Abstract
Genome-wide association (GWA) studies offer a powerful unbiased method for the identification of multiple susceptibility genes for complex diseases. Here we report the results of a GWA study for Crohn's disease (CD) using family trios from the Quebec Founder Population (QFP). Haplotype-based association analyses identified multiple regions associated with the disease that met the criteria for genome-wide significance, with many containing a gene whose function appears relevant to CD. A proportion of these were replicated in two independent German Caucasian samples, including the established CD loci NOD2 and IBD5. The recently described IL23R locus was also identified and replicated. For this region, multiple individuals with all major haplotypes in the QFP were sequenced and extensive fine mapping performed to identify risk and protective alleles. Several additional loci, including a region on 3p21 containing several plausible candidate genes, a region near JAKMIP1 on 4p16.1, and two larger regions on chromosome 17 were replicated. Together with previously published loci, the spectrum of CD genes identified to date involves biochemical networks that affect epithelial defense mechanisms, innate and adaptive immune response, and the repair or remodeling of tissue.
Keywords: haplotype, complex disease, IL23R
Crohn's disease (CD) is a chronic inflammatory bowel disease characterized by transmural inflammatory lesions that can affect the entire gastrointestinal tract (1). The lifetime prevalence is 0.5–1% in Caucasian populations (2) and reflects the combined effects of genetic predisposition and environmental factors (3). Genetic linkage and candidate gene approaches (4–14) have contributed to the elucidation of loci influencing genetic susceptibility to CD. More recently, genome-wide association (GWA) studies (15–21) have provided further insight into the molecular pathogenesis of the disease. The top candidate genes or loci that consistently replicate include NOD2, IL23R, ATG16L1, the IBD5 region on chromosome 5q31, and a region on 5p13.1 near the PTGER4 gene. The nature of these genes suggests that the major genetic risk factors for CD are involved in the innate immune response and destruction of intracellular bacteria.
It is now clear that GWA studies provide a powerful and robust new tool for the identification of the multiple susceptibility alleles involved in complex diseases. Importantly, these types of studies have the ability to identify genes that impart only moderate increases in risk (21, 22). However, most studies performed to date have identified only a few top signals, and the validation of true association among signals with lower statistical significance remains a challenge. In addition, most of the GWA studies to date have been performed by using general populations, for which very large sample sizes are required for success. They have also largely relied on single-marker analysis, with genome-wide haplotype-based association analyses receiving little attention.
In early 2004, we conducted a GWA study for CD using 382 trio samples from the Quebec Founder Population (QFP). This population descended in genetic isolation from several thousand founders who emigrated from France in the 17th century (23). The demographic history of the QFP, which is characterized by a population bottleneck, rapid population expansion, and little admixture, makes it a valuable resource for use in genetic studies (24). The population has been well characterized as having reduced genetic heterogeneity for Mendelian diseases (25).
We first used a haplotype-based GWA study to detect regions associated with CD. Several regions with association signals were then followed up by fine mapping at a greater marker density in a larger QFP sample of 477 trios. A selection of the most promising regions was then tested for replication in two independent German samples. In summary, this study identified multiple CD-associated loci, many of which were replicated in an independent Caucasian sample. We report details for several of these, including the previously reported NOD2, IBD5, and IL23R loci, along with four replicated regions on 3p21, 4p16.1, 17q11.1, and 17q22-q23.
Results
Genome-Wide Scan.
Disease association was systematically tested across the genome for both single markers and multimarker window haplotypes (three, five, seven, and nine contiguous markers). Sixteen regions with nominal P values <10−5 were identified (Table 1). The genome-wide significance of nominal P values was determined by permutation tests. The previously identified NOD2, IBD5, and IL23R gene regions were among the top seven loci identified in this study.
Table 1.
Regions identified in the QFP GWA
| Chromosome | Peak RefSNP ID | P values | Marker window | Lower bound | Upper bound |
|---|---|---|---|---|---|
| 16q12.1 | 2076756 | <1e-9 | 5 | 0.000 | 0.000 |
| 15q21.3 | 2414476 | 8.71e-09 | 5 | 0.012 | 0.138 |
| 5q15 | 255969 | 1.41e-07 | 9 | 0.038 | 0.164 |
| 5q31.1 | 272888 | 4.79e-07 | 9 | 0.056 | 0.190 |
| 17q23.2 | 8077981 | 5.13e-07 | 7 | 0.026 | 0.070 |
| 17q11.1 | 4794986 | 6.92e-07 | 9 | 0.012 | 0.044 |
| 1p31.3 | 17129659 | 8.32e-07 | 5 | 0.006 | 0.068 |
| 10q26.11 | 10886462 | 9.55e-07 | 5 | 0.004 | 0.038 |
| 11q21 | 12576495 | 1.00e-06 | 9 | 0.002 | 0.042 |
| 8q22.3 | 2386922 | 2.04e-06 | 5 | 0.000 | 0.200 |
| 8q11.23 | 11775611 | 4.07e-06 | 5 | 0.070 | 0.474 |
| 13q21.32 | 17837226 | 4.37e-06 | 7 | 0.042 | 0.360 |
| Xp21.1 | 6632039 | 6.03e-06 | 9 | 0.090 | 0.460 |
| 3q13.33 | 12695416 | 6.31e-06 | 5 | 0.028 | 0.258 |
| 6q14.1 | 13201732 | 8.32e-06 | 7 | 0.046 | 0.276 |
| 12q12 | 11181674 | 8.71e-06 | 3 | 0.034 | 0.216 |
Regions identified from the GWA with a minimum nominal P value <1 × 10−5 are shown with the chromosome band location. The National Center for Biotechnology Information dbSNP ID of the peak SNP, the nominal P value at the peak marker window, and the number of consecutive analyzed SNPs defining the haplotype (one, three, five, seven, or nine) are shown. The permuted P value of the lower and upper bounds of genome wide significance (see Methods) are also shown.
Fine Mapping, Ultrafine Mapping, and Replication.
A selection of regions was followed up by fine mapping in an expanded set of 477 QFP by trios using an increased marker density (approximately one SNP per 2–5 kb). Regions were prioritized for fine mapping based upon both the strength of the genetic evidence (P values, allele frequencies, etc.) and the presence of functionally interesting candidate genes. After fine mapping, a selection of regions was chosen for replication studies in two samples from the German population (521 trios and 750 additional cases and 828 independent controls). Eight regions from among the top 16 genome-wide scan results, as well as eight more modestly associated regions containing functional candidates, were selected for replication studies using the same SNPs previously used in the QFP fine-mapping studies. The significance of the replication results was determined based on permutation tests. Seven regions demonstrated significant replication in either one or both of the German samples; five of these (excluding NOD2 and IBD5) are presented in Table 2. Five of six regions tested with P values <10−6 in the GWA study were replicated in the German samples. We next combined genotypes from the QFP and German samples for the seven replicated regions and reanalyzed the merged data sets [supporting information (SI) Table 5 A–C]. Five of the seven regions showed substantial increases in statistical significance, with one region remaining virtually unchanged and one exhibiting reduced significance.
Table 2.
Fine mapping in the QFP and replication in German samples
| Chromosome | 477 QFP trios |
German trios |
German cases and controls |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P value | Marker window | Peak RefSNP ID | P value | Marker window | Peak RefSNP ID | Significance P value | P value | Marker window | Peak RefSNP ID | Significance P value | |
| 1p31.3 | 4.20e-06 | 9 | rs1925411 | 2.64e-02 | 1 | rs1925411 | 0.238 | 3.58e-06 | 5 | rs1983860 | 0.002† |
| 1p31.3 | 6.13e-06 | 7 | rs4620509 | 3.42e-02 | 1 | rs12131222 | 0.154 | 5.47e-05 | 5 | rs4486425 | 0.002† |
| *1p31.3 | 1.51e-08 | 5 | rs11208994 | 3.42e-02 | 1 | rs2755250 | 0.334 | 1.23e-05 | 5 | rs1024228 | <0.002† |
| 1p31.3 | 6.18e-08 | 7 | rs7518660 | 1.68e-05 | 5 | rs9988642 | 0.008† | 3.06e-08 | 5 | rs7539625 | <0.002† |
| 3p21.31 | 8.06e-05 | 5 | rs6997 | 7.67e-05 | 5 | rs9863142 | 0.014† | 8.61e-05 | 5 | rs1568661 | 0.008† |
| *3p21.31 | 1.99e-06 | 5 | rs1131095 | 1.97e-04 | 5 | rs6446285 | 0.016† | 1.21e-06 | 9 | rs9875617 | 0.004† |
| 3p21.31 | 2.57e-04 | 1 | rs9821675 | 2.34e-03 | 1 | rs2352967 | 0.044† | 3.28e-03 | 1 | rs695238 | 0.106 |
| *4p16.1 | 3.65e-06 | 5 | rs10003892 | 4.89e-04 | 9 | rs4234723 | 0.020† | 2.93e-03 | 1 | rs10470721 | 0.108 |
| 17q11.1 | 5.85e-04 | 5 | rs2948527 | 5.56e-03 | 7 | rs2948529 | 0.066 | 3.66e-02 | 9 | rs11656620 | 0.277 |
| 17q11.1 | 7.34e-04 | 1 | rs4796052 | 3.67e-01 | 1 | rs1113283 | 0.942 | 9.04e-05 | 5 | rs231480 | 0.002 |
| *17q11.1 | 5.15e-05 | 5 | rs4435306 | 2.52e-02 | 1 | rs4420579 | 0.428 | 5.02e-03 | 1 | rs11654637 | 0.154 |
| 17q23.2 | 5.57e-04 | 5 | rs2645482 | 2.22e-01 | 1 | rs1292037 | 0.878 | 1.10e-03 | 9 | rs4968401 | 0.031 |
| *17q23.2 | 5.39e-04 | 1 | rs6504016 | 1.34e-02 | 9 | rs9303437 | 0.092 | 9.16e-03 | 7 | rs9652858 | 0.106 |
Multiple peaks (rows) are present for some regions; each corresponding peak in the German samples was analyzed for replication significance (see Methods). The minimum nominal P value from fine mapping of 477 QFP trios, the number of consecutive analyzed SNPs defining the allele or haplotype (one, three, five, seven, or nine) and the RefSNP ID of the central marker are shown for each peak. Asterisks denote the peak with the most significant P value in the QFP for that region. Similar data are shown for the German trios and cases/controls along with the P value, indicating the significance of replication.
†Peaks that exhibit significant replication (P < 0.05).
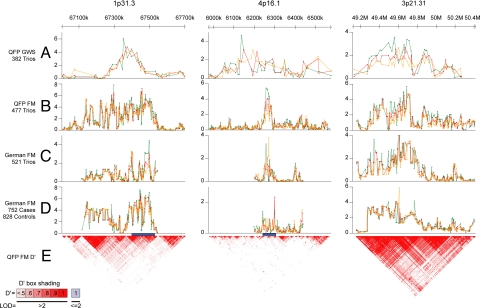
Not surprisingly, both NOD2 and IBD5 (SI Figs. 2 and 3 and SI Tables 6 A–C and 7 A–C) were replicated in the German samples. The IL23R region on 1p31.3, a region near the JAKMIP1 gene on 4p16.1, and a region with multiple genes on 3p21.3 were also significantly replicated. Association plots for these three regions are shown in Fig. 1 and are discussed in more detail below (also see SI Fig. 4 and SI Tables 8 A–F, 9 A–F, and 10 A–C). Two additional regions on chromosome 17 were also significantly replicated (SI Figs. 5 and 6).
Fig. 1.
Overview of GWA study, fine-mapping, and replication studies in regions on chromosomes 1p31.3, 4p16.1, and 3p21.31. A shows the results of the GWA analysis for three regions (1p31.3, 700 kb; 4p16.1, 600 kb; and 3p21.31, 1.2 Mb). Plots indicate the nominal −log10 P values for haplotypes corresponding to five (green), seven (red), and nine (orange) consecutive marker windows. B shows the FM results for the QFP, C shows the FM results for the German trios, and D shows the FM results for the German cases/controls. Regions sequenced in the 1p31.3 and 4p16.1 regions are shown as a blue bar. E shows the LD structure in controls from the QFP based on the D′ algorithm as implemented in Haploview 3.32 (36).
The complete genomic intervals for the IL23R gene (130 kb) and the 4p16.1 region (60 kb), including all introns, were sequenced in 16 QFP individuals selected to represent multiple copies of representative haplotypes. A total of 167 SNPs (119 in the IL23R gene) were genotyped and analyzed in the QFP and German trio samples (SI Table 8 A–F). A total of 147 SNPs in the 4p16.1 region were genotyped in the QFP and German trio samples (SI Table 9 A–F).
Region 1p31.3.
The fine-mapping data for the QFP samples in the IL23R region revealed two independent association signals (r2 = 0.001), one within the SLC35D1 gene (P < 10−6 for a five-marker haplotype) and a second within IL23R (P < 10−5 for a seven-marker haplotype). Significant replication in both German samples was observed (five-marker haplotypes with P values <10−4). The ultrafine mapping of the IL23R gene region in the QFP and German trio samples revealed multiple significantly associated SNPs within the IL23R gene itself or within linkage disequilibrium (LD) blocks extending from the gene into 3′ and 5′ intergenic regions (SI Fig. 4).
Although the lowest nominal P values occurred for haplotype associations, single SNPs within IL23R were associated with CD at nominal P values <10−5 in the QFP trios and German cases/controls (SI Fig. 7). Single-marker association results for 17 of the most significantly associated SNPs in all populations are presented in Table 3. Exact nominal P values, odds ratios (OR), and case/control allele frequencies for these markers are presented in SI Table 8 A–F. Analyses of the combined QFP and German data sets resulted in substantially increased significance for many markers, with multiple SNPs associated at P values <10−7. Although many of the most significant SNPs were in high LD, there appear to be at least two distinct association signals in the 5′ and 3′ regions of the gene (SI Table 12 A and B). Several two-marker haplotypes consisting of nonadjacent SNPs demonstrated an increase in significance by several orders of magnitude over either single marker alone. These included pairs of SNPs from the 5′ and 3′ region of the gene, suggesting the presence of an epistatic interaction between distinct risk alleles located in separate LD blocks.
Table 3.
Association of 17 Selected SNPs across IL23R
Single-marker association results for 17 representative SNPs that span the IL23R gene region and that were significantly associated in the QFP or replication samples. Each column corresponds to data for 1 of 17 SNPs. Color scale indicates nominal P value for single marker association. More detailed information for each SNP can be found in SI Table 11A.
Extended haplotypes spanning IL23R for all 17 SNPs in Table 3 were constructed (SI Table 11 A and B and Table 4). Two risk and two protective haplotypes were significantly associated with CD in all population samples. These four haplotypes have a combined frequency of ≈45%. ORs for the significant risk and protective haplotypes were similar for the QFP and German samples. ORs for the most frequent risk haplotype ranged from 1.33 [confidence interval (C.I.) 1.07–1.66] in the German trios to 1.69 (C.I. 1.36–2.05) in the QFP sample. Risk haplotype 1 was significantly associated in all data sets, as was protective haplotype 2, which contains the opposite allele from risk haplotype 1 at each SNP position. Markers 1 and 3 (which are in high LD, r2 = 0.84) and Marker 6 (which is in moderate LD with Markers 1 and 3, r2 = 0.30–0.31) show consistent alternate alleles on both risk and both protective haplotypes, suggesting they are tagging disease mutations. Other SNPs, including the previously reported Marker 10 (R381Q) (15), may play functional roles in the disease, but their effect depends upon other alleles in the haplotype upon which they reside.
Table 4.
Association of haplotypes formed by the 17 markers in IL23R
| Population | Statistic | Risk | Protective | Risk | Protective |
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| Qfp trios | P | 1.86e-06 | 2.57e-03 | 0.04 | 0.05 |
| F | 29.39 | 19.80 | 1.63 | 3.94 | 3.69 | 2.08 | 10.20 | 13.13 | |
| OR | 1.69 (1.36–2.09) | 0.40 (0.22–0.74) | 1.80 (1.02–3.19) | NS | |
| German trios | P | 0.01 | 1.15e-03 | 0.60 | 0.13 |
| F | 26.48 | 21.28 | 0.91 | 3.09 | 3.65 | 3.20 | 9.13 | 11.33 | |
| OR | 1.33 (1.07–1.66) | 0.29 (0.13–0.64) | NS | NS | |
| Combined trios | P | 1.95e-07 | 1.12e-05 | 0.07 | 0.01 |
| F | 27.98 | 20.93 | 1.28 | 3.52 | 3.67 | 2.63 | 9.68 | 12.25 | |
| OR | 1.50T(1.29–1.75) | 0.35 (0.22–0.57) | NS | 0.77 (0.62–0.95) | |
| German case controls | P | 2.05e-03 | 6.31e-03 | 0.62 | 3.98e-03 |
| F | 30.40 | 25.43 | 1.21 | 2.55 | 3.76 | 3.42 | 10.20 | 13.56 | |
| OR | 1.28 (1.09–1.50) | 0.47 (0.27–0.82) | NS | 0.72 (0.58–0.90) | |
| All combined | P | 1.00e-08 | 2.04e-07 | 0.05 | 5.75e-04 |
| F | 29.53 | 23.34 | 1.25 | 3.11 | 4.17 | 3.26 | 10.40 | 13.12 | |
| OR | 1.38 (1.23–1.53) | 0.39 (0.27–0.57) | NS | 0.77 (0.66–0.89) |
The four risk and protective haplotypes described in SI Table 11B are shown at the top. P values (P, significance of association of each haplotype tested against all other haplotypes in a 2 × 2 case control table using Pearson's χ2), allele frequences (F; cases/controls), and OR (95% C.I.) are shown for each of the analyses. NS, haplotype not significantly associated (C.I. for OR includes 1.0).
Immunohistochemical methods were used to examine the expression of IL23R in CD patients and controls. IL23R expression was detected on mononuclear cells within the colonic lamina propria of unaffected individuals but was significantly up-regulated in CD patients, primarily within epithelial cells (SI Fig. 8). The IL23R gene is known to give rise to multiple splice variants (26). Interestingly, the observed up-regulation seems to affect the shorter isoforms of the protein.
Region 4p16.1.
The 30-kb replicated region located on chromosome 4p16.1 contains two genes, JAKMIP1 and LOC285484. The maximum association signal for both the QFP and German trio fine-mapping (FM) data sets was located upstream of JAKMIP1, within LOC285484 (Fig. 1). The entire region spanning JAKMIP1 and LOC285484 exhibits very low LD and contains several LD blocks, one of which spans the 5′ ends of both JAKMIP1 and LOC285484. Many of the SNPs within this block were associated at P values <0.001 (see SI Fig. 9 for single-marker association plots).
The most significantly associated SNPs (all in nearly complete LD, r2 = 0.996) in the QFP (P values <10−5) were located within an intron of LOC285484. The most significant SNP within the JAKMIP1 gene itself lies within the first intron (rs9991241, P = 4.17 × 10−4). The risk allele was present in 86.5% of cases and 80.3% of controls (OR of 1.56, 95% C.I. 1.22–2.01). In the German trios, the most significantly associated SNPs were located 3′ to LOC285484. Four SNPs with P values <0.001 in the German trios lie within a 2-kb region and are in complete LD, with risk allele frequencies of 53% in the cases and 45% in the controls (OR 1.41, 95% C.I. 1.16–1.72). These SNPs were also associated in the QFP with similar P values. The risk and protective alleles, however, are reversed in the two trio samples. This phenomenon of association with opposite risk and protective alleles has been observed for other genes associated with complex disease in multiple populations. Although this lack of consistency can be the result of type 1 error, it has been argued that such replications may indeed be real, and that the “flip-flop” may be due to complex interactions with other disease loci (27).
Region 3p21.3.
A replicated region is located on chromosome 3p21.3 (Fig. 1). Although this region was not among the most highly significant regions identified from the GWA study, it was selected for FM and replication studies based on the presence of strong functional candidates. The lowest nominal P value from the GWA study within this region (9.55 × 10−5) occurred at marker rs11718165, within an intron of the bassoon (BSN) gene. Two distinct association signals were observed in the FM analysis of QFP samples in two adjacent LD blocks spanning ≈800 kb, one centered at rs4855873 (P = 8.13 × 10−5), and one centered at rs1131095 (P = 1.99 × 10−6). These two associated regions were replicated in the German trios (P = 2.09 × 10−4 and P = 1.0 × 10−4 for five-marker haplotypes) and appear to be independent (r2 between haplotypes = 0.1830 in QFP trios, r2 = 0.2972 in German trios). Multiple single-marker associations (P values <10−5) were observed within both of these regions in the combined QFP and German trio analysis. In the full combined analysis, 17 single-marker associations at P values <10−5 were observed in the first telomeric region (minimum P value = 3.16 × 10−7 at rs6997). Five single-marker associations significant at P < 10−7 were observed in the second centromeric region (minimum P value = 1.26 × 10−8 at rs9822268, SI Fig. 10). This region, which exhibited only moderate statistical evidence in the GWA study (9.55 × 10−5), became the third-highest rank in the combined analyses with P values for haplotype association of 1.82 × 10−9 (all trios) and 3.24 × 10−15 (all three samples).
Additional Regions.
Two additional regions demonstrated significant replication in the German case/control sample. Both of these regions are relatively large and may span multiple independent signals. The first, on 17q11, spans three relatively large blocks of LD and exhibits multiple signals with minimum P values of 5.13 × 10−5 in the QFP and 1.29 × 10−7 in the German case/control sample (SI Fig. 5). The second region, on chromosome 17q23, spans at least three large LD blocks. Again, multiple signals were observed in this region, with minimum P values of 5.39 × 10−4 in the QFP and 1.10 × 10−5 in the German cases/controls (SI Fig. 6).
Discussion
Recent GWA studies, including the one presented here, are beginning to reveal a clear picture of the major susceptibility genes and loci for CD (15–21). We identified three of the most replicated loci within our top seven GWA signals (NOD2, IBD5, and IL23R). Other loci that have been reported, such as the ATG16L1 gene, the IRGM gene, a locus on 1q24, and an intergenic region near the PTGER4 gene, are also observed in this study but exhibit lower statistical significance (≈0.001). One of our most convincing replicated regions (3p21.3) was also observed in the most recent published GWA study (16, 21). The lack of complete overlap in association signals from the recent GWA studies may be due to genetic heterogeneity, differences in phenotype definition, sample sizes, and SNP distribution in the various scans.
We have also presented the most extensive analysis to date of the association of the IL23R region with CD. Our results suggest that it is highly unlikely that the nonsynonymous SNP rs11209026 (R381Q) fully explains the functional role of this gene in CD etiology. This SNP does not occur consistently in all risk and protective haplotypes. In addition, there is strong evidence for an epistatic interaction between polymorphisms located in the 5′ and 3′ end of the gene. It is possible that variants controlling isoform expression of IL23R are the major causative mutations. Interestingly, we have shown that a short isoform is up-regulated in CD patients.
The region on 4p16.1 contains two candidate genes (JAKMIP1 and LOC285484). JAKMIP1 is involved in IL23 signaling and binds to TYK2, a member of the Janus (tyrosine) kinase (Jak) family, which is associated with the IL12RB chain and mediates STAT-4 activation (28). The alternative disease gene at this locus, LOC285484, contains a characteristic high-mobility group (HMG)-box domain. The prototypic family member, HMGB1, is a DNA-binding molecule that binds single-stranded DNA and unwinds double-stranded DNA; however, HMGB1 is also a secreted cytokine and may serve as a danger signal during acute and chronic inflammation after release by damaged cells and activated macrophages (29). The similarity of LOC285484 to HMGB1 suggests that it could be involved in aspects of intestinal inflammation and remodeling.
The region on 3p21.3 contains two adjacent but apparently independent replicated regions with strong candidate CD genes, including GPX1, RHOA, DAG1 BSN, APEH, and MST1. GPX1, located in the more telomeric region, encodes the powerful antioxidant, glutathione peroxidase isoform 1. Double-knockout mice lacking both GPX1 and GPX2 activity were found to have an inflammatory bowel disease phenotype (30). RHOA, also in the telomeric region, encodes a small GTPase involved in blockage of stress fiber formation in the innate cellular immune response in the presence of effector proteins released by pathogenic Yersinia species (31). The more centromeric region contains the BSN gene, which encodes a scaffolding protein expressed in axons. MST1, macrophage stimulatory protein 1, is involved in inflammation and tissue remodeling for wound healing. APEH (APH) encodes a serine peptidase with a reported functional role in the degradation of bacterial peptide breakdown products in the gut to prevent excessive immune response (32).
The results presented here illustrate the power of GWA studies to identify networks of disease susceptibility genes. We believe the success of this study is due in large part to the utilization of genome-wide haplotype association analyses, in addition to the use of unique population samples from the QFP. The biochemical pathways associated with CD susceptibility genes suggest a model of CD that appears to converge on pathophysiological pathways central to epithelial defense mechanisms, the interplay between innate and adaptive immune response, and the repair or remodeling of tissue as part of a response to inflammatory and damage-induced stimuli. Such a model provides multiple potential targets for new therapies for CD.
Methods
Details of the methods used in this study are available in SI Text.
Population Samples.
The population sample used for the GWA study consisted of 382 CD patients collected within parent–parent–child trios sampled from the QFP. Recruitment into the study was subject to ethical review by various affiliated ethics review committees (SI Table 13), and all subjects gave written informed consent for participation in the study. Two additional samples were used for replication studies. These consisted of 521 affected child trios, 752 cases, and 828 independent controls, all from Northern Germany. German control individuals were obtained from the POPGEN biobank (33).
CD was diagnosed by colonoscopy, barium radiological examination during abdominal surgery, or exploratory biopsy, with patients showing signs of CD in either the colon or ileum or in both locations (SI Table 14). The expression of CD included fistulizing, stenosing, and inflammatory forms. Ulcerative colitis, acute infectious colitis, and indeterminate colitis were excluded.
Genotyping.
The genome-wide scan SNP map was designed and genotyping performed by Perlegen Sciences, Mountain View, CA. Markers were chosen to maximize uniformity of coverage and were distributed approximately evenly throughout the genome. After removal of insufficiently polymorphic markers, genotypes for 164,279 markers were analyzed. For fine-mapping and replication studies, additional equally spaced SNPs to a density of one SNP per 2–5 kb within these regions were genotyped by using the Illumina GoldenGate platform.
Statistical Analyses.
Genotype data were cleaned before analysis by removing markers or individuals that did not fit defined tolerances of genotype quality. These thresholds included a maximum of 1% missing values, conformity to Hardy–Weinberg equilibrium for markers, and conformity to Mendelian segregation patterns within trios.
Phase information was first deduced from the trio genotype information with the remaining unresolved phase assignments estimated using a modified version of the PL-EM algorithm (34). Haplotypes were estimated within 11-marker overlapping blocks, which advanced in one-marker increments across the chromosome. Estimated haplotypes were then ligated into chromosome long haplotypes by using trio information. Final phase assignment for any ambiguous marker was determined from consensus within the 21 sliding blocks within which it was contained. For the German case/control samples, phase was estimated directly by the PL-EM algorithm using 11 marker blocks.
Because ≈25% of the trios had affected parents, the data were not analyzed by using the transmission disequilibrium test. Instead, for the parent-affected trios, spousal chromosomes were used as controls. For child-affected trios, parental nontransmitted chromosomes were used as controls. Case-control analyses were performed by using a sliding window of one, three, five, seven, or nine markers. Case/control allele or haplotype frequency tables were constructed at each window position and size and, from these, a χ2 value was calculated. A Pearson's χ2 statistic with 1 degree or 2 degrees of freedom was calculated and nominal asymptotic P values determined for significance of association for single markers and for three-marker haplotypes, respectively. For five-, seven-, and nine-marker haplotypes, the expected values and variances of the square root of calculated χ2 values were determined by the method of Smith (35) and used to construct a Z score. A nominal P value was calculated from this Z score.
Nominal P values were used to define candidate regions identified by the GWS. Peaks were identified as regions containing a maximum nominal−log10 P value >3.0 (P ≤ 0.001) and extending to the left and right until all haplotype or single marker−log10 P values dropped below 1.5. P values for multimarker haplotypes were also calculated exclusively by permutation of case control status.
Genome-Wide Significance.
The genome-wide significance of the observed nominal asymptotic P value at peaks was determined by performing random permutations of case and control status of haplogenotypes within trios, keeping haplotypes intact. For each permuted data set, a new GWA analysis was performed, and peaks were identified and ranked. Five hundred permutations were performed to construct the null distribution of the first, second, third, etc., order statistics for the −log10 P values across the genome. The observed peaks in the original GWA study, ranked by their nominal −log10 P values, were compared with the null distribution of their corresponding order statistics to determine genome-wide significance. A conservative upper bound and a range for the genome-wide significance of individual nominal P values were obtained by successively removing (peeling), one at a time, the highest remaining signal that was genome-wide significant (P ≤ 0.05) and measuring nominal P values of the remaining peaks against the distribution of order statistics shifted by one.
Significance in the Replication Study.
Statistical significance of replication was determined by permutation studies in which case and control status of the German samples was randomly permuted to produce the null distribution of nominal P values within the German samples. Peak-wide replication significance in the German samples was estimated by first extracting the peak region from the QFP GWA analysis and then comparing it with peaks from the analysis of the German samples. An overlap of peaks would occur if a peak in the QFP study and a peak in the German sample both reside in the intersection of the two candidate peak regions. The peak-wide replication significance (based on 500 random permutations of case-control status) measures how often by chance alone an overlap occurs with a P value as significant as was observed in the replication sample. The permutation procedure corrects for all SNPs and SNP combinations considered to define a peak and its surrounding peak region and therefore represent a true peak-wide replication probability. The permutations were performed without taking the phenotypic status of the parents into account.
Sequencing.
Sixteen QFP samples were selected to represent at least two copies of all major haplotypes (≥5% frequency) and one copy of minor haplotypes (1–5% frequency) observed. A region of 130 kb spanning the IL23R locus was sequenced and 339 SNPs, 60 microsatellites, and 18 insertions/deletions were identified (SI Table 8D). A region of 60 kb spanning the JAKMIP1/LOC285484 association locus was sequenced, and 226 SNPs, 16 microsatellites, and 20 insertions/deletions were identified (SI Table 9D).
Immunohistochemistry.
Paraformaldehyde-fixed paraffin-embedded biopsies were prepared from five normal controls and five patients with confirmed colonic CD. Two slides of each biopsy were stained with hematoxylin/eosin for routine histological evaluation. The other slides were subjected to a citrate-based antigen retrieval procedure, permeabilized by incubation with 0.1% Triton X-100 in 0.1 M PBS, washed three times in PBS, and blocked with 0.75% BSA in PBS for 20 min. Sections were subsequently incubated with the primary antibody (anti-IL23R, Abcam, Cambridge, MA) at a 1:200 dilution in 0.75% BSA for 1 h at room temperature. After washing in PBS, tissue-bound antibody was detected by using biotinylated goat anti-rabbit (Vector Laboratory, Burlingame, CA) and followed by HRP-conjugated avidin, both diluted at 1:100 in PBS. Controls were included by using irrelevant primary antibodies and omitted the primary antibodies by using only secondary antibodies and/or HRP-conjugated avidin. No significant staining was observed with any of these controls (data not shown).
Supplementary Material
Acknowledgments
We thank the patients and their families and physicians for contributions to the collection. Valuable contributions to this work were made by S. Briand, V. Bruat, O. Gingras, E. Hardy, N. Henderson, J. F. Levesque, T.-V. Nguyen, V. Pinchuk, C. Prive, V. Serre, D. St. Louis, B. Stojkovic, G. te Meerman, N. Malo, T. Kaacksteen, R. Vogler, T. Wesse, I. E. Baumgartner, and S. Ehlers. We thank Perlegen Sciences, Inc., for the high-throughput genotyping used in the GWA study and T. Behrens and N. Ghilardi for comments on the manuscript. The German population collection was financed by grants from the German Crohn's and Colitis Foundation (DCCV), the National Genome Research Network (NGFN), the Federal Ministry of Education and Research (BMBF), the European Commission, and the German Research Foundation (DFG).
Abbreviations
- GWA
genome-wide association
- CD
Crohn's disease
- QFP
Quebec Founder Population
- FM
fine mapping
- C.I.
confidence interval
- OR
odds ratio
- LD
linkage disequilibrium.
Footnotes
Conflict of interest statement: Genizon BioSciences, Inc., has received all commercial rights in the collaboration from the University Hospital Schleswig–Holstein. S.S. is a member of the scientific advisory board of Applied Biosystems, Inc., and has consulted for Abbott, Centocor, and Schering–Plough.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706645104/DC1.
References
- 1.Podolsky DK. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Lashner BA. Gastroenterol Clin North Am. 1995;24:467–474. [PubMed] [Google Scholar]
- 3.Halme L, Paavola-Sakki P, Turunen U, Lappalainen M, Farkkila M, Kontula K. World J Gastroenterol. 2006;12:3668–3672. doi: 10.3748/wjg.v12.i23.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hampe J, Grebe J, Nikolaus S, Solberg C, Croucher PJ, Mascheretti S, Jahnsen J, Moum B, Klump B, Krawczak M, et al. Lancet. 2002;359:1661–1665. doi: 10.1016/S0140-6736(02)08590-2. [DOI] [PubMed] [Google Scholar]
- 5.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, et al. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 6.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, et al. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 8.Brant SR, Panhuysen CI, Nicolae D, Reddy DM, Bonen DK, Karaliukas R, Zhang L, Swanson E, Datta LW, Moran T, et al. Am J Hum Genet. 2003;73:1282–1292. doi: 10.1086/379927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho GT, Soranzo N, Nimmo ER, Tenesa A, Goldstein DB, Satsangi J. Hum Mol Genet. 2006;15:797–805. doi: 10.1093/hmg/ddi494. [DOI] [PubMed] [Google Scholar]
- 10.McGovern DP, Hysi P, Ahmad T, van Heel DA, Moffatt MF, Carey A, Cookson WO, Jewell DP. Hum Mol Genet. 2005;14:1245–1250. doi: 10.1093/hmg/ddi135. [DOI] [PubMed] [Google Scholar]
- 11.Panwala CM, Jones JC, Viney JL. J Immunol. 1998;161:5733–5744. [PubMed] [Google Scholar]
- 12.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, Gu X, Newman B, Van Oene M, Cescon D, Greenberg G, et al. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- 13.Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S, et al. Nat Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- 14.Stoll M, Corneliussen B, Costello CM, Waetzig GH, Mellgard B, Koch WA, Rosenstiel P, Albrecht M, Croucher PJ, Seegert D, et al. Nat Genet. 2004;36:476–480. doi: 10.1038/ng1345. [DOI] [PubMed] [Google Scholar]
- 15.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, et al. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, et al. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, et al. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, et al. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 19.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, et al. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T, et al. Hum Mol Genet. 2005;14:3499–3506. doi: 10.1093/hmg/ddi379. [DOI] [PubMed] [Google Scholar]
- 21.Wellcome Trust Case Control Consortium. Nature. 2007;447:661–678. [Google Scholar]
- 22.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, et al. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charbonneau H, Desjardins B, Guillemette A, Landry Y, Legare J, Nault F. The First French Canadians: Pioneers in the St. Lawrence Valley. Newark: Univ of Delaware Press; 1993. [Google Scholar]
- 24.Gagnon A, Heyer E. Am J Phys Anthropol. 2001;114:30–41. doi: 10.1002/1096-8644(200101)114:1<30::AID-AJPA1003>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Betard C, Kessling AM, Roy M, Chamberland A, Lussier-Cacan S, Davignon J. Hum Genet. 1992;88:529–536. doi: 10.1007/BF00219339. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XY, Zhang HJ, Zhang Y, Fu YJ, He J, Zhu LP, Wang SH, Liu L. Immunogenetics. 2006;57:934–943. doi: 10.1007/s00251-005-0067-0. [DOI] [PubMed] [Google Scholar]
- 27.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watford WT, Hissong BD, Bream JH, Kanno Y, Muul L, O'Shea JJ. Immunol Rev. 2004;202:139–156. doi: 10.1111/j.0105-2896.2004.00211.x. [DOI] [PubMed] [Google Scholar]
- 29.Yang D, Chen Q, Yang H, Tracey KJ, Bustin M, Oppenheim JJ. J Leukocyte Biol. 2007;81:59–66. doi: 10.1189/jlb.0306180. [DOI] [PubMed] [Google Scholar]
- 30.Esworthy RS, Aranda R, Martin MG, Doroshow JH, Binder SW, Chu FF. Am J Physiol. 2001;281:G848–G855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- 31.Aepfelbacher M, Zumbihl R, Heesemann J. Curr Top Microbiol Immunol. 2005;291:167–175. doi: 10.1007/3-540-27511-8_9. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen KT, Pei D. Biochemistry. 2005;44:8514–8522. doi: 10.1021/bi050191o. [DOI] [PubMed] [Google Scholar]
- 33.Krawczak M, Nikolaus S, von Eberstein H, Croucher PJ, El Mokhtari NE, Schreiber S. Commun Genet. 2006;9:55–61. doi: 10.1159/000090694. [DOI] [PubMed] [Google Scholar]
- 34.Qin ZS, Niu T, Liu JS. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith CA. Ann J Hum Genet. 1986;50:163–167. doi: 10.1111/j.1469-1809.1986.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.