added to each well. Colonies producing active REGα became highly fluorescent in about 30 min of incubation at 37°C; those producing inactive REGα remained dark. After the activity assay, the reaction mixtures were transferred to a nitrocellulose membrane and anti-REGα specific serum was used to detect the expression of each clone after a standard Western blot protocol (21). Clones that were inactive in the enzymatic assay and positive in the antibody test were rescreened using the same procedures. The clones that survived two rounds of screening were induced, and plasmids from the clones that expressed full-length REG proteins as judged by SDS/PAGE were purified and sequenced.
Purification and Characterization of Inactive REGα Mutants.
Each REGα mutant was overexpressed in E. coli and purified to homogeneity by using a combination of DEAE and gel filtration chromatography (14). Each purified protein was concentrated and dialyzed against 0.5× TSD (Tris-salts-DTT) (14) before biochemical characterization as described below. The proteasome stimulatory activity of REGα mutants was assayed by adding 1.5 μg of purified protein to human red cell proteasomes (170 ng) in 10 mM Tris, pH 7.5, and incubating at 37°C. After 10 min, 50 μl of 200 μM Suc-LLVY-MCA in 10 mM Tris, pH 7.5, was added to produce a 100-μl reaction mixture. After an additional 10 min, the reaction was quenched with 200 μl of ice-cold ethanol and the fluorescence was measured as described (14). To measure the activity of mtREGα/REGβN135Y heterooligomers, each mutant protein was incubated at 4°C overnight with an equal amount of purified REGβN135Y. Then 1.5 μg of protein from each mixture was used to measure activity as described above. To determine the oligomeric state of a REGα mutant, 1.5 ml of 0.6 mg/ml protein was loaded on a Superdex 200 gel filtration column (Hiload 26/60, Pharmacia) and eluted with TSD, 175 ml KCl. Under these conditions, more than 95% wild-type REGα elutes as a heptamer.
A direct proteasome binding assay (14) was used to determine whether REGα mutants bind the proteasome. Typically, 60 μg/ml and 20 μg/ml of rREGs were incubated with human proteasomes tethered to ELISA plates by antibodies. After washing away unadsorbed protein, the bound proteins were eluted with 0.5 M KCl, transferred to nitrocellulose membranes, and then probed with rabbit anti-REGα specific serum. Bound antibodies were detected by using peroxidase-conjugated goat anti-rabbit IgG and chemiluminescence detection (NEN). Wild-type REGα exhibited binding at both concentrations, with less binding to the proteasome at 20 μg/ml (see Fig. 3c). MtREGα/REGβN135Y mixtures were used to determine proteasome binding at 60 μg/ml and 20 μg/ml. After elution in 0.5 M KCl, the proteins were transferred to nitrocellulose, and the membrane was probed with anti-REGα antiserum to detect the bound REGα mutants. The membrane was then stripped and reprobed with anti-REGβ antisera to detect bound REGβN135Y. All the mixtures tested exhibited equal amounts of REGs α and β bound to the proteasome at both concentrations confirming the higher affinity of α/β heterooligomers for the proteasome as reported in ref. 14.
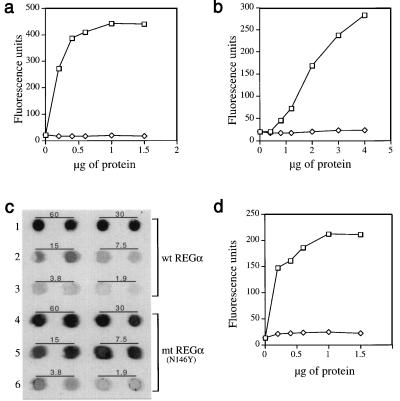
Figure 3.
Importance of a conserved asparagine for proteasome activation by REG homologs. (a) Proteasome stimulation by recombinant REGα (squares) and rREGαN146Y (diamonds). Human red cell proteasome (170 ng) was mixed with increasing amounts of REGα or rREGαN146Y to a final volume of 50 μl, incubated at 37°C for 10 min, and then 50 μl of 200 μM Suc-LLVY-MCA was added. After 10 min the reaction was quenched by using 200 μl of 100% ethanol, and enzyme activity is reported as fluorescence units. (b) Proteasome stimulation by REGβ (squares) and REGβN135Y (diamonds). This experiment was performed as described in a. (c) Proteasome binding properties of rREGα and rREGαN146Y. After tethering proteasomes to ELISA plates with anti-proteasome antibody, different concentrations of rREGα or rREGαN146Y were added to each well and incubated. Bound REG was eluted with 0.5 M KCl, blotted onto nitrocellulose, and detected with rabbit anti-REGα (see Methods). The number above each pair of dot blots indicates the incubation concentration of REGs in μg/ml. The apparent tighter binding exhibited by REGα(N146Y) is due to the fact that anti REGα serum detects the mutant protein better than it detects wild-type REGα (data not shown). (d) Proteasome stimulation by REGγ (squares) and REGγN151Y (diamonds). The assays were done in a except that Suc-LRR-MCA was used as substrate. Each data point in a, b, and d represents the mean of three measurements from a single experiment. But equivalent results were observed in at least two experiments by using different preparations of the various REG proteins.
Site-Directed Mutagenesis.
REGβN135Y and REGγN151Y were constructed by site-directed mutagenesis as described (22, 23).
RESULTS AND DISCUSSION
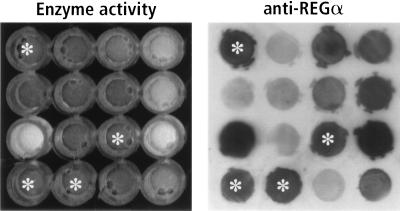
PCR was used to produce mutations in a REGα cDNA contained in the prokaryotic expression plasmid pAED4. The PCR products were ligated into pET11a, which was used to transform E. coli cells. Inactive REGα variants were identified by transferring bacteria from individual colonies to microtiter wells that contained 0.3% Triton X-100, proteasomes, and a fluorogenic peptide substrate. The plates were incubated and viewed under near-UV illumination. Active REGα released from the bacteria markedly increases the yield of fluorescent aminomethylcoumarin (MCA) produced by the proteasome, whereas wells lacking REGα or containing inactive variants remain dark (Fig. 1 Left). To distinguish bacteria expressing inactive REGα subunits from nonproducers, each reaction mixture was transferred to a nitrocellulose filter and immunostained with rabbit anti-REGα serum (Fig. 1 Right).
Figure 1.
Activity screen for REGα mutants. The two panels show a small section of a 96-well ELISA plate that illustrates the enzyme assay (Left) and the antibody screen for REGα expression (Right). Wells marked with white asterisks identify inactive REGα variants. The fluorescence shown in the positive wells is similar to that in wells containing bacteria that express wild-type REGα as control. In addition, the use of antibodies to detect REGα variants may account for the paucity of folding mutants in our collection.
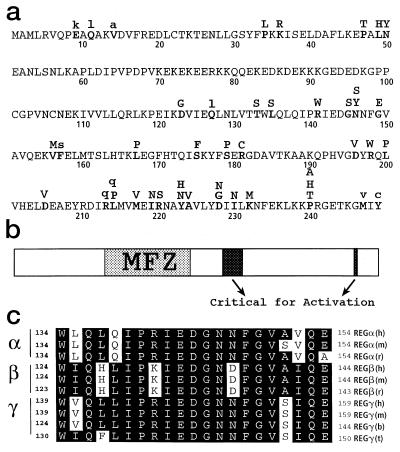
Plasmids were isolated from bacteria producing inactive REGα proteins, and DNA sequencing identified the positions of 36 inactive and 9 partially active single-site mutants. These amino acid substitutions are presented in the one-letter amino acid code above the wild-type REGα sequence (Fig. 2a). A striking feature of the distribution of inactivating mutations is the large gap between residues 51 and 122 in the REGα sequence (see Fig. 2b). This mutation-free zone (MFZ) encompasses the homolog-specific “inserts”—the amino acid sequences unique to each REG homolog (14). Lack of inactivating mutations in the 72-aa interval indicates that the insert region is not important for oligomerization of REGα, its binding to the proteasome, or subsequent activation of catalysis. This inference is supported by the REGα crystal structure, in which the “insert” region is located within a large, disordered loop that is distant from the apparent proteasome-binding surface (24). Moreover, deletion of residues 70 through 97 from REGα produces a recombinant protein able to form heptamers and activate the proteasome to the same extent as wild-type REGα (Z.Z., C.R., A.C., S. Endicott, and M.R., unpublished data). These findings clearly demonstrate that the homolog-specific insert of REGα functions not in proteasome activation per se, but most likely in biological processes, such as intracellular localization and/or association with other cellular components, possibly chaperonins in the endoplasmic reticulum membrane as previously suggested (25).
Figure 2.
Inactivating single-site mutations in REGα. (a) Single-site changes that produce completely inactive variants are indicated by bold capital letters placed above the wild-type sequence, which is presented in the one-letter code (50 residues per line). Nine amino acid substitutions that produce partially active variants are denoted by lowercase letters. (b) The sequence of REGα is depicted as an elongated rectangle in which the mutation free zone (MFZ) is identified by stippling, and the positions of residues important for proteasome activation are shown in black. (c) Sequence alignments of the region most highly conserved among REG homologs. These sequences contain the blackened activation region, Arg-141 to Gly-149, shown near the center of b and which appears to possess a suitable geometry for interaction with the proteasome (24). The letters at the far right identify the species: h, human; m, mouse; r, rat; t, tick; b, bovine.
All of the inactive mutant proteins shown in Fig. 2a were purified to homogeneity and subjected to gel filtration. Thirty-one of the mutants appear to be monomeric by gel filtration and four mutants form heptamers (see Table 1). The ability of each mutant to bind proteasomes was determined by using the assay described in Fig. 3c. Thirty-one of the REGα monomers and two of the four inactive heptamers failed to bind the proteasome ( Table 1, column 2). Thus, oligomerization of REGα subunits is important for their association with the proteasome but, as shown by the heptameric mutants, A224V and S175F, not necessarily sufficient.
Table 1.
Biochemical properties of inactive REGα mutants
| Position | Oligomeric state | Binding to proteasome | Activity when mixed with REGβn135y | Proteasome binding by REGβn135y Heterooligomers |
|---|---|---|---|---|
| wtREGα | H | ++ | + | +++ |
| P34L | M | − | + | +++ |
| K36R | M | − | + | ND |
| P47T | M | − | + | ND |
| L49H | M | − | + | +++ |
| N50Y | M | − | + | +++ |
| D123G | M | − | + | +++ |
| T133S | M | − | + | +++ |
| L135S | M | − | + | ND |
| R141W | M/h | − | − | +++ |
| G145S | H | + | − | +++ |
| N146Y | H | ++ | − | +++ |
| N146S | M | − | − | +++ |
| G149E | M/D | − | − | +++ |
| V156M | M | − | + | ND |
| L167P | M/h | − | + | +++ |
| S175F | H | − | + | +++ |
| S179P | M | + | + | +++ |
| R181C | M | − | − | − |
| D196V | M | − | + | ND |
| R198W | M | − | + | +++ |
| L200P | M | − | + | ND |
| D205V | M | − | − | − |
| L214P | M | − | + | +++ |
| M217V | M | − | + | +++ |
| I219N | − | − | + | ND |
| R220S | M | − | + | ND |
| Y223N | M | − | + | ND |
| Y223H | M | − | + | ND |
| A224V | H | − | + | ND |
| D228G | M | − | + | ND |
| D228N | M | − | + | +++ |
| I230N | M | − | + | +++ |
| K232M | M | − | −/+ | ++ |
| P240A | M | − | − | +++ |
| P240H | M | − | − | +++ |
| P240T | M | − | − | +++ |
The positions of 36 single-site inactive REGα mutants are indicated in the column at the left. Second column: Each mutant protein was purified and its extent of oligomerization determined by gel filtration. H, ≥95% of protein eluted as heptamer; M, monomer; any heptamer was below detection limit of the UV monitor; M/h, predominantly monomer, trace heptamer; M/D, about 60% monomer, 40% dimer. Third column: Each protein was tested for its ability to associate with the proteasome by the direct binding assay. REGαs at 60 μg/ml and 20 μg/ml were incubated with proteasomes tethered to ELISA wells by antibodies. After washing away any unadsorbed REGα, the bound proteins were eluted with high salt, transferred to a nitrocellulose membrane, and probed with anti-REGα specific antiserum. Wild-type REGα exhibited binding at both concentrations, but less protein was bound at 20 μg/ml (++). Most mutants failed to bind at either concentration (−). Those that exhibited binding at 60 μg/ml but not at 20 μg/ml are scored (+). Fourth column: Each mutant REGα protein was mixed with an equal amount of purified REGβn135y and incubated at 4°C overnight. The activity of mtREGα/REGβn135y was measured by adding 1.5 μg of protein from each mixture to 170 ng of proteasome and 100 μM Suc-LLVY-MCA. Stimulation by the active mixtures (+) was at least 5-fold compared with REGβn135y plus proteasome alone. A minus sign designates those mixtures unable to stimulate the proteasome under the experimental conditions. Fifth column: All combinations of mutant REGα and REGβn135y that remained inactive in the peptide hydrolysis assay were tested for proteasome binding concentrations of 60 μg/ml and 20 μg/ml. The bound proteins were detected with either anti-REGα or anti-REGβ specific antiserum. +++, amount of bound REG proteins was virtually identical at these two concentrations, and it is known that α/β heterooligomers bind the proteasome tighter than REGα alone (14) (++). Thirteen active combinations were tested for their proteasome binding properties; all bound the proteasome so we did not test the rest of the active mtREGα/REGβn135y combinations, which are designated ND for not determined. Note: REGα mutants that bind the proteasome when mixed with REGβn135y but fail to activate the enzyme are presented in bold type.
One of the REGα heptamers, N146Y, is particularly interesting. It does not activate the proteasome (see Fig. 3a), but it binds the enzyme as tightly as wild-type REGα (Fig. 3c). Asparagine 146 in REGα is located near the center of the sequence most highly conserved among REG homologs (Fig. 2c). Therefore, we converted the corresponding asparagines in REGβ (N135) or REGγ (N151) to tyrosines, expecting that this might produce inactive homologs that would also bind the proteasome. This expectation was clearly fulfilled because neither REGβN135Y nor REGγN151Y activated the proteasome (Fig. 3 b and d). Direct binding assays showed that REGβN135Y and REGγN151Y bind the proteasome to the same extent as their wild-type counterparts do (data not presented). And, enzyme competition assays demonstrated that each REG homolog bearing the Asn-to-Tyr substitution inhibited proteasome activation by the corresponding recombinant wild-type REG protein (see Fig. 4 a–c). Indeed, the heterooligomer produced by mixing REGαN146Y and REGβN135Y was a potent inhibitor of the 11S REG isolated directly from human red blood cells (Fig. 4d). This observation raises the possibility that REG molecules bearing critical Asn-to-Tyr mutations will inhibit REG function in vivo and could, therefore, prove to be valuable experimental reagents for studies on antigen presentation.
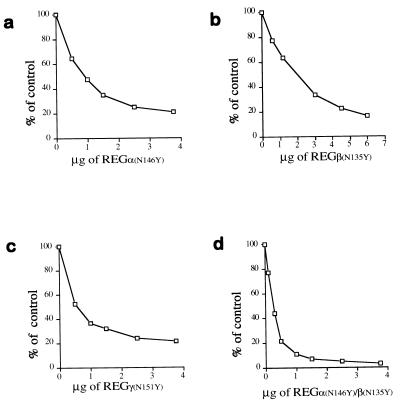
Figure 4.
Dominant negative properties of REG homologs with Asn-to-Tyr substitutions. (a) Inhibition of REGα-activated peptide hydrolysis by REGαN146Y. Proteasomes (170 ng) and 1.5 μg of REGα were mixed with increasing amounts of REGαN146Y, incubated at 37°C for 10 min, and then 50 μl of 200 μM Suc-LLVY-MCA was added to start the reactions. Product formation was measured as fluorescence units, and the results are expressed as a percentage of the activity in the absence of REGαN146Y. (b) Inhibition of REGβ’s activity by REGβN135Y. The assay was performed as described in a except that 3 μg of REGβ was added to each reaction. (c) Inhibition of REGγ’s activity by REGγN151Y. The assay was performed as described in a except that Suc-LRR-MCA was the substrate. (d) Inhibition of the activity of REG from human red blood cells by REGαN146Y/REGβN135Y. REGαN146Y/REGβN135Y heterooligomers were preformed by overnight incubation at 4°C and purified by gel filtration chromatography. Hrbc REG (1.5 μg) was added to each reaction, and increasing amounts of REGαN146Y/REGβN135Y were added prior to substrate. The enzymatic reaction was then performed as in a. Less inhibition was observed when recombinant wild-type REGα/β oligomers were challenged with REGαN146Y/REGβN135Y. Each point in all the figures represents the mean of three measurements. All inhibition experiments were performed independently at least twice by using REG proteins from different purifications.
Recombinant REGβ molecules are monomers that, at low concentration, barely activate the proteasome. Active heterooligomers can form, however, when REGβ monomers are mixed with mutant REGα monomers or heptamers (ref. 14; Z.Z. et al., unpublished data). The ability of REGβ molecules to “rescue” REGα mutants allowed us to test whether active heterooligomers would form upon mixing REGβN135Y with each inactive REGα mutant. Twenty-six of the REGα mutants produced heterooligomers able to activate the proteasome (Table 1, column 3). Of the 10 REGα mutants that produced inactive heterooligomers, 5 were located between Arg-141 and Gly-149, three resulted from mutation of Pro-240, one was Asp-205 to Val, and one was Arg-181 to Cys. As mentioned, residues 141 to 149 are present in the region most highly conserved among REG homologs (Fig. 2c), and they form a loop on the presumed proteasome binding face of each REGα subunit (24). Proline 240 contacts this loop directly. An additional mutant, K232M, exhibited only a trace of activity. In the REGα crystal structure Lys-232 is a solvent-exposed residue that is adjacent to Arg-141 of the neighboring subunit. Thus, the REGα/REGβN135Y mixing experiments identify a small area on the surface of REGα subunits that is important for proteasome activation.
Conceivably the inactive heterooligomers were unable to bind the proteasome. This was tested by using the tethered proteasome assay described in Fig. 3c. Neither R181C nor D205V bound the proteasome after being mixed with REGβN135Y. In fact, they do not form heterooligomers with the mutant REGβ subunit. The other eight mutants formed complexes with REGβN135Y that associated with the proteasome as tightly as heterooligomers formed from wild-type REGα (see Table 1, column 4). Clearly then, formation of inactive REGα/REGβ complexes from REGα molecules bearing mutations in the Arg-141 to Gly-149 loop or at Pro-240 cannot be explained by their impaired association with the proteasome.
The mutants described here were characterized before determination of the REGα crystal structure. It is, therefore, satisfying that their properties can be readily interpreted in light of the structure (see Fig. 5 a and b). For example, the absence of mutants in the homolog-specific insert region correlates both with its disorder in the crystal and its position opposite the presumed proteasome binding face (24). Proteasome crystal structures (5, 6) reveal that access to the central proteolytic chamber is virtually blocked by the proteasome α subunits. The importance for proteasome activation of REGα residues Arg-141 to Gly-149 and Pro-240 can be rationalized by their proximity to each other, their location on the presumed proteasome binding face, and the assumption that they promote conformation changes that open a channel to the proteasome interior and/or activate the proteasome β subunits. The observation that the vast majority of REGα mutants are monomeric by gel filtration correlates with their locations either at subunit interfaces or in buried positions close to interfaces, where a change in packing could indirectly alter the geometry and stability of association. Formation of active oligomers from these mutants upon mixing with REGβ presumably results because stronger REGα/REGβ interactions compensate for the partially destabilizing mutation. Mutating Arg-181 to Cys or Asp-205 to Val produced the only REGα mutants unable or nearly unable to coassemble with REGβ. The severity of these changes can be explained by the observation that the Arg-181 guanidinium participates in two buried salt bridge interactions, one to Asp-205 of the same subunit, and the other to Asp-195 of a neighboring subunit (24). Therefore, it appears that for these two mutants the additional REGα/REGβ binding energy is not sufficient to stabilize an active heptamer.
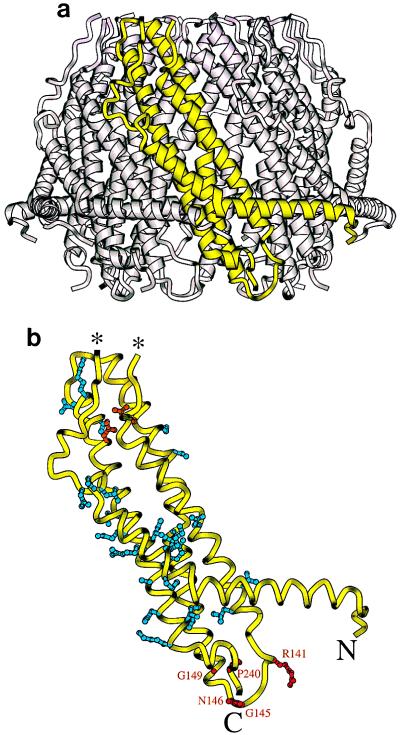
Figure 5.
Location of inactive mutations in the rREGα crystal structure. (a) One rREGα monomer is shown in yellow within the assembled heptamer. (b) The monomer backbone is shown in yellow with the side chains of mutated residues. This model stops at residue 241 because the last 8 residues are disordered in the crystal structure. Residues that block proteasome binding and/or activation are shown in red and labeled explicitly. Mutants that block heptamer formation are in blue. Also in blue are residues Ser-175 and Ala-224, which, when mutated, produce heptamers that fail to bind the proteasome; we speculate that these mutations result in a distorted heptameric conformation. The more severe “folding” mutant residues, Arg-181 and Asp-205, which form a buried salt bridge, are shown in orange. The ends of the disordered 39-residue loop are indicated with asterisks.
In summary, the mutants described here demonstrate that a loop and a neighboring proline residue on one face of REGα subunits are critical for proteasome activation. Mutation of a highly conserved asparagine in this loop to tyrosine produces variants of REGα, β, and γ that are not only inactive but also inhibit proteasome activation by their wild-type counterparts. These “dominant-negative” mutants should prove useful in studies on antigen presentation by major histocompatability complex class I molecules. Moreover, our studies clearly indicate that activation of the proteasome by REG involves an event besides association of the two protein complexes. Whether malfunction of REGαN146Y heptamers results from their failure to open a channel into the proteasome or their inability to induce conformation changes in the catalytically active proteasome β subunits remains to be determined. Finally, the demonstrated importance of REGα residues 141 to 149 and 240 offers an attractively small target for future site-directed mutagenesis aimed at elucidating the mechanism by which REG homologs activate the proteasome.
Acknowledgments
These studies were supported by grants from the National Institutes of Health, the American Cancer Society, and the Lucille P. Markey Charitable Trust. We thank Wes Sundquist, Venki Ramakrishnan, and Vicenca Ustrell for comments on the manuscript and Frank Whitby for preparation of Fig. 5.
Footnotes
A commentary on this article begins on page 2727.
References
- 1.Yewdell J W, Bennink J R. Adv Immunol. 1992;52:1–123. doi: 10.1016/s0065-2776(08)60875-5. [DOI] [PubMed] [Google Scholar]
- 2.Heemels M-H, Ploegh H. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- 3.Groettrup M, Soza A, Kuckelhorn U, Kloetzel P-M. Immunol Today. 1996;17:429–435. doi: 10.1016/0167-5699(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K, Tanahashi N, Tsurumi C, Yokota K Y, Shimbara N. Adv Immunol. 1997;64:1–38. doi: 10.1016/s0065-2776(08)60885-8. [DOI] [PubMed] [Google Scholar]
- 5.Gross M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik H D, Huber R. Nature (London) 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 6.Löwe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 7.Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Science. 1995;268:579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman L, Pratt G, Rechsteiner M. J Biol Chem. 1992;267:22362–22368. [PubMed] [Google Scholar]
- 9.Udvardy A. J Biol Chem. 1993;268:9055–9062. [PubMed] [Google Scholar]
- 10.Peters J M, Franke W W, Kleinschmidt J A. J Biol Chem. 1994;269:7709–7718. [PubMed] [Google Scholar]
- 11.Ma C-P, Vu J H, Proske R J, Slaughter C A, DeMartino G N. J Biol Chem. 1994;269:3539–3547. [PubMed] [Google Scholar]
- 12.Ma C-P, Slaughter C A, DeMartino G N. J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 13.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 14.Realini C, Jensen C, Zhang Z, Johnston S, Knowlton J R, Hill C P, Rechsteiner M. J Biol Chem. 1997;272:25483–25492. doi: 10.1074/jbc.272.41.25483. [DOI] [PubMed] [Google Scholar]
- 15.Realini C, Dubiel W, Pratt G, Ferrell K, Rechsteiner M. J Biol Chem. 1994;269:20727–20732. [PubMed] [Google Scholar]
- 16.Min W, Pober J S, Johnson D R. J Immunol. 1996;156:3174–3182. [PubMed] [Google Scholar]
- 17.Groettrup M, Soza A, Eggers M, Kuehn L, Dick T P, Schild H, Rammensee H G, Koszinowski U H, Kloetzel P-M. Nature (London) 1996;381:166–168. doi: 10.1038/381166a0. [DOI] [PubMed] [Google Scholar]
- 18.Dick T P, Ruppert T, Groettrup M, Kloetzel P-M, Kuehn L, Koszinowski U H, Stevanovic S, Schild H, Rammensee H-G. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 19.Leung D W, Chen E, Goeddel D V. Technique. 1989;1:11–15. [Google Scholar]
- 20.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–83. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 21.Harlow E, Lane D. Antibodies. Cold Spring Harbor Lab. Press; 1988. pp. 493–506. [Google Scholar]
- 22.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A, Roberts J D, Zakour R A. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 24.Knowlton R, Johnston S, Whitby F, Realini C, Zhang Z, Rechsteiner M, Hill C. Nature (London) 1997;390:639–643. doi: 10.1038/37670. [DOI] [PubMed] [Google Scholar]
- 25.Realini C, Rogers S W, Rechsteiner M. FEBS Lett. 1994;348:109–113. doi: 10.1016/0014-5793(94)00569-9. [DOI] [PubMed] [Google Scholar]