Abstract
The infrared spectroscopy of elastin-like polypeptides and the relation to the inverse thermal transition are discussed. To correlate the spectroscopic observations with structure a density function theory model was created that captures the essential hydrogen bonding and packing of the β-spiral structure proposed for elastin and elastin-like polypeptides. The infrared spectrum was calculated using periodic boundary conditions and a method for estimating the difference dipole moment permits both frequencies and intensities to be obtained for the modeling of spectra. The two observed amide I bands at 1615 cm−1 and 1656 cm−1 are shown to arise from the β-spiral structure. The increase in intensity of these bands with increasing salt concentration and temperature is assigned to the closer association of strands of the β-spiral. The sharp inverse temperature transition is observed within 1°C and involves a change in secondary structure that involves formation of interstrand β-sheets for ∼25% of the amino acids. This conclusion is consistent with available data and simulations that have been reported to date.
INTRODUCTION
Elastin-like proteins (ELPs) have been studied for many years because of their important properties of extensibility and controlled aggregation, both of which have utility in biology as well as biotechnology. ELPs exhibit an unusual inverse temperature transition that produces a reversible aggregated state of the polypeptide above the transition temperature. This transition has utility for applications such as protein purification (1–10) and drug delivery (11–15). The inverse temperature transition also known as coacervation, first observed in elastin (16,17), is associated with a collapse of extended polymer chains and eventually with segregation from solution. The nature of the transition is difficult to probe by structural methods since the aggregate is too large for NMR spectroscopy and is noncrystalline. The inverse temperature transition that leads to a decrease in the volume of ELPs produces a gel state that has been the subject of a number of theoretical studies (18–22).
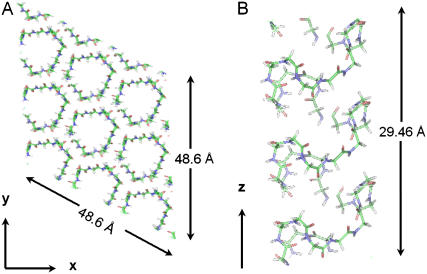
The major objective of this study is to be able to obtain structural information on the coacervation transition using attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy combined with the density function theory (DFT) calculations of the vibrational spectrum derived from the β-spiral and β-sheet models for the low temperature and high temperature forms of elastin, respectively. Application of DFT models to β-turn structures is precedented (6,23,24), however, this study focuses on the unusual lineshape of the β-spiral. The unusual shape of the amide I band of the ELP (25) and peptide models (26–28) raises an important issue of spectroscopic interpretation. There is a large literature using the amide I vibration of proteins as a marker for secondary structure components. For example, it is common to interpret 1633–1640 cm−1 as indicative of β-sheet structure and 1650–1655 cm−1 as α-helix structure among other assignments (29–38). The correlation of spectra with structure is based on protein libraries. Small peptides and large aggregates are two extremes that have important differences in hydration compared to the typical proteins used to derive secondary structure correlation. The amide I band of ELP has an unusual line shape with a low frequency component around 1615 cm−1 and a high frequency component near 1650 cm−1. The common interpretation in terms of the secondary structure components of globular proteins would be to associate these two components with an aggregated and α-helical structure, respectively (25,26,39). However, given the hypothesized β-spiral structure of elastin and ELPs (21) it does not appear reasonable to assume that the 1650 cm−1 component corresponds to an α-helical component of secondary structure (25). To understand the origin of this vibrational mode, the sequence of β-spiral in ELP was used to generate a model that could be geometry optimized using periodic boundary conditions for a vibrational frequency calculation using density functional theory (DFT) methods. The geometry optimized structure based on the repeated sequence (VPGVG)n is shown in Fig. 1, which is a 15 amino acid fragment from the structure used for molecular dynamics simulations of ELP (21). Fig. 1 shows 27 unit cells to illustrate the periodicity along the z axis and packing of ELP strands in the x,y plane. The vibrational frequencies calculated from this structure were used to generate an infrared spectrum using methods previously reported for extended β-sheets. The use of DFT methods supports the application of secondary structure interpretations in ELP and helps in understanding the structural change that is associated with coacervation in ELP.
FIGURE 1.
The calculated model β-spiral is shown. (A) To illustrate the periodicity and packing 27 unit cells are shown. (B) The geometry of a single β-spiral chain is depicted.
Two ELPs were used in this study. The first was a 61-kDa oligomeric repeat of pentapeptide VPGVG. The second was a 59-kDa oligomeric repeat of the same pentapeptide VPGVG with both natural abundance and 13C labeled protein. Studies were performed using ATR-FTIR to observe the change in spectral features as a function of both temperature and NaCl concentration. According to previous studies ELP undergoes coacervation and structural transition with increasing temperature (16,40), whereas with increasing concentration of NaCl it undergoes coacervation with little change in overall structure (this study). The ATR-FTIR method shows that the structural change involved with the collapse is a β-spiral → β-sheet transition. This assignment is supported by DFT calculations of a model for the β-spiral and β-sheet that reproduces key features in the observed infrared spectra.
MATERIALS AND METHODS
ELP expression and purification
The 59-kDa ELP consisting of 150 pentapeptides of the repetitive sequence VPGXG, where X consists of valine, alanine, and glycine in a 5:2:3 ratio was expressed from a modified pET25b vector (Novagen, Madison, WI) resulting in the fully expressed amino acid sequence (M)SKGPG-(VPGXG)150-WP. The 61-kDa ELP consisting of 160 pentapeptides of the repetitive sequence VPGXG, where X consists of valine, alanine, and glycine in a 1:8:7 ratio was expressed from a modified pET25b vector (Novagen) resulting in the fully expressed amino acid sequence (M)SKGPG-(VPGXG)160-WP. The synthesis of this gene has been described previously (7,41).
13C labeling of ELP
The 13C labeling of ELP1-150 was performed as described elsewhere (42). Briefly, Escherichia coli (BLR DE3), transformed with a plasmid encoding an ∼59-kDa ELP[V5A2G3-150] gene (41) (also known as ELP1-150; (43)), were grown in 250-ml Erlenmeyer flasks (50-ml medium) at 37°C, aerated by orbital shaking at 250 rpm. The bacterial cell density was monitored by the medium's optical density at 600 nm. A nutrient rich medium, CircleGrow (CG) (Qbiogene, Carlsbad, CA) was used in the adaptation of E. coli for growth in M9 minimal medium. The fully adapted E. coli to M9 medium were inoculated into 50 ml of modified M9 medium consisted of 6 g/L Na2HPO4, 3 g/L KH2PO4, 1 g/L NH4Cl, 0.5 g/L NaCl, 4 g/L glucose, and 100 mg/L ampicillin, and cultured overnight to serve as a starter culture. Five milliliters of the overnight culture were centrifuged and resuspended in 5 ml of modified M9 medium, then inoculated into 1 L of modified M9 medium supplemented with 0.4% (w/v) [U-13C]-D-glucose (Cambridge Isotope Laboratories, Andover, MA) as only carbon source to completely substitute for glucose in the medium and cultured until an OD600 of 2.0 was reached (∼6 h). At this time, IPTG was added to the culture at a final concentration of 0.5 mM. The cells were harvested 2 h later, and the ELP was purified by ITC. The purity of ELP was determined by SDS- PAGE analysis.
ELP characterization
The thermal properties of [13C]ELP were evaluated by measuring the OD350 from an upward thermal ramp from 25 to 50°C on a Cary Bio300 ultraviolet-visible spectrophotometer (Varian Instruments, Walnut Creek, CA). The Tt was defined as the maximum in dOD/dT. These experiments were performed with an ELP concentration of 25 μM in either phosphate buffer saline (PBS) or mock serum consisting of PBS supplemented with 1 mM bovine serum albumin. The molecular weight of [13C]ELP was measured by matrix-assisted laser desorption/ ionization mass spectrometry (MALDI-MS) using a PE Biosystems Voyager-DE spectrometer equipped with a nitrogen laser (337 nm). The MALDI-MS samples were prepared in 50:50 (v/v) water/acetonitrile solution containing 0.1% trifluoroacetic acid using sinapinic acid as a matrix.
ATR-FTIR spectroscopy
Spectra were collected on a Digilab FTS 6000 FTIR spectrometer, equipped with a Pike Technologies (Madison, WI) ATR attachment and with a liquid nitrogen cooled mercury cadmium telluride-detector, operating in both microscope and bench mode. Ge crystal was used for collection of spectra of ELP protein solution. In a bench mode operation 10 μl sample was pipetted into Teflon well placed on top of Ge crystal, and spectra were collected from 4000 to 400 cm−1, at 2 cm−1 resolution. Sixty-four scans were accumulated per spectrum. To eliminate contributions due to atmospheric water the instrument was continually purged with dry air. Spectra were collected at room temperature and protein concentration was 1 mM in solution containing 10 mM Na phosphate at pH 7.2. All spectra were acquired using the software package Win-IR-Pro v2.97 (Varian, Randolph, MA). The protein spectra were corrected for buffer and NaCl contributions by subtracting the two spectra acquired under identical experimental conditions. The spectra were processed using the software packages Origin 7.5 (Microcal, Northhampton, MA) and IgorPro 5.0 (Wavemetrics, Lake Oswego, OR), whereby the positions of the peaks were assigned to secondary structural elements by second derivative analysis of amide I band, and the structure content was determined by band deconvolution using Gaussian band fitting profile (2,10).
The ELP coacervation temperature experiments were performed by varying temperature at a rate of 1°C/min, with AutoPro temperature controller (Pike Technologies) equipped bench mode Ge crystal setup, and collecting FT-IR spectra after each temperature change. FT-IR spectra were collected at 1–2°C temperature intervals. Coacervation induced with NaCl was determined by adding increasing concentrations of NaCl to the protein solution, and monitoring the NaCl effect on the protein by acquiring the FT-IR spectra at each NaCl concentration. These spectra were collected at room temperature.
Computational methods
A β-spiral structure in periodic boundary conditions consisting of three repeats of the VPGVG sequence and comprising 183 atoms per unit cell was constructed based on the coordinates that were used for molecular dynamics simulations (21). The unit cell was a hexagonal cell with dimensions a = 16.2 Å, b = 16.2 Å, c = 9.82 Å. The hexagonal cell provides close packing of the cylindrical β-spiral structure as shown in Fig. 1. Fig. 1 shows 27 unit cells to illustrate the periodicity of the structure.
The optimized ground state geometries were obtained using both the generalized gradient approximation of Perdew and Wang (44) as implemented in DMol3 (45,46) (Accelrys). All calculations were carried out on the cluster at the High Performance Computing center at North Carolina State University. A numerically tabulated basis set of double-ζ plus quality was employed. The geometry optimizations were carried out until the energy difference was <10−6 a.u. on subsequent iterations. Following geometry optimization the Hessian matrix was constructed by finite difference. The isotopic labeling was calculated by manual substitution of mass 13 carbon atoms into the inverse mass matrix followed by rediagonalization of the mass-weighted Hessian matrix. Analogous methods have been employed to study other secondary structures by DFT methods (21,47). The intensities of the vibrations cannot be calculated directly in periodic boundary conditions calculations. The intensities were estimated by calculation of the difference dipole moments as described previously (48). The infrared frequencies and intensities were used to create model spectra using a 20 cm−1 Gaussian broadening to provide a realistic spectral shape.
RESULTS AND DISCUSSION
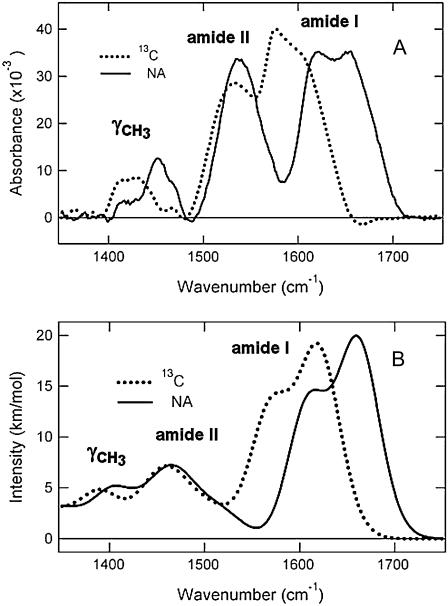
We investigated structural transitions of an elastin-like-polypeptide, by ATR-FTIR spectroscopy using natural abundance as well as 13C isotopically labeled proteins. A DFT calculation of the vibrational spectrum was used to aid in the interpretation of the data. The proposed β-spiral structure for ELP (49) is not common to other proteins that have been studied by FTIR spectroscopy (29–38), thus comparisons with known structural libraries of FTIR spectra do not allow for unequivocal assignment of structural content. The frequencies were calculated for β-spiral structure in periodic boundary conditions shown in Fig. 1. The calculated frequencies for the natural abundance and the 13C isotopically labeled sample were calculated and compared to experiment in the region of the FTIR spectrum (1450–1750 cm−1) relevant to the data obtained on ELP proteins (Table 1). The experimental and calculated spectra are compared in Fig. 2. The calculated β-spiral amide I band (Fig. 2) consists of two major components at 1609 cm−1 and 1662 cm−1, which can be compared to the experimental amide I bands at 1615 cm−1 and 1656 cm−1. The relative errors in the wavenumbers are <0.4% for both components. The relative intensities of the two components are nearly the same in the experimental spectra, whereas the 1609 cm−1 (calculated) component is 73% as large as the higher wavenumber 1656 cm−1 component. The differences between calculated and observed spectra are due to differences in hydration and anharmonicity, neither of which is included in the theoretical calculations of the β-spiral spectra. The calculated 13C isotope shifts are −38 cm−1 and −40 cm−1 lower energy for the respective amide I bands. Again these values compare well to the experimentally measured 13C isotope shifts of −39 cm−1 and −45 cm−1 in the 13C-labeled 59 kDa ELP protein (data not shown). The fit parameters for the β-spiral components of amide I band shown in Fig. 2 are reported in Table 1. The absolute position of the amide II band is 1486 cm−1 in the calculated spectrum compared to 1538 cm−1 in the experimental spectrum. The discrepancy is 4.8%, which is much larger than that observed for the amide I band. The absence of waters of solvation and anharmonic coupling are three factors that likely account for the difference between DFT calculation and experiment. These factors are larger for amide II due to the large contribution of the C-N-H in-plane bending in that mode. On the other hand, both the calculation and the experiment show a much smaller shift of −7 cm−1 for the amide II band. The calculated methyl deformation mode (γCH3) at 1406 cm−1 shifts down by −19 cm−1 and the experimental γCH3 mode at 1406 cm−1 shifts down by −23 cm−1. The computational model is remarkably consistent with the assignment of the double-peaked spectrum with bands at 1618 cm−1 and 1654 cm−1 as a signature of the β-spiral. However, these components would have a different interpretation in the standard approach of fitting of the bands to multiple components. In the following, the conformational changes are considered using the fitting to multiple bands to obtain a comparison between the method developed here using DFT and more traditional methods using fitting to multiple (usually five) bands.
TABLE 1.
Gaussian parameters for the calculated and fitted experimental amide I bands of 59-kDa ELP for both natural abundance (NA) and 13C isotopically labeled proteins
| Experimental
|
Calculated
|
|||
|---|---|---|---|---|
| Parameter | NA | 13C | NA | 13C |
| ω1 (cm−1) | 1615 | 1576 | 1609 | 1571 |
| σ1 (cm−1) | 15.1 | 15.1 | 21.3 | 25.7 |
| A1 | 0.34 | 0.45 | 0.39 | 0.48 |
| ω2 (cm−1) | 1656 | 1611 | 1662 | 1623 |
| σ2 (cm−1) | 22.8 | 18.9 | 22.2 | 21.1 |
| A2 | 0.66 | 0.55 | 0.61 | 0.52 |
Two Gaussians were used in the band fitting.
FIGURE 2.
Comparison of experimental and calculated infrared spectra for NA and 13C-labeled ELP. (A) The experimental infrared spectra obtained from the 59-kDa ELP protein are shown. (B) The calculated infrared spectra are presented. The solid line represents the NA data and calculation. The dotted line represents the 13C isotopomer.
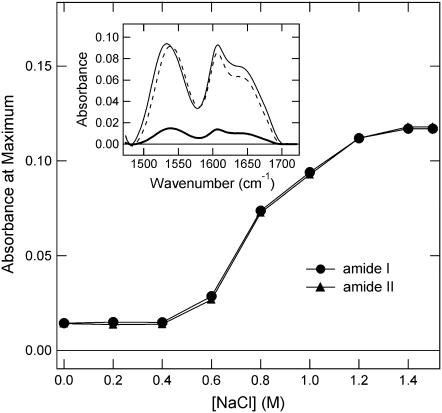
The effect of NaCl concentration on the FTIR spectra of the 61-kDa ELP is shown in Fig. 3. The 61-kDa ELP was titrated with increasing amounts of NaCl near the surface of an ATR internal reflecting element to characterize the effect of NaCl on the structure. The spectra were acquired at room temperature and in buffer containing 10 mM sodium phosphate at pH 7.2. Fig. 3 shows the change in absorbance intensity of amide I band (absorbance maxima at 1615 cm−1 and 1656 cm−1) and amide II band (maximum absorbance at 1540 cm−1) with increasing concentrations of NaCl. The inset of Fig. 3 represents the amide I and II region of FTIR spectrum of the 61-kDa ELP. Amide I arises from C=O stretching vibrations of peptide bond and is the most studied infrared band of proteins for the estimation of the secondary structure. The percentage of secondary structure for proteins has been estimated from the relative area of deconvolved bands of this spectral region. However, the correlation of spectra with structure is based on a set of folded proteins and these correlations are not necessarily applicable to peptides or protein aggregates. In all of the correlations of amide I bands in proteins it is not common to find a wavenumber as low as 1615 cm−1 observed in the lowest wavenumber band of the ELP, although there are reports that aggregated β-sheets produce such low frequencies of amide I (50).
FIGURE 3.
Change of absorbance at the maximum of amide I (circles) and amide II (triangles) region of ATR-FTIR spectra of the 61-kDa ELP (1 mM in 10 mM phosphate buffer, pH 7.2) as a function of NaCl concentration. (Inset) Amide I and amide II frequency region of the FTIR spectra of the 61-kDa ELP at 0.4, 0.4 trace multiplied by 8 for clarity of comparison, and 1.4 M NaCl (darker, dashed, and lighter lines, respectfully).
It is evident from Fig. 3 that both amide I and II spectral regions increase in intensity by the same extent as the NaCl concentration is increased with little change in structure. The intensity increases by 10-fold as the NaCl concentration is increased to 1.5 M. The increase in sample concentration is caused by aggregation of the chains without an apparent change in secondary structure. This causes the sample to be in closer contact with the Ge internal reflecting element. The higher protein concentration implies that water is excluded and that the β-spiral packing density increases. The data suggest that the β-spiral strands are forming an aggregate without changing their internal hydrogen-bonding pattern. In other words, the data appear to be consistent with formation of more closely packed β-helices without formation of interstrand rearrangements such as the formation of β-sheet aggregates, since we observed only increase in the absorbance, and no peak shifts as NaCl concentration was increased.
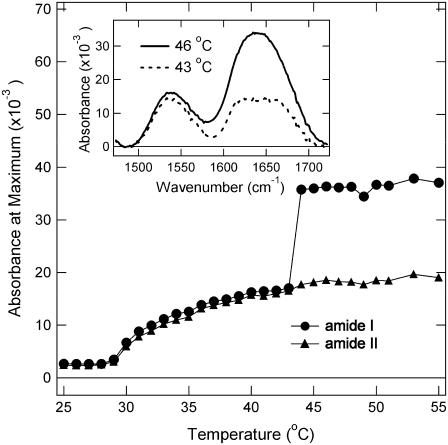
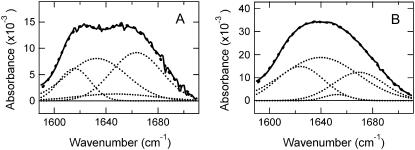
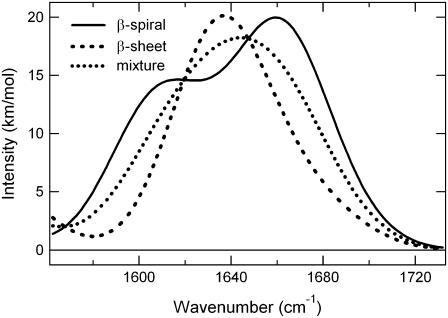
The temperature effect on the 61-kDa ELP is shown in Fig. 4 as a function of the intensity of the amide I and II infrared bands. The inset in Fig. 4 shows the absorbance of the amide I and II region at temperatures below (43°C) and above (46°C) the inverse transition temperature. From Fig. 4 it is evident that the amide I and II bands both increase with temperature to the same extent up to 43°C. Between 43°C and 44°C there is a large change in the shape and intensity of the amide I band with almost no change in amide II. The amide I band of the 61-kDa ELP both above and below the thermal transition temperature was fit to four Gaussian components (determination aided by the second derivative analysis; see Materials and Methods section), shown in Fig. 5. Fitting to components is only an approximation to the chemical information available in the lineshape. The novel method developed here explains the features observed at 1615 cm−1 and 1656 cm−1 without resorting to a spectral library. The components reported in Table 2 show that there is a significant increase at 1624 cm−1 indicative of an increase β-sheet structure as the temperature is raised. This aspect of the structural change in ELP is well represented by the traditional band fitting to components shown in Table 2. We show in the following that the DFT calculations complement the band fitting analysis (Fig. 6). The β-sheet component in Fig. 6 was reported previously (48). The model in Franzen (48) uses periodic boundary conditions to determine the frequencies for an antiparallel β-sheet using the same level of DFT used in this work. The comparison in Fig. 6 using DFT methods is consistent with traditional assignments of amide I in support of a β-spiral → β-sheet transition as a major secondary structural change that accompanies the sharp transition of ELP at 43°C. The spectrum observed above the thermal transition is quite similar to a linear combination of 50% β-spiral and 50% β-sheet. The calculation suggests that ELP undergoes a conformational transition in which part of the β-spiral structure melts to form intrastrand β-sheet contacts.
FIGURE 4.
Temperature-induced structural changes in ELP. Absorbance change at the maximum of amide I (circles) and amide II (triangles) region of ATR-FTIR spectra of the 61-kDa ELP is monitored as a function of temperature. Experimental conditions as in Fig. 1. (Inset) Amide I and amide II section of the FTIR spectra at two selected temperatures, 43°C and 46°C, immediately below and above the coacervation temperature.
FIGURE 5.
Fits to amide I band of the 61-kDa ELP (lighter solid lines) at (A) 43°C and (B) 46°C. Experimental conditions are those in Fig. 4. Band positions and percentage area are reported in Table 2.
TABLE 2.
Peak positions and relative integrated intensities of deconvolved amide I peak components at 30°C (I) and 50°C (II) for 1 mM 61-kDa ELP in 10 mM Na phosphate buffer, pH 7.2
| I
|
II
|
|||
|---|---|---|---|---|
| Assignment | position (cm−1) | area (%) | position (cm−1) | area (%) |
| β-sheet aggregation | 1616 | 15.5 | – | – |
| β-sheet | – | – | 1624 | 25.1 |
| β-sheet/β-turn | 1633 | 38.8 | 1640 | 50.3 |
| α-helix | 1647 | 8.3 | 1653 | 2.5 |
| β-turn | 1665 | 37.0 | 1669 | 22.1 |
FIGURE 6.
Comparison of the calculated β-sheet and β-spiral models. The mixture is a linear combination of 50% β-spiral and 50% β-sheet.
Two observations that stem from this study are: i), NaCl exerts similar effect on both amide I and amide II spectral bands, whereas temperature changes affect primarily band I; and ii), although both increasing concentration of NaCl and increasing temperature lead to coacervation of ELP, they do not affect the posttransition structure of this polypeptide in the same fashion. One possible explanation of these differences is that, perhaps, NaCl concentration promotes aggregation, but does not provide sufficient excess energy to allow formation of new hydrogen bonds in a β-sheet structure.
CONCLUSION
A spectral analysis based on first principles calculation of the infrared spectra DFT calculations with periodic boundary conditions is consistent with the proposed β-spiral structure for ELP for both natural abundance and 13C isotopomer samples. The signature of the β-spiral spectrum consists of two bands at 1615 cm−1 and 1656 cm−1. The assignment of the double-band line shape of the β-spiral represents a new basis spectrum for protein structure-spectroscopy correlation by FTIR spectroscopy. This view supercedes a more traditional view of the secondary structure correlation presented in Table 2.
The effects of NaCl and temperature on the structural transitions of ELP have been investigated using FTIR spectroscopy. We have determined that a high concentration of NaCl causes no major shifts in positions of the FT-IR absorbance bands, and no significant changes in the relative percent area under these bands, indicating that high NaCl concentration does not induce changes in secondary structure, but does affect the packing of β-spiral strands. On the other hand, the effect of increased temperature involves both the change in strand packing seen at high salt concentration and an additional sharp transition that is associated with a change in secondary structure. This change is highly cooperative, judging by sharp increase in absorbance of amide I spectral band that occurs within a 1°C temperature span. Comparison of tradition spectral analysis and DFT calculations using periodic boundary conditions both suggest that the transition involves a decrease β-spiral structure in favor of a β-sheet structure.
Acknowledgments
The authors thank Ashutosh Chilkoti for helpful discussions.
Editor: Brian R. Dyer.
References
- 1.Frey, W., D. E. Meyer, and A. Chilkoti. 2003. Thermodynamically reversible addressing of a stimuli responsive fusion protein onto a patterned surface template. Langmuir. 19:1641–1653. [Google Scholar]
- 2.Frey, W., D. E. Meyer, and A. Chilkoti. 2003. Dynamic addressing of a surface pattern by a stimuli-responsive fusion protein. Adv. Mater. 15:248–251. [Google Scholar]
- 3.Wang, Y. L., P. S. Belton, H. Bridon, E. Garanger, N. Wellner, M. L. Parker, A. Grant, P. Feillet, and T. R. Noel. 2001. Physicochemical studies of caroubin: a gluten-like protein. J. Agric. Food Chem. 49:3414–3419. [DOI] [PubMed] [Google Scholar]
- 4.Hyun, J., W. K. Lee, N. Nath, A. Chilkoti, and S. Zauscher. 2004. Capture and release of proteins on the nanoscale by stimuli-responsive elastin-like polypeptide “switches”. J. Am. Chem. Soc. 126:7330–7335. [DOI] [PubMed] [Google Scholar]
- 5.Trabbic-Carlson, K., D. E. Meyer, L. Liu, R. Piervincenzi, N. Nath, T. LaBean, and A. Chilkoti. 2004. Effect of protein fusion on the transition temperature of an environmentally responsive elastin-like polypeptide: a role for surface hydrophobicity? Protein Eng. Des. Sel. 17:57–66. [DOI] [PubMed] [Google Scholar]
- 6.Kim, J., R. Huang, J. Kubelka, P. Bour, and T. A. Keiderling. 2006. Simulation of infrared spectra for beta-hairpin peptides stabilized by an Aib-Gly turn sequence: correlation between conformational fluctuation and vibrational coupling. J. Phys. Chem. B. 110:23590–23602. [DOI] [PubMed] [Google Scholar]
- 7.Meyer, D. E., and A. Chilkoti. 1999. Purification of recombinant proteins by fusion with thermally responsive polypeptides. Nat. Biotechnol. 17:1112–1115. [DOI] [PubMed] [Google Scholar]
- 8.Meyer, D. E., K. Trabbic-Carlson, and A. Chilkoti. 2001. Protein purification by fusion with an environmentally responsive elastin-like polypeptide: effect of polypeptide length on the purification of thioredoxin. Biotechnol. Prog. 17:720–728. [DOI] [PubMed] [Google Scholar]
- 9.Shimazu, M., A. Mulchandani, and W. Chen. 2003. Thermally triggered purification and immobilization of elastin-OPH fusions. Biotechnol. Bioeng. 81:74–79. [DOI] [PubMed] [Google Scholar]
- 10.Sun, X. L., C. A. Haller, X. Y. Wu, V. P. Conticello, and E. L. Chaikof. 2005. One-pot glyco-affinity precipitation purification for enhanced proteomics: the flexible alignment of solution-phase capture/release and solid-phase separation. J. Proteome Res. 4:2355–2359. [DOI] [PubMed] [Google Scholar]
- 11.Chilkoti, A., M. R. Dreher, and D. E. Meyer. 2002. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv. Drug. Deliv. Rev. 54:1093–1111. [DOI] [PubMed] [Google Scholar]
- 12.Chilkoti, A., M. R. Dreher, D. E. Meyer, and D. Raucher. 2002. Targeted drug delivery by thermally responsive polymers. Adv. Drug. Deliv. Rev. 54:613–630. [DOI] [PubMed] [Google Scholar]
- 13.Dreher, M. R., M. Elas, K. Ichikawa, E. D. Barth, A. Chilkoti, G. M. Rosen, H. J. Halpern, and M. Dewhirst. 2004. Nitroxide conjugate of a thermally responsive elastin-like polypeptide for noninvasive thermometry. Med. Phys. 31:2755–2762. [DOI] [PubMed] [Google Scholar]
- 14.Dreher, M. R., D. Raucher, N. Balu, O. M. Colvin, S. M. Ludeman, and A. Chilkoti. 2003. Evaluation of an elastin-like polypeptide-doxorubicin conjugate for cancer therapy. J. Control. Release. 91:31–43. [DOI] [PubMed] [Google Scholar]
- 15.Furgeson, D. Y., M. R. Dreher, and A. Chilkoti. 2006. Structural optimization of a “smart” doxorubicin-polypeptide conjugate for thermally targeted delivery to solid tumors. J. Control. Release. 110:362–369. [DOI] [PubMed] [Google Scholar]
- 16.Long, M. M., D. W. Urry, B. A. Cox, T. Ohnishi, and M. Jacobs. 1975. Coacervation of repeat sequences of elastin. Biophys. J. 15:A71 (Abstr.). [Google Scholar]
- 17.Urry, D.W. 1976. Molecular mechanisms of elastin coacervation and coacervate calcification. Faraday Discuss. Chem. Soc. 61:205–212. [DOI] [PubMed] [Google Scholar]
- 18.Hoeve, C. A. J., and P. J. Flory. 1974. Elastic properties of elastin. Biopolymers. 13:677–686. [DOI] [PubMed] [Google Scholar]
- 19.Gosline, J. M., and C. J. French. 1979. Dynamic mechanical-properties of elastin. Biopolymers. 18:2091–2103. [DOI] [PubMed] [Google Scholar]
- 20.Chang, D. K., and D. W. Urry. 1988. Molecular-dynamics calculations on relaxed and extended states of the polypentapeptide of elastin. Chem. Phys. Lett. 147:395–400. [Google Scholar]
- 21.Kubelka, J., and T. A. Keiderling. 2001. The anomalous infrared amide I intensity distribution in C-13 isotopically labeled peptide beta-sheets comes from extended, multiple-stranded structures. An ab initio study. J. Am. Chem. Soc. 123:6142–6150. (Erratum in J. Am. Chem. Soc. 123:8163). [DOI] [PubMed] [Google Scholar]
- 22.Bellingham, C. M., M. A. Lillie, J. M. Gosline, G. M. Wright, B. C. Starcher, A. J. Bailey, K. A. Woodhouse, and F. W. Keeley. 2003. Recombinant human elastin polypeptides self-assemble into biomaterials with elastin-like properties. Biopolymers. 70:445–455. [DOI] [PubMed] [Google Scholar]
- 23.Setnicka, V., R. Huang, C. L. Thomas, M. A. Etienne, J. Kubelka, R. P. Hammer, and T. A. Keiderling. 2005. IR study of cross-strand coupling in a beta-hairpin peptide using isotopic labels. J. Am. Chem. Soc. 127:4992–4993. [DOI] [PubMed] [Google Scholar]
- 24.Hilario, J., J. Kubelka, and T. A. Keiderling. 2003. Optical spectroscopic investigations of model beta-sheet hairpins in aqueous solution. J. Am. Chem. Soc. 125:7562–7574. [DOI] [PubMed] [Google Scholar]
- 25.Lin, S. Y., T. F. Hsieh, and Y. S. Wei. 2005. PH- and thermal-dependent conformational transition of PGAIPG, a repeated hexapeptide sequence from tropoelastin. Peptides. 26:543–549. [DOI] [PubMed] [Google Scholar]
- 26.Nicolini, C., R. Ravindra, B. Ludolph, and R. Winter. 2004. Characterization of the temperature- and pressure-induced inverse and reentrant transition of the minimum elastin-like polypeptide GVG(VPGVG) by DSC, PPC, CD, and FT-IR spectroscopy. Biophys. J. 86:1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petty, S. A., and S. M. Decatur. 2005. Experimental evidence for the reorganization of beta-strands within aggregates of the a beta(16–22) peptide. J. Am. Chem. Soc. 127:13488–13489. [DOI] [PubMed] [Google Scholar]
- 28.Starzyk, A., W. Barber-Armstrong, M. Sridharan, and S. M. Decatur. 2005. Spectroscopic evidence for backbone desolvation of helical peptides by 2,2,2-trifluoroethanol: an isotope-edited FTIR study. Biochemistry. 44:369–376. [DOI] [PubMed] [Google Scholar]
- 29.Krimm, S., and Y. Abe. 1972. Intermolecular interaction effects in the amide I vibrations of β polypeptides. Proc. Natl. Acad. Sci. USA. 69:2788–2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wi, S., P. Pancoska, and T. A. Keiderling. 1998. Predictions of protein secondary structures using factor analysis on Fourier transform infrared spectra: effect of Fourier self-deconvolution of the amide I and amide II bands. Biospectroscopy. 4:93–106. [DOI] [PubMed] [Google Scholar]
- 31.Douseeau, F., and M. Pezolet. 1990. Determination of the secondary structure content of proteins in aqueous solutions from their amide I and amide II infrared bands. Comparison between classical and partial least-squares methods. Biochemistry. 29:8771–8779. [DOI] [PubMed] [Google Scholar]
- 32.Byler, D., and H. Susi. 1986. Examination of the secondary structure of proteins by deconvolved FTIR spectra. Biopolymers. 25:469–487. [DOI] [PubMed] [Google Scholar]
- 33.Levitt, M. 1977. Automatic identification of secondary structure in globular proteins. J. Mol. Biol. 114:181–239. [DOI] [PubMed] [Google Scholar]
- 34.Pancoska, P., E. Bitto, V. Janota, M. Urbanova, V. Gupta, and T. Keiderling. 1995. Comparison of and limits of accuracy for statistical analyses of vibrational and electronic circular dichroism spectra in terms of correlations to and predictions of protein secondary structure. Protein Sci. 4:1384–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pribic, R. 1994. Principal component analysis of Fourier transform infrared and/or circular dichroism spectra of proteins applied in a calibration of protein secondary structure. Anal. Biochem. 223:26–34. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, C. T., and R. Zhang. 2000. A graphic approach to evaluate algorithms of secondary structure prediction. J. Biomol. Struct. Dyn. 17:829–842. [DOI] [PubMed] [Google Scholar]
- 37.Smith, B. M., and P. J. Gemperline. 2000. Wavelength selection and optimization of pattern recognition methods using the genetic algorithm. Anal. Chim. Acta. 423:167–177. [Google Scholar]
- 38.Smith, B. M., and S. Franzen. 2002. Single-pass attenuated total reflection Fourier transform infrared spectroscopy for the analysis of proteins in H2O solution. Anal. Chem. 74:4076–4080. [DOI] [PubMed] [Google Scholar]
- 39.Debelle, L., A. J. P. Alix, S. M. Wei, M. P. Jacob, J. P. Huvenne, M. Berjot, and P. Legrand. 1998. The secondary structure and architecture of human elastin. Eur. J. Biochem. 258:533–539. [DOI] [PubMed] [Google Scholar]
- 40.Cox, B. A., B. C. Starcher, and D. W. Urry. 1973. Coacervation of alpha-elastin results in fiber formation. Biochim. Biophys. Acta. 317:209–213. [DOI] [PubMed] [Google Scholar]
- 41.Meyer, D. E., and A. Chilkoti. 2002. Genetically encoded synthesis of protein-based polymers with precisely specified molecular weight and sequence by recursive directional ligation: examples from the elastin-like polypeptide system. Biomacromolecules. 3:357–367. [DOI] [PubMed] [Google Scholar]
- 42.Liu, W., M. Dreher, and A. Chilkoti. 2006. Tracking the invivo fate of recombinant polypeptides by isotopic labeling. J. Controlled Release. 114:184–192. [DOI] [PubMed] [Google Scholar]
- 43.Meyer, D. E., G. A. Kong, M. W. Dewhirst, M. R. Zalutsky, and A. Chilkoti. 2001. Targeting a genetically engineered elastin-like polypeptide to solid tumors by local hyperthermia. Cancer Res. 61:1548–1554. [PubMed] [Google Scholar]
- 44.Perdew, J. P., J. A. Chevary, S. H. Vosko, K. A. Jackson, M. R. Pederson, D. J. Singh, and C. Fiolhais. 1992. Atoms, molecules, solids and surfaces: applications of the generalized gradient approximation for exchange and correlation. Phys. Rev. B. 46:6671–6687. [DOI] [PubMed] [Google Scholar]
- 45.Delley, B. 1990. An all-electron numerical method for solving the local density functional for polyatomic-molecules. J. Chem. Phys. 92:508–517. [Google Scholar]
- 46.Delley, B. 2000. From molecules to solids with the DMol(3) approach. J. Chem. Phys. 113:7756–7764. [Google Scholar]
- 47.Silva, R., S. C. Yasui, J. Kubelka, F. Formaggio, M. Crisma, C. Toniolo, and T. A. Keiderling. 2002. Discriminating 3(10)- from alpha helices: vibrational and electronic CD and IR absorption study of related AIB-containing oligopeptides. Biopolymers. 65:229–243. [DOI] [PubMed] [Google Scholar]
- 48.Franzen, S. 2003. Use of periodic boundary conditions to calculate accurate beta-sheet frequencies using density functional theory. J. Phys. Chem. A. 107:9898–9902. [Google Scholar]
- 49.Venkatachalam, C. M., and D. W. Urry. 1981. Development of a linear helical conformation from its cyclic correlate: beta-spiral model of the elastin poly(pentapeptide) (Vpgvg)N. Macromolecules. 14:1225–1229. [Google Scholar]
- 50.Fandrich, M., and C. M. Dobson. 2002. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 21:5682–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]