Abstract
Hepatocyte growth factor (HGF) activates the Met receptor tyrosine kinase by binding and promoting receptor dimerization. Here we describe a mechanistic basis for designing Met antagonists based on NK1, a natural variant of HGF containing the N-terminal and the first kringle domain. Through detailed biochemical and structural analyses, we demonstrate that both mouse and human NK1 induce Met dimerization via a conserved NK1 dimer interface. Mutations designed to alter the NK1 dimer interface abolish its ability to promote Met dimerization but retain full Met-binding activity. Importantly, these NK1 mutants act as Met antagonists by inhibiting HGF-mediated cell scattering, proliferation, branching, and invasion. The ability to separate the Met-binding activity of NK1 from its Met dimerization activity thus provides a rational basis for designing Met antagonists. This strategy of antagonist design may be applicable for other growth factor receptors by selectively abolishing the receptor activation ability but not the receptor binding of the growth factors.
Keywords: cancer therapy, heparin, hepatocyte growth factor, Met antagonist, tumor metastasis
Hepatocyte growth factor (HGF) (also known as scatter factor) and its receptor, Met, a receptor tyrosine kinase (RTK), mediate a network of signaling pathways that control cell proliferation, survival, and motility (1). Proper signaling of HGF–Met is essential for normal embryonic development and organ formation (2) but aberrant activation of HGF–Met signaling is closely associated with tumor growth, invasion, and metastasis. Hyperactivation of Met is found in >50% of solid tumors and is correlated with poor prognosis (www.vai.org/met). Thus, anticancer therapy based on Met antagonists has emerged as a prominent and rational goal of pharmaceutical discovery.
HGF contains two polypeptide chains linked by a disulfide bond (3, 4). The N-terminal α chain comprises a heparin-binding domain (N) followed by a repeat of four kringle domains (K1–K4). The C-terminal β chain is a serine protease-homology (SPH) domain devoid of protease activity (Fig. 1A). NK1 is a fragment containing only the N-terminal heparin-binding domain and the first kringle domain (K1) that occurs naturally through alternative splicing of the primary HGF transcript (5). NK1 binds Met and is described either as a receptor antagonist or agonist in vitro depending on the context of assay formats and cell types (6). In vivo studies in transgenic mice, however, have clearly established that NK1 is a potent Met activator (7), and other studies have clarified that the agonist activity of NK1 depends on the presence of glycosaminoglycans such as heparan sulfate (8, 9). Whereas the detailed interactions between HGF and Met remain poorly characterized, mutagenesis data have pointed out that the fragment corresponding to NK1 is responsible for the high-affinity binding of HGF to Met (6).
Fig. 1.
Binding of the human and mouse NK1 to Met. (A) A schematic representation of the domain arrangement of HGF and Met. (B) The purified proteins used in this study are depicted. Proteins shown are the mouse and human NK1 (lanes 1 and 2), the human NK1 with R134G mutation (lane 3), biotinylated human NK1 (lane 4), and the Met sema domain (lane 5). The asterisk in lane 2 indicates the truncated N domain of human NK1. The dashed line indicates that lanes 4 and 5 were run in a separate gel. (C) A diagram of AlphaScreen assay for detecting NK1-Met interactions. (D) Binding of biotinylated NK1 to Met in the absence or presence of heparin as measured by AlphaScreen assay is depicted. (E) Binding affinity (IC50 values) of various NK1s to Met as determined by the inhibition of the binding of the biotinylated NK1 to Met with dose competition of unlabeled NK1s is shown.
Met, the receptor for HGF, was originally identified as an oncogene produced by chromosomal rearrangement (10). The 170-kDa Met receptor contains a large extracellular domain (ectodomain), a transmembrane domain, and an intracellular tyrosine kinase domain (Fig. 1A). The mature Met ectodomain (residues 25–932) is cleaved into two chains between residues 307 and 308 by furin protease (11). The first 514 amino acids of Met form the so-called sema domain, a seven-bladed β-propeller structure tightly linked with a cysteine-rich domain (amino acids 519–561) that are shared among the semaphorin and plexin protein families (12, 13). The sema domain contains the major binding site for HGF and is required for HGF-induced Met dimerization (12, 14). Following the sema domain is a tandem repeat of four Ig-like domains.
Met is thought to be activated by HGF through ligand-induced receptor dimerization, because HGF and the Met sema domain can form a complex with 2:2 stoichiometry (15). Furthermore, Met on the cell surface is readily cross-linked into dimers or higher-order oligomers upon treatment with HGF (14–16). However, the role of Met dimerization in its activation remains unclear, because the isolated full-length Met ectodomain in complex with HGF is predominately monomeric (12). In this study, we demonstrate that HGF NK1 activates Met by inducing Met dimerization through a conserved NK1 dimer interface. Mutations in this dimer interface specifically retain the NK1 Met binding activity but abolish its ability to promote Met dimerization and activation. The ability to separate Met binding of NK1 from its Met activation function thus provides a mechanistic basis for designing Met antagonists and has conceptual implications for antagonist design of other growth-factor-activated tyrosine kinase receptors.
Results
Direct Binding of the Human and Mouse NK1 to the Met Extracellular Domain.
To understand the mechanisms of Met binding and activation by NK1, we purified mouse and human NK1, the human NK1 mutant R134G, and the Met ectodomain, which contains the sema- and cystine-rich domains (Fig. 1B). The purified Met ectodomain (residues 25–567 with a C-terminal histidine tag) consists of a 35-kDa α-chain (residues 25–307) and a 32-kDa β chain (residues 308–567) (12). Both mouse and human NK1 run as 21-kDa bands, but the human NK1 expressed in Escherichia coli was consistently contaminated with an N-domain truncation product (Fig. 1B, lane 2) because of the presence of arginine 134 (R134) in the human NK1 that is sensitive to cleavage by proteases (17). Replacement of R134 with a glycine, the residue present in mouse NK1, removed the protease site (Fig. 1B, lane 3) and improved the yield of the full-length NK1 product. A biotinylated version of the human NK1 was also expressed in bacteria and purified for Met binding assays (Fig. 1B, lane 4).
To determine the functional activity of the above purified proteins, we measured the direct interactions of Met with the biotinylated NK1 by using AlphaScreen assays as illustrated in Fig. 1C. In this assay, streptavidin donor beads and nickel-chelated acceptor beads were attached to biotinylated NK1 and histidine-tagged Met, respectively. When NK1 interacts with Met, excitation of a laser beam at 680 nm causes the donor beads to emit singlet oxygen molecules to activate the fluorophores in the acceptor beads, and a light signal is detected at 520–620 nm. As shown in Fig. 1D, incubation of NK1 with Met plus heparin yielded ≈80,000 photon counts versus <300 photon counts produced by either NK1 or Met alone. Importantly, interaction of NK1 with Met depended on the presence of heparin. Addition of heparin increased the NK1-Met binding signal by >200-fold, consistent with the fact that Met activation by NK1 requires the presence of heparin as a Met coreceptor (8, 9, 18, 19).
To determine the binding affinities of the various NK1s for Met, we performed competition experiments by using unlabeled NK1 proteins (Fig. 1E). Both mouse NK1 and human R134G mutant bind to Met with a similar affinity (IC50 of ≈30 nM), whereas the human NK1 binds to Met with a 3-fold weaker affinity (IC50 of ≈100 nM). We also measured the binding affinity of the full-length HGF, which binds to Met with an IC50 of 0.5–5.0 nM (data not shown), which is close to the range of HGF potency as measured in cell-based assays (20, 21). These quantitative measurements thus establish that the affinities of the NK1 fragments are 5- to 20-fold weaker than the full-length HGF.
Met Dimerization Promoted by NK1 Binding.
To probe the mechanism of Met activation by NK1, we designed a Met dimerization assay based on AlphaScreen technology (Fig. 2A). In this assay, both nickel chelate donor and acceptor beads were attached to Met via its C-terminal histidine tag. When Met formed an NK1-mediated dimer, the dimerization signal was recorded. As shown in Fig. 2B, the Met was monomeric regardless of the presence of heparin (<500 photo counts; Fig. 2B). Addition of human NK1 and heparin dramatically increased the Met dimerization signal to >40,000 photo counts. In the absence of heparin, NK1 only promotes a basal Met dimerization signal. Similar results were obtained with the mouse NK1 and the human R134G NK1. Furthermore, in a dose titration experiment (Fig. 2C), we found that NK1 induced Met dimerization with an effective concentration (EC50) ranging from 50 to 200 nM, which is closely related to the binding affinities of NK1 for Met from Fig. 1E. Together, these results suggest that NK1 is capable of binding and inducing Met dimerization in a heparin-dependent manner.
Fig. 2.
Met dimerization induced by NK1 binding. (A) A diagram of AlphaScreen assay for detecting Met dimerization promoted by NK1 binding. (B) Met dimerization induced by 1 μM NK1 in the presence and absence of 10 μM heparin as measured by AlphaScreen assays. (C) Dose curves of various NK1 to induce Met dimerization in the presence of 10 μM heparin.
The AlphaScreen is a proximity assay which detects signals arising from Met dimerization or oligomerization. To determine the nature of the Met dimer/oligomer, we performed dynamic light-scattering analysis to determine the hydraulic diameter of the Met/NK1 complex. In this assay, NK1 was used as a positive control because it has been shown to form a dimer in a heparin-dependent manner (22). In the absence of heparin, dynamic light scattering revealed that NK1 forms a monodispersed monomer with a hydraulic diameter of 54 Å (Table 1). The referred molecular weight from this measurement for the NK1 monomer is 33 kDa, which is overestimated from the real molecular mass of NK1 at 21 kDa because of the elongated shape of NK1 monomer structure (see Fig. 3B). Addition of heparin increased the hydraulic diameter of NK1 to 66 Å, with a referred molecular mass of 51 kDa, which is better correlated with the expected molecular mass of 48 kDa for a 2:2 NK1 dimer/heparin complex, in which the heparin that was used had an average molecular mass of 3 kDa. Regardless of the presence of heparin, the Met sema domain is monomeric with a monodispersed diameter of 82–84 Å and an estimated molecular mass of 95 kDa, which is also slightly overestimated from its expected molecular mass of 63 kDa because of the elliptical structure of the Met sema domain (13). Addition of NK1 plus heparin to Met resulted in a monodispersed complex (error of sum of square difference between the measured and calculated autocorrelation curves (SOS) <1.0) with its hydraulic diameter increased to 114 Å. The estimate molecular mass from this measurement for this complex is 194 kDa, which approximates the expected molecular mass of 174 kDa for a 2:2:2 Met/NK1/heparin complex, suggesting that, in the presence of heparin, Met and NK1 form a uniform dimer rather than a higher oligomeric complex. In the absence of heparin, NK1 and Met also form a dimeric complex which, however, is polydispersed and highly unstable as indicated by the high SOS value (Table 1). The Met sema domain used in our experiments has been shown to be necessary for Met dimerization in cell-based cross-linking experiments (14). Our results here further showed that the Met sema domain is sufficient to form a dimeric complex with NK1 in vitro.
Table 1.
NK1-induced Met dimerization as determined by dynamic light scattering
| Samples | Diameter, Å | Molecular mass, kDa | SOS values |
|---|---|---|---|
| NK1 | 54 | 33 ± 1 | 0.284 |
| NK1 + heparin | 66 | 51 ± 1 | 0.453 |
| Met | 82 | 94 ± 2 | 0.813 |
| Met + heparin | 84 | 95 ± 2 | 0.777 |
| Met + NK1 | 116 | 205 ± 3 | 5.545 |
| Met + NK1 + heparin | 114 | 194 ± 3 | 0.838 |
Except for the NK1-Met without heparin, all measurements have SOS values <1.0, indicating monodispersion of the complex. The high SOS value in the NK1-Met without heparin indicate a polydispersed complex, reflecting the unstable nature of the Met-NK1 complex in the absence of heparin. SOS, error of sum of square difference between the measured and calculated autocorrelation curves.
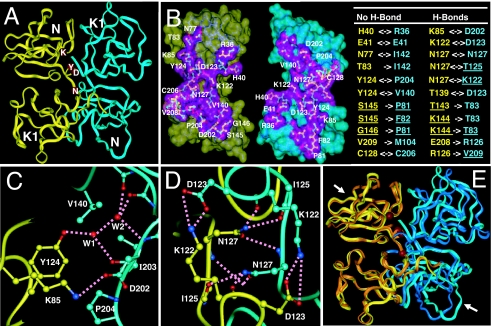
Fig. 3.
Crystal structures of the mouse and human NK1. (A) An overall view of the head-to-tail dimer of the mouse NK1. The letters (K, D, Y, and N) indicate the position of residues K85, D123, Y124, and N127, respectively. Only yellow monomer is labeled. (B) The NK1 dimer interface and the intermolecular interactions. The interface within 6.0 Å of the other monomer is shown as a purple surface and inner residues are gold, green, and red if they are within 4.0, 5.0, and 6.0 Å, respectively, of the other NK1. The list of interactions includes non-H-bond packing and H-bond interactions within 4.0 Å between the two NK1 monomers. Double-headed arrows indicate reciprocal interactions between two monomers and single-headed arrows indicate interactions from one monomer to the other. Underlining indicates that the residues' protein backbone atoms are involved in dimer interactions. (C) Interactions mediated by Y124 and K85 (yellow monomer) with V140, D202, I203, and P204 (cyan monomer). Hydrogen bonds are indicated by dashed lines. Water-mediated interactions are indicated by W1 and W2. (D) Interactions mediated by K122, D123, and N127 (yellow monomer) with K122, D123, and N127 (cyan monomer). Hydrogen bonds are indicated by dashed lines. (E) Comparison of the mouse and human NK1 (superposition of the Cα atoms). The two mouse monomers are in yellow and cyan, and the human monomers are in gold and blue. Arrows indicate the heparin binding sites identified in previous studies (23).
Crystal Structures of the Mouse and Human NK1.
To investigate the basis by which NK1 induces Met dimerization, we have determined a 2.4-Å structure of mouse NK1 and a 1.8-Å structure of human NK1. The overall structure of mouse NK1 forms a disk-shaped head-to-tail dimer with approximate dimensions of 60 × 56 × 33 Å (Fig. 3A). Each monomer comprises two globular domains, the N-terminal heparin binding domain and the C-terminal kringle domain, connected by a two-residue linker (residues 126 and 127). The shape of the NK1 monomer is relatively elongated with an approximate size of 50 × 27 × 22 Å (Fig. 3B). The maximum dimensions of the NK1 dimer and monomer structures (60 and 50 Å, respectively) are remarkably correlated with their hydraulic diameters measured by dynamic light scattering (Table 1).
The head-to-tail NK1 dimer is formed by the intertwined packing between the N domain and the C-terminal kringle domain of the two monomers that mediated an intricate network of interactions (Fig. 3B). The core hydrophobic interface comprises reciprocal interactions of Y124 with V140 and P204 (Fig. 3C) and the packing of the C128–C206 disulfide bond from both monomers (Fig. 3B). The extensive NK1 dimer interface includes two pairs of charged interactions of K85–D202 and K122–D123 (Fig. 3 C and D) and a network of hydrogen bonds mediated by N127 (Fig. 3D). Together, these hydrophilic interactions are the key determinants of the overall specificity and stability of the NK1 dimer complex.
The mouse NK1 is 90.0% identical in sequence to the human NK1 [supporting information (SI) Fig. 6], and its monomer structure and dimer arrangement resemble those of the human NK1 structure reported here (Fig. 3E), as well as previously reported structures (22–24). Despite different crystallization conditions and species of proteins, all NK1 structures determined to date contain a highly similar head-to-tail dimer arrangement. Furthermore, all residues involved in the formation of the NK1 dimer interface are conserved in species from chicken to human (SI Fig. 6), suggesting that the observed NK1 dimer may perform an evolutionarily conserved function in HGF–Met signaling.
Effects of Mutations in the NK1 Dimer Interface.
To determine the role of the NK1 dimer configuration in the binding and activation of Met, we made single or combined mutations in four key residues (Y124, K85, D123, and N127) that form the dimer interface (highlighted in Fig. 3A). These mutations were made in the human R134G background to facilitate purification. Because the dimer interface is far from the heparin binding site located in the N domain (Fig. 3E), these dimerization mutants retained full heparin binding activity and were purified by a heparin affinity column to homogeneity for Met dimerization and binding assays. NK1 again promoted Met dimerization in a heparin-dependent manner (Fig. 4A). By contrast, all other NK1 mutants dramatically reduced the ability to induce Met dimerization regardless of the presence of heparin. Specifically, three mutants (Y124A, K85A/D123A, and K85A/N127A) abolished Met dimerization activity.
Fig. 4.
Effects of the NK1 dimerization mutants. (A) The ability of various NK1 mutants to induce Met dimerization in the presence and absence of 10 μM heparin. The D123A and N127A single mutants behave similarly to the K85A mutant (data not shown). (B) The ability of NK1 mutants (1 μM) to inhibit NK1-mediated Met dimerization.
We also measured the Met binding activity of these NK1 mutants by using the same competition conditions as in Fig. 1E. In contrast to the Met dimerization activity, the Met binding activity of these NK1 mutants were little affected (Table 2). The Met binding affinity for two NK1 mutants (Y124A and K85A) was even slightly better than that for the wild-type NK1, suggesting that the Met-binding activity of NK1 can be separated from its ability to induce Met dimerization. Furthermore, the above NK1 dimerization mutants can dominantly inhibit Met dimerization induced by wild-type NK1 (Fig. 4B), implying that they may potentially function as Met antagonists.
Table 2.
Binding affinity (IC50 values) of NK1 mutants to Met as determined by competition of the binding of the biotinylated NK1 to Met
| NK1 mutants | IC50, nM |
|---|---|
| NK1 | 49 ± 3 |
| Y124A | 39 ± 4 |
| K85A | 32 ± 2 |
| K85A/D123A | 75 ± 9 |
| K85A/N127A | 97 ± 14 |
NK1 Dimerization Mutants Are Met Antagonists.
To determine whether the NK1 dimerization mutants can function as Met antagonists, we first performed protease assays for the urokinase-type plasminogen activator (uPA) because it is induced by HGF-mediated Met activation in Madin–Darby canine kidney (MDCK) cells and various cancer cell lines (21). In MDCK cells, uPA activity is consistently elevated by 3- to 4-fold by treatment with HGF (Fig. 5A). Treatment with the mouse and human NK1 or the human R134G NK1 mutant also induced uPA activity by 3- to 4-fold, whereas addition of the four dimerization mutants showed little effect (Fig. 5A). Furthermore, whereas wild-type NK1 showed no inhibitory effects on HGF-mediated uPA activation, the four NK1 dimerization mutants significantly blocked HGF-induced uPA activity, demonstrating that they can function as dominant HGF antagonists in this assay. Remarkably, the agonist and antagonist properties of these NK1 mutants are closely correlated with their ability to induce Met dimerization in AlphaScreen assays, as shown in Fig. 4A.
Fig. 5.
Effects of NK1 dimerization mutants in cell-based assays. (A) Effects of NK1 mutants (1 μM) on uPA assays in the presence and absence of HGF. (B) Effects of NK1 mutants (1 μM) on proliferation of MDCK cells in the presence and absence of HGF. (C) Effects of NK1 (1 μM) on MDCK cell-scattering assays in the presence and absence of HGF. (D) Effects of NK1 mutants (1 μM) on branching morphogenesis of the prostate cancer cell line DU145. Arrows indicate the cell branching induced by HGF. (E) Effects of NK1 mutants (1 μM) on invasion of the glioblastoma cell line DBTRG. (F) Inhibition of Met dimerization on the DU145 cell surface by the Y124A mutant as detected by Western blot analysis of cross-linking proteins. The positions of Met monomer (p145) and dimer as well as the unprocessed Met (p175) and the nonspecific band are indicated. (G) Inhibition of Akt and Erk phosphorylation by the Y124A mutant. All assays were performed with 60–100 ng/ml HGF when indicated and 2 μM heparin except for F and G, where heparin used is 1.0 mg/ml.
Because Met activation by HGF also induces cell proliferation, we used thymidine incorporation to measure HGF-stimulated DNA synthesis. Both HGF and wild-type NK1 stimulated thymidine incorporation by 2- to 4-fold (Fig. 5B). The four NK1 mutants on their own had little effect but efficiently blocked HGF-mediated DNA synthesis, again demonstrating the antagonist properties of these NK1 dimerization mutants.
HGF-mediated Met activation also affects the mobility behavior of cells, e.g., cell scattering, branching, and invasion, thus, we tested whether the NK1 mutants can inhibit the above HGF-mediated effects. As shown in Fig. 5C, the untreated MDCK cells grow as tightly packed colonies. Addition of HGF promotes dispersion of these MDCK colonies. Whereas addition of the Y124A mutant did not disperse the cells, it dominantly inhibited HGF-induced cell scattering. HGF also induces branching morphogenesis of DU145 cells, a prostate cancer cell line (Fig. 5D), and cell invasion of DBTRG, a glioblastoma cell line (Fig. 5E). The Y124A mutant effectively inhibits both HGF-mediated activities, further demonstrating the antagonist properties of this dimerization mutant.
To determine the basis of the Y124A antagonist property on DU145 cells, we performed cell-surface covalent cross-linking experiments. Regardless of the presence of HGF or heparin, Met is readily cross-linked into dimer [Fig. 5F, lanes 2–4 and supporting information (SI) Fig. 7], consistent with previous reports (14–16). Treatment with Y124A abolishes the formation of the Met dimer (Fig. 5F, lanes 5 and 6), indicating that the Y124A mutant indeed meditates its antagonistic activity by preventing Met dimerization. Together, these results indicate that Met dimerization itself is not sufficient, but is required, for the receptor activation. The Y124A mutant also inhibits the downstream signaling of Met activation by preventing phosphorylation of Akt and Erk in DU145 cells (Fig. 5G). Remarkably, even at 10 nM concentration, the Y124A antagonist achieves >80% inhibition of Akt or Erk activation. Similar results were observed with DBTRG cells (data not shown). The activities of the Y124A mutant on DU145 and DBTRG imply that NK1-based antagonists might have potential application in anticancer therapy.
Discussion
To serve as an RTK agonist, growth factors like HGF must have at least two functions, namely receptor binding and activation. Thus, a mutated growth factor with selective disruption of its receptor activation ability but not its receptor binding may function as an RTK antagonist. In this study, we have provided a proof of concept for the above idea by designing NK1-based Met antagonists. Through detailed structural and functional analyses, we demonstrate that NK1 mutants with selective loss of their ability to induce Met dimerization, but not their Met binding activity, are HGF antagonists. These results provide important insights into the molecular mechanism of Met activation by NK1 and a rational basis for design and optimization of Met antagonists for cancer therapy.
Mechanism of Met Activation.
Activation of Met was shown to be mediated through receptor dimerization as first demonstrated by the fusion protein of the Met kinase domain with a TPR leucine zipper dimerization motif (25). However, the mechanism of HGF-mediated Met dimerization remains unclear. Recent data shows that the Met sema domain is sufficient for HGF binding (12) and is required for receptor dimerization and activation (14). Our binding data are consistent with these previous observations and further demonstrate that the Met sema domain is sufficient for NK1-mediated receptor dimerization. Even though the binding affinity of NK1 for Met is 5- to 20-fold weaker than that of HGF, the ability of NK1 to induce Met dimerization is readily observed and consistent with its agonistic activity in transgenic mice and in cell-based assays (7–9, 18, 19).
Receptor dimerization is a general paradigm for activation of RTKs (26), and diverse structural mechanisms of ligand-induced receptor dimerization have been revealed by crystallographic studies of ligand–receptor complexes of RTKs. Dimerization of the receptors for vascular endothelial growth factor (VEGF) and nerve growth factor (NGF) is mediated by preformed dimeric ligands (27, 28). In fibroblast growth factor receptor (FGFR), heparin drives dimerization of FGF ligand and the receptor (29, 30). In contrast, dimerization of epidermal growth factor receptor (EGFR) is through the receptor itself, independent of direct ligand–ligand interactions (31, 32). The EGF binding site in the receptor is distinct from the receptor dimer interface, but the binding of EGF induces a conformational change of the receptor to expose the dimerization loop, thus enabling the receptor to dimerize (33). How does NK1 induce Met dimerization? One plausible mechanism is that dimerization of Met is mediated by the NK1–NK1 dimer interface, in a manner similar to receptor/ligand complexes of VEGF and NGF (SI Fig. 8A). In this model, NK1 is monomeric in the absence of heparin, and addition of heparin drives NK1 dimerization and the formation of a stable NK1/Met complex that is followed by receptor activation. Although the validation of such a model waits for a structure of the NK1/Met/heparin complex, this model is supported by strong experimental evidences. The first evidence is that NK1 forms a dimer in a heparin-dependent manner, and this is directly correlated with the requirement of heparin for NK1 to bind and promote Met dimerization (Figs. 1 and 2). The consistent requirement of heparin for NK1 dimerization and for the formation of a stable NK1/Met complex suggests that the NK1–NK1 dimer plays a direct role in mediating Met dimerization and activation. Secondly, the crystal structures of both mouse and human NK1 reveal a conserved NK1–NK1 interface, and mutations that are designed to alter the NK1 dimer interface (Y124A, K85A, K85A/D123A, and K85A/N127A) abolish the ability of NK1 to induce Met dimerization (Fig. 4A). This finding also provides strong support that the NK1 dimer interface observed in the crystal structures is critical for Met dimerization and activation. The highly conserved nature of residues involved in the formation of the NK1 dimer interface further indicate that this mode of Met dimerization and activation by NK1 is preserved across species from chicken to man. Finally, the recently determined low-resolution structure of an HGF–Met complex by cryoelectron microscopy and small angle x-ray scattering suggests that the packing of the HGF–Met complex may be mediated by the NK1–NK1 dimer interface (15), and, interestingly, the K85A and/or N127A mutations in the context of the full-length HGF abolish the NK1–NK1 dimer interface's ability to scatter MDCK cells, further supporting the importance of the observed NK1 dimer interface in HGF-mediated Met activation (15).
A Basis for Designing Met Antagonists.
The above mechanism of Met activation by HGF via NK1 dimerization thus provides a rational basis for designing NK1-based Met antagonist by selectively disrupting the ability of NK1 to dimerize (SI Fig. 8B). Because these NK1 mutants retain full Met-binding activity (Table 2), they act as effective Met antagonists (Fig. 5). The ability to disrupt NK1-induced Met dimerization by a number of NK1 mutations indicates that receptor dimerization is very sensitive to the perturbation of the NK1 dimer interface and suggests a way to make and design other NK1 dimer mutants that could serve as Met antagonists. In fact, we have made mutations in several other residues involved in NK1 dimer formation (listed in Fig. 3B). Most of these NK1 dimer mutants lose their ability to induce Met dimerization (data not shown). One exception is the V140R mutant, which actually increases its ability to promote Met dimerization over the wild-type NK1 (SI Fig. 9A). Cell-based experiments indicate that the V140R mutant acts as a Met agonist as indicated by induction of uPA activity and scattering of MDCK cells (SI Fig. 9 B and C). In the structure, V140 is loosely packed against Y124 in the NK1 dimer interface (Fig. 3C). An arginine residue can be easily modeled into the V140 position, consistent with the V140R mutant phenotype. The spectrum of our NK1 dimer interface mutants indicates that NK1 dimerization can be tuned according to the purpose of modulating its ability to activate or inhibit Met.
Because aberrant activation of the HGF–Met signaling is closely correlated with tumor proliferation, progression, invasion, and metastasis (1), targeting Met activation has become an intense area of anticancer therapeutical research. Current methods of Met inhibition include small molecules targeting the Met intracellular kinase domain (34), antibodies against HGF or Met (35), decoy receptors using the Met extracellular domain (14), and HGF-based Met antagonists like single-chain HGF derivatives (36). Although these methods demonstrate various degrees of Met inhibition, they have displayed limitations with respect to their application, e.g., limited bioavailability and specificity of small molecule inhibitors, the large size and costly production of antibody and decoy Met receptors, protein stability, and mixed agonist/antagonist properties of single chain HGF derivatives. The NK1-based Met antagonists described here can serve as an alternative method with several distinct advantages. First, NK1 is a naturally circulated HGF variant that targets the extracellular domain of Met. Second, NK1 is a much smaller protein than antibody, decoy Met receptor, or single-chain HGF and NK4. Third, because of its small size, NK1 can be easily produced in large quantities at high purity. Finally, the potency of our NK1-based antagonists, which is currently within one order of magnitude of HGF, can be further improved by protein engineering.
The family of ligand-activated receptor tyrosine kinases has a widespread role in tumorigenesis and metastasis (37). Our mechanistic-based design of Met antagonists has important implications for other growth factor/tyrosine kinase receptor systems, including VEGF-, NGF-, and EGF receptors. Like Met, VEGF, NGF, and EGF receptors are activated through growth factor-induced receptor dimerization. To activate their receptors, NGF and VEGF contain separate receptor-binding and dimerization surfaces (27, 28). Based on our work with NK1 and Met, it is conceivable that one can design VEGF and NGF antagonists by separating their receptor-binding activity from their receptor dimerization activity. In the case of EGF, ligand binding induces conformational changes of the receptor that mediates direct receptor/receptor dimerization (31, 32). It is conceptually possible that a mutated EGF may induce a distinct conformational change of the receptor that is incompatible with receptor dimerization and activation. Although experimental validation for the antagonist design remains to be demonstrated for these growth factor receptors, these ideas are conceptually intriguing. Given the relatively small size of VEGF and EGF, such growth factor-based antagonists may provide an exciting alternative to current antibody-based approaches.
In summary, our biochemical and structural analyses reveal that activation of Met is mediated through the conserved NK1–NK1 dimer interface. The ability to design specific mutations in the NK1 dimer interface that selectively disrupts NK1's ability to induce Met dimerization but retains its Met-binding activity provides a mechanistic basis for designing Met antagonists for therapeutic applications. Because activation of RTK is proposed to be mediated through a conserved mechanism of receptor dimerization, our design of NK1-based Met antagonists also provides a generalized concept for designing antagonists of other ligand-activated tyrosine kinase receptors.
Methods
Protein Production, Binding Assays, and Crystallization.
Both mouse and human HGF NK1 (residues 28–209) were expressed as 6xHis-thioredoxin fusion proteins from the expression vector pET-Duet1 in the E. coli strain Rosetta/gami (DE) (Novagen, Madison, WI) to promote disulfide-bond formation. The biotinylated NK1 was produced by fusing the 20-aa biotin acceptor peptide sequence from the pDW464 plasmid (38) to the N terminus of NK1. The Met protein (residues 25–567, containing the sema domain and the cysteine-rich domain) was expressed as a C-terminal hexahistidine tag fusion protein from Lec 3.2.8.1 cells (12). All proteins were purified to homogeneity for binding assays and crystallization with details described in SI Methods.
Data Collection and Structure Determination.
Diffraction data were collected at beamline 5-ID (DND-CAT) at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) with details described in SI Methods. The structure was solved by molecular replacement with the Protein Data Bank coordinates 1NK1 (22). Molecular replacement and model refinement were performed with CNS, where twin fraction was incorporated for the refinement for the mouse structure, and manual model building was done with the program O (39). Statistics of data and the refined structures are listed in SI Table 3.
Met Activation Assays.
Cell-based Met activation assays, including scattering of MDCK cells, uPA activation, cell proliferation, invasion, and branching morphogenesis assays followed published protocols (20, 21) with details described in SI Methods.
Supplementary Material
Acknowledgments
We thank J. S. Brunzelle and Z. Wawrzak for assistance in data collection at sector 5-ID-B of the Advanced Photo Source. Use of the Advanced Photon Source was supported by the Office of Science of the U. S. Department of Energy. This work was supported in part by the Jay and Betty Van Andel Foundation (H.E.X., G.V.W. and C.M.), Department of Defense Grant W81XWH0510043 (to H.E.X.), National Institutes of Health Grants DK071662 and DK066202 (to H.E.X.), Michigan Economic Development Corporation Grant 085P1000817 (to H.E.X.), Medical Research Council Program Grant G9704528 (to E.G.).
Abbreviations
- HGF
hepatocyte growth factor
- RTK
receptor tyrosine kinase
- uPA
urokinase-type plasminogen activator
- MDCK
Madin–Darby canine kidney
- NGF
nerve growth factor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2QJ4 and 2QJ2).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704290104/DC1.
References
- 1.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 2.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 3.Gherardi E, Gray J, Stoker M, Perryman M, Furlong R. Proc Natl Acad Sci USA. 1989;86:5844–5848. doi: 10.1073/pnas.86.15.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottaro DP, Rubin JS, Faletto DL, Chan AM, Kmiecik TE, Vande Woude GF, Aaronson SA. Science. 1991;251:802–804. doi: 10.1126/science.1846706. [DOI] [PubMed] [Google Scholar]
- 5.Cioce V, Csaky KG, Chan AM, Bottaro DP, Taylor WG, Jensen R, Aaronson SA, Rubin JS. J Biol Chem. 1996;271:13110–13115. doi: 10.1074/jbc.271.22.13110. [DOI] [PubMed] [Google Scholar]
- 6.Lokker NA, Presta LG, Godowski PJ. Protein Eng. 1994;7:895–903. doi: 10.1093/protein/7.7.895. [DOI] [PubMed] [Google Scholar]
- 7.Jakubczak JL, LaRochelle WJ, Merlino G. Mol Cell Biol. 1998;18:1275–1283. doi: 10.1128/mcb.18.3.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyon M, Deakin JA, Lietha D, Gherardi E, Gallagher JT. J Biol Chem. 2004;279:43560–43567. doi: 10.1074/jbc.M408510200. [DOI] [PubMed] [Google Scholar]
- 9.Rubin JS, Day RM, Breckenridge D, Atabey N, Taylor WG, Stahl SJ, Wingfield PT, Kaufman JD, Schwall R, Bottaro DP. J Biol Chem. 2001;276:32977–32983. doi: 10.1074/jbc.M105486200. [DOI] [PubMed] [Google Scholar]
- 10.Cooper CS, Park M, Blair DG, Tainsky MA, Huebner K, Croce CM, Vande Woude GF. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 11.Komada M, Hatsuzawa K, Shibamoto S, Ito F, Nakayama K, Kitamura N. FEBS Lett. 1993;328:25–29. doi: 10.1016/0014-5793(93)80958-w. [DOI] [PubMed] [Google Scholar]
- 12.Gherardi E, Youles ME, Miguel RN, Blundell TL, Iamele L, Gough J, Bandyopadhyay A, Hartmann G, Butler PJ. Proc Natl Acad Sci USA. 2003;100:12039–12044. doi: 10.1073/pnas.2034936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamos J, Lazarus RA, Yao X, Kirchhofer D, Wiesmann C. EMBO J. 2004;23:2325–2335. doi: 10.1038/sj.emboj.7600243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong-Beltran M, Stamos J, Wickramasinghe D. Cancer Cell. 2004;6:75–84. doi: 10.1016/j.ccr.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 15.Gherardi E, Sandin S, Petoukhov MV, Finch J, Youles ME, Ofverstedt LG, Miguel RN, Blundell TL, Vande Woude GF, Skoglund U, et al. Proc Natl Acad Sci USA. 2006;103:4046–4051. doi: 10.1073/pnas.0509040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faletto DL, Tsarfaty I, Kmiecik TE, Gonzatti M, Suzuki T, Vande Woude GF. Oncogene. 1992;7:1149–1157. [PubMed] [Google Scholar]
- 17.Pediaditakis P, Monga SP, Mars WM, Michalopoulos GK. J Biol Chem. 2002;277:14109–14115. doi: 10.1074/jbc.M112196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwall RH, Chang LY, Godowski PJ, Kahn DW, Hillan KJ, Bauer KD, Zioncheck TF. J Cell Biol. 1996;133:709–718. doi: 10.1083/jcb.133.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catlow K, Deakin JA, Delehedde M, Fernig DG, Gallagher JT, Pavao MS, Lyon M. Biochem Soc Trans. 2003;31:352–353. doi: 10.1042/bst0310352. [DOI] [PubMed] [Google Scholar]
- 20.Gao CF, Xie Q, Su YL, Koeman J, Khoo SK, Gustafson M, Knudsen BS, Hay R, Shinomiya N, Vande Woude GF. Proc Natl Acad Sci USA. 2005;102:10528–10533. doi: 10.1073/pnas.0504367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Q, Gao CF, Shinomiya N, Sausville E, Hay R, Gustafson M, Shen Y, Wenkert D, Vande Woude GF. Oncogene. 2005;24:3697–3707. doi: 10.1038/sj.onc.1208499. [DOI] [PubMed] [Google Scholar]
- 22.Chirgadze DY, Hepple JP, Zhou H, Byrd RA, Blundell TL, Gherardi E. Nat Struct Biol. 1999;6:72–79. doi: 10.1038/4947. [DOI] [PubMed] [Google Scholar]
- 23.Lietha D, Chirgadze DY, Mulloy B, Blundell TL, Gherardi E. EMBO J. 2001;20:5543–5555. doi: 10.1093/emboj/20.20.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ultsch M, Lokker NA, Godowski PJ, de Vos AM. Structure (London) 1998;6:1383–1393. doi: 10.1016/s0969-2126(98)00138-5. [DOI] [PubMed] [Google Scholar]
- 25.Park M, Dean M, Cooper CS, Schmidt M, O'Brien SJ, Blair DG, Vande Woude GF. Cell. 1986;45:895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- 26.Weiss A, Schlessinger J. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 27.Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, de Vos AM. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 28.Wiesmann C, Ultsch MH, Bass SH, de Vos AM. Nature. 1999;401:184–188. doi: 10.1038/43705. [DOI] [PubMed] [Google Scholar]
- 29.Pellegrini L, Burke DF, von Delft F, Mulloy B, Blundell TL. Nature. 2000;407:1029–1034. doi: 10.1038/35039551. [DOI] [PubMed] [Google Scholar]
- 30.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 31.Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, et al. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 32.Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, et al. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 33.Schlessinger J. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 34.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, Chen J, Wang X, Ruslim L, Blake R, et al. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 35.Cao B, Su Y, Oskarsson M, Zhao P, Kort EJ, Fisher RJ, Wang LM, Vande Woude GF. Proc Natl Acad Sci USA. 2001;98:7443–7448. doi: 10.1073/pnas.131200498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto K, Nakamura T. Cancer Sci. 2003;94:321–327. doi: 10.1111/j.1349-7006.2003.tb01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blume-Jensen P, Hunter T. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 38.Duffy S, Tsao KL, Waugh DS. Anal Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- 39.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.