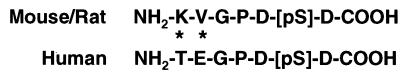Abstract
The tumor suppressor p53 is a nuclear phosphoprotein in which DNA-binding activity is increased on exposure to DNA-damaging agents such as UV or γ radiation by unknown mechanisms. Because phosphorylation of p53 at the casein kinase (CK) II site activates p53 for DNA-binding function in vitro, we sought to determine the in vivo relevance of phosphorylation at this site after UV and γ radiation. A polyclonal antibody was generated that binds to bacterially expressed p53 only when phosphorylated in vitro by CK II. Using this antibody, we showed that p53 is phosphorylated at the CK II site upon UV treatment of early passage rat embryo fibroblasts and RKO cells. In addition, DNA-binding assays indicated that phosphorylated p53 bound to a p53-responsive element, suggesting functional activation. However, γ radiation, which also stabilizes p53, did not result in phosphorylation at the CK II site. These results indicate that phosphorylation at the CK II site is one of the post-translational mechanisms through which p53 is activated in response to UV radiation and that different mechanisms activate p53 after DNA damage by γ radiation.
The p53 tumor suppressor, mutated in over 50% of human cancers (1), suppresses oncogene-mediated cell transformation (2, 3) and inhibits proliferation of transformed cells (4, 5). After DNA damage, increased levels of p53 block cell cycle progression, leading to growth arrest or apoptosis (6, 7). The DNA-binding ability and the transcriptional activity of p53 are induced on exposure to several DNA-damaging agents such as UV and γ radiation (6, 7).
p53 is phosphorylated at multiple sites by several kinases such as DNA-dependent protein kinase (8), protein kinase C (PKC; ref. 9), mitogen-activated protein kinase (10), CK I (11), and CK II (12). In vitro, the DNA-binding function of p53 is activated by phosphorylation of the C-terminal serine (serine 389 in mouse) by purified CK II (13). A glutamic acid mutant at the C-terminal serine in murine p53 yields a functional p53 in transient transfection assays in which wild-type p53 is unable to activate transcription or bind DNA (14). In addition, mutation of the C-terminal serine to alanine has been shown to abolish the antiproliferative activity of murine p53 (15). A C-terminal deletion of p53 (including the terminal serine) or binding to a C-terminal-specific mAb, pAb421, also activates the sequence-specific DNA-binding activity of p53 (13). Furthermore, the DNA-binding function of latent p53 can be activated by incubation of latent p53 tetramers with synthetic peptides derived from the C-terminal region of p53 (16). Taken together, these data suggest that the C-terminal region of p53 contains a negative regulatory domain that may be suppressed by modification or deletion of this domain. Phosphorylation of the C-terminal serine may alter the conformation of p53 such that it becomes active for DNA-binding and transactivation.
However, conflicting reports have emerged concerning the role of phosphorylation at this site for the function of p53. Several studies suggest that phosphorylation by CK II is not required for transcriptional activity of p53. Rat and murine p53 retained DNA-binding activity when the C-terminal serine was mutated to alanine (17, 18). Fiscella et al. (19) reported that human p53, with the serine in the CK II site mutated to alanine, retained the ability to block cell cycle progression in transient transfection experiments.
Thus, the role of phosphorylation at the CK II site in p53 function remains unclear. Using a polyclonal antibody that specifically binds to p53 phosphorylated at the CK II site, we showed that p53 is activated by phosphorylation at this site upon UV but not γ radiation, suggesting that different kinds of DNA lesions activate p53 through different mechanisms. Further, phosphorylation at the CK II site activated p53 for DNA-binding. This report shows direct in vivo evidence for activation of p53 via phosphorylation at the CK II site upon DNA damage.
MATERIALS AND METHODS
Cell Culture.
Rat embryo fibroblasts, RKO cells, and A1–5 cells were cultured in DMEM supplemented with 10% fetal calf serum. Cells were grown to 80% confluency in 10-cm plates before UV or γ radiation. Cells were treated with UV-C radiation using a Stratalinker model (Stratagene) or γ radiation from a Cs137 source.
Generation of Ab389.
Ab389 was raised against a C-terminal phosphopeptide of murine p53 by Quality Controlled Biochemicals, Hopkington, MA. For affinity purification, the crude serum was first bound to thiol-coupling gel (Quality Controlled Biochemicals) linked to a nonphosphorylated version of the peptide. The flow-through was passed through a second column with thiol-coupling gel linked to the phosphopeptide. The phosphopeptide-specific antibody was eluted in 100 mM glycine, pH 2.5.
Enzymatic Assays.
Calf intestinal alkaline phosphatase (New England Biolabs) treatment was done in buffer containing 50 mM Tris⋅Cl (pH 7.9), 10 mM MgCl2, 100 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM DTT at 37°C for 1 h. In vitro phosphorylation reactions were performed at 30°C for 30 min using CK I (in buffer containing 50 mM Tris⋅Cl (pH 7.5), 10 mM MgCl2, 5 mM DTT, 1 mM PMSF, and 100 μM ATP; Calbiochem); CK II (in buffer containing 20 mM Tris⋅Cl (pH 7.5), 50 mM KCl, 10 mM MgCl2, 1 mM PMSF, and 100 μM ATP; Calbiochem), or PKC (in buffer containing 20 mM Mops, 25 mM glycerophosphate, 1 mM sodium orthovanadate, 1 mM CaCl2, 1 mM DTT, 1 mM PMSF, 0.5 mg/ml phosphatidyl serine, 0.5 mg/ml diglycerides, and 100 μM ATP; Upstate Biotechnology, Lake Placid, NY). All reactions were supplemented with 200 μCi/mmol of [γ-32P]ATP (DuPont).
DNA Binding Assay.
Five micrograms of extract and 1 × 105 cpm of a [32P]-end-labeled fragment of DNA containing the p21 promoter were used for each reaction (20). The immunoprecipitated DNA-protein complexes were digested with 10 μg of proteinase K (Upstate Biotechnology), and the bound DNA was analyzed on 5% nondenaturing polyacrylamide gel followed by autoradiography.
RESULTS AND DISCUSSION
To determine the in vivo significance of phosphorylation for p53 function, we synthesized a polyclonal antibody against the C-terminal phosphopeptide of murine p53, encompassing the phosphorylated serine 389. This peptide sequence is identical to the C terminus of rat p53 and has high homology with the C terminus of human p53 (Fig. 1).
Figure 1.
Sequence comparison of the C-terminal phosphopeptide of murine p53 used for generating Ab389 (QCB) with the C-terminal sequence of human p53.
Ab389 Is Specific to p53 Phosphorylated at CK II site.
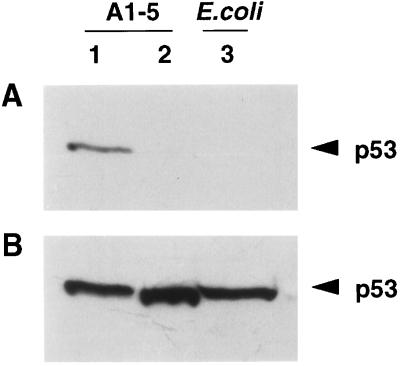
To determine whether the phosphopeptide antibody (Ab389) is specific to phosphorylated p53, A1–5 cells, which express a temperature-sensitive murine p53 (21), were shifted to permissive temperature and extracts from these cells were treated with calf intestinal alkaline phosphatase (CIP). Ab389 bound p53 from untreated extracts (Fig. 2A, lane 1) but not from CIP-treated extracts (Fig. 2A, lane 2). In addition, Ab389 did not recognize Escherichia coli-expressed human p53 (Fig. 2A, lane 3). To ensure that CIP treatment had not degraded the protein, the Western blot was stripped and reprobed with the p53 mAb, pAb421, which indicated that all lanes had similar amounts of p53 (Fig. 2B). These data suggest that Ab389 specifically recognizes a phosphorylated p53.
Figure 2.
Ab389 does not bind to dephosphorylated murine p53. (A) Extracts from A1–5 cells expressing a wild-type p53 at the permissive temperature of 32°C were treated with CIP at 37°C for 1 h. Untreated (lane 1) and CIP-treated extracts (lane 2) were then resolved on 10% SDS/PAGE followed by Western blotting using Ab389. Enhanced chemiluminescence kit (Amersham) was used to detect antibody-specific proteins. Extracts from E. coli cells expressing human p53 were run in lane 3. (B) The Western blot from a was stripped and reprobed with p53-specific mAb, pAb421.
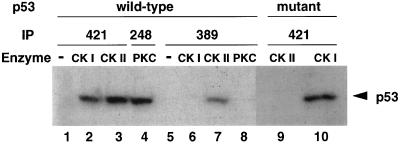
To test whether Ab389 specifically recognizes p53 phosphorylated at the CK II site, crude extracts from E. coli cells expressing human p53 were phosphorylated in vitro using either CK II, which phosphorylates the C-terminal serine (392 in human; ref. 12), CK I, which phosphorylates N-terminal serine residues (11), or protein kinase C, which phosphorylates serine 378 (9). p53 was immunoprecipitated using either pAb421 or Ab389 (Fig. 3). In addition, pAb248, a mAb that binds to the N-terminal region of p53, was used to immunoprecipitate p53 phosphorylated by PKC because pAb421 is unable to bind p53 phosphorylated by this kinase (22). Ab389 bound p53 only when phosphorylated by CK II (Fig. 3, lane 7) and not when phosphorylated by CK I (Fig. 3, lane 6) or PKC (Fig. 3, lane 8), suggesting that this antibody is specific to p53 phosphorylated at the CK II site. Controls (Fig. 3, lanes 2–4) showed that pAb421 or pAb248 bound p53 when phosphorylated by CK I, CK II, or PKC.
Figure 3.
Ab389 binds human p53 when phosphorylated in vitro by CK II. Extracts from E. coli cells expressing wild-type (lanes 1–8) or mutant (serine to alanine substitution at 392; lanes 9 and 10) human p53 were used for in vitro phosphorylation reactions. Extracts were treated with CK I (lanes 2, 6, and 10), CK II (lanes 3, 7, and 9), or with PKC (lanes 4 and 8). All reactions were supplemented with 200 μCi/mmol of [γ-32P]ATP and were performed at 30°C for 30 min followed by immunoprecipitation by using pAb421 (lanes 1–3 and 9–10), pAb248 (lane 4), or Ab389 (lanes 5–8) and then SDS/PAGE analysis. Lanes 1 and 5 are controls in which no enzyme was added in the phosphorylation reactions.
To verify that the C-terminal serine is the only site phosphorylated by CK II, we generated a mutant p53 with a serine to alanine substitution at this site. This mutant form of p53 could be phosphorylated by CK I (Fig. 3, lane 10) but not by CK II (Fig. 3, lane 9). Taken together, these data indicate that Ab389 specifically binds to p53 when phosphorylated at the C-terminal serine.
Ab389 Binds to p53 After UV but not γ Radiation.
Next, we used Ab389 as a tool to determine the in vivo significance of phosphorylation of p53 at the CK II site. Because the DNA binding activity of p53 is increased upon treatment with DNA-damaging agents, we sought to determine the phosphorylation status of the C-terminal serine of p53 upon UV and γ radiation. Although both UV and γ radiation cause DNA damage and an increase in p53 levels, γ radiation results in single- and double-strand breaks, whereas UV causes DNA to form bulky adducts on individual bases, leading to mismatches, insertions, and deletions. Although p53 is activated in both cases, the types of lesions to be repaired are very different.
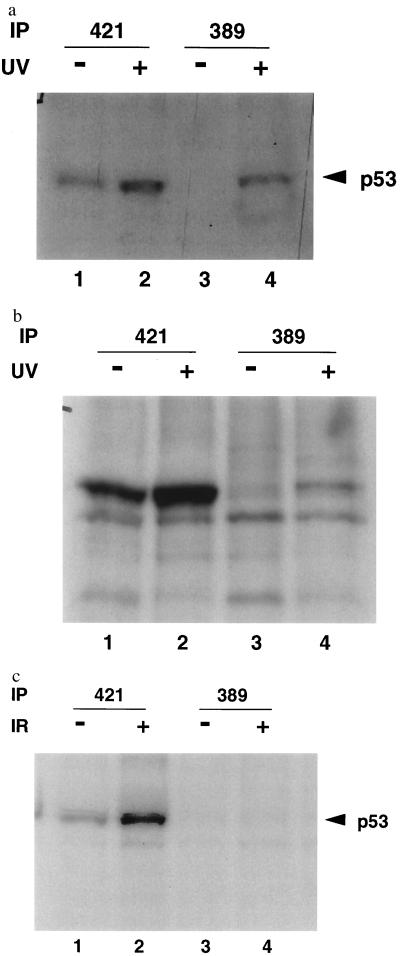
A human colorectal carcinoma cell line, RKO (23) was treated with 30 J/m2 of UV radiation. Immunoprecipitation with pAb421 showed that this treatment caused an increase in p53 levels (Fig. 4a, cf. lanes 1 and 2) as has been demonstrated (24). Ab389 showed enhanced binding to p53 in UV-treated extracts (Fig. 4a, cf. lanes 3 and 4) in multiple experiments. Consistently, only a fraction of the p53 protein bound to Ab389 upon UV treatment (Fig. 4a, cf. lanes 2 and 4). Increasing the amount of Ab389 used for immunoprecipitation in these experiments did not change the level of p53 immunoprecipitated. To ensure that phosphorylation at the C-terminal serine was not limited to RKO cells, a tumor derived cell line, we treated normal rat embryo fibroblasts with 10 J/m2 of UV radiation. As with RKO cells, rat embryo fibroblasts showed increased binding to Ab389 upon UV treatment (Fig. 4b, lane 4). These results suggest that p53 is phosphorylated at the CK II site upon UV treatment.
Figure 4.
Ab389 binds to p53 from UV-treated cells. RKO cells (a) and early passage rat embryo fibroblasts (b) were treated with 10 J/m2 (30 J/m2 for RKO cells) and metabolically labeled with [S35]methionine and [S35]cysteine for 30 min, 24 h (2 h for RKO cells) after radiation. Cells were harvested and lysed as described (14). Extracts from untreated (lanes 1 and 3) and UV-treated (lanes 2 and 4) cells were used for immunoprecipitation with pAb421 (lanes 1 and 2) or Ab389 (lanes 3 and 4) followed by SDS/PAGE and autoradiography. (c) RKO cells were treated with 6 Gy of γ radiation. Four hours after radiation, cells were metabolically labeled and harvested as described above. Extracts from these cells were used for immunoprecipitation with pAb421 (lanes 1 and 2) or Ab389 (lanes 3 and 4) followed by SDS/PAGE and autoradiography.
To determine whether phosphorylation at the CK II site also occurred upon γ radiation, RKO cells were treated with 6 Gy of γ radiation. p53 was induced as can be seen in pAb421 immunoprecipitates (Fig. 4c, cf. lanes 1 and 2). However, Ab389 did not bind to p53 upon activation by γ radiation (Fig. 4c, cf. lanes 3 and 4). These data suggest that p53 becomes phosphorylated at the CK II site upon UV but not γ radiation.
p53 Bound to Ab389 is Functionally Active.
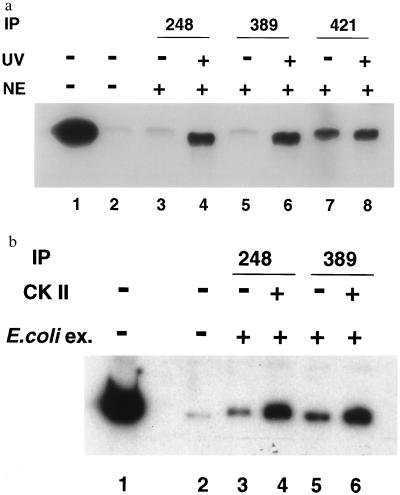
To determine whether Ab389 bound a functional p53, we performed DNA-binding assays (Fig. 5a). p53 from untreated and UV treated rat embryo fibroblasts was bound to a radiolabeled DNA fragment containing the p53-responsive element of the p21 promoter (25). The DNA-protein complex was immunoprecipitated with pAb248, Ab389, or pAb421. The ability of p53 to bind the p21 promoter was enhanced after UV treatment as can be seen in pAb248 immunoprecipitates (Fig. 5a, cf. lanes 3 and 4). Ab389 bound p53 from UV-treated cells, which suggests that Ab389 recognizes only an active p53 that can bind DNA (Fig. 5a, cf. lanes 5 and 6). pAb421, which activates the latent DNA-binding activity of p53, bound well to p53 in extracts from both UV-treated and untreated cells (Fig. 5a, lanes 7 and 8). Of importance, whereas pAb421 bound and activated p53 irrespective of UV treatment, the binding of Ab389 did not in and of itself activate p53 for binding DNA.
Figure 5.
p53 bound to DNA is immunoprecipitated by Ab389. (a) Nuclear extracts (NE) were prepared from UV-treated and untreated rat embryo fibroblasts, and DNA-binding assays were performed as described (20). Extracts from untreated (lanes 3, 5, and 7) and UV-treated cells (lanes 4, 6, and 8) were incubated with the radiolabeled DNA containing the p21 promoter (20) for 30 min at 4°C. DNA-protein complexes were immunoprecipitated by either pAb248 (lanes 3 and 4), Ab389 (lanes 5 and 6), or pAb421 (lanes 7 and 8), and the bound DNA was analyzed on 5% nondenaturing polyacrylamide gel followed by autoradiography. Lanes 1 and 2 contain free probe and no nuclear extract, respectively. (b) Bacterially expressed p53 was phosphorylated in vitro with CK II (as described in Fig. 3). Untreated and phosphorylated extracts were used for DNA-binding assays as described above. Immunoprecipitations were performed with pAb248 (lanes 3 and 4) or Ab389 (lanes 5 and 6), and bound DNA was analyzed as above. Lanes 1 and 2 contain free probe and no nuclear extract, respectively.
The data presented above do not address the question whether phosphorylation of p53 by CK II is sufficient for activation of its DNA binding activity. Therefore, to determine whether phosphorylation at the CK II site is activating p53 for DNA-binding, we used bacterially expressed p53 for in vitro phosphorylation with CK II and performed DNA-binding assays (Fig. 5b). CK II treatment activated p53 for binding to the p21 promoter (Fig. 5b, lanes 4 and 6), whereas unphosphorylated p53 was incapable of binding DNA. Thus in our system, as has been shown (13), phosphorylation by CK II was sufficient for activating the DNA binding activity of p53.
In summary, we have shown that Ab389 is a phosphoserine-specific antibody that recognizes p53 from several mammalian species (mouse, human, and rat) when phosphorylated at the CK II site and that p53 is phosphorylated at this site upon treatment with UV but not γ radiation. DNA-binding assays indicate that the p53 bound to Ab389 is active. This is the first in vivo evidence for phosphorylation at the CK II site and activation of p53 upon UV treatment. However, not all of the active p53 is phosphorylated suggesting that phosphorylation of the C-terminal serine may be one of the mechanisms through which p53 is activated upon exposure to UV radiation. Our data support the existence of other mechanisms of p53 activation upon UV treatment because only a subset of active p53 bound Ab389 (Fig. 4 a and b).
UV and γ radiation cause different kinds of DNA lesions and trigger different repair pathways. Whereas UV radiation activates nuclear excision repair, γ radiation results in the activation of the base excision repair machinery. Our data suggest that UV and γ radiation activate p53 through different mechanisms. This hypothesis is bolstered by the recent finding that phosphorylation of p53 serine 15 occurs after γ radiation and disrupts the binding to an inhibitor of p53, MDM2 (26). Thus, it is possible that site-specific modification of p53 allows differential interaction with proteins involved in a specific repair pathway. The fact that p53 is phosphorylated at the C-terminal serine by UV, but not γ radiation, may explain also why different assay conditions have resulted in conflicting reports as to the importance of phosphorylation at the CK II site.
Our data show that p53 is activated via phosphorylation at the CK II site in response to UV radiation. However, this does not exclude the possibility that concomitant phosphorylation and/or dephosphorylation at other sites also play a role in the activation of p53 upon UV treatment. p53 has been shown to be phosphorylated by mitogen-activated protein kinase (10) and c-Jun kinase (27) in response to UV radiation. However the significance of phosphorylation by these kinases for p53 function is not yet known.
Other mechanisms that have been shown to activate p53 include glycosylation, binding to p53 antibody pAb421, C-terminal deletion of p53, binding to specific interacting proteins, and alternate splicing of p53 mRNA (28, 13, 29, 30). Specifically, DNA damage activates p53 in some systems by binding poly (ADP ribose) polymerase, by alternative splicing, and by proteolysis (31, 32, 33). In addition, both the conformation of p53 and the sequence-specific DNA-binding activity are modulated also by metal chelators and oxidizing agents (34).
The possibility that multiple pathways activate p53 function stresses the critical role of this tumor suppressor in regulating cell growth. Activation of p53 upon DNA damage appears to be a complicated process composed of several pathways converging to yield a functional p53. Whereas some of these pathways may activate p53 through phosphorylation of the C-terminal serine, others may involve phosphorylation at other sites or may be independent of phosphorylation.
Acknowledgments
We thank Dr. J. Little for the gift of RKO cells, Dr. V. Reinke for helpful discussions, and G. Zong for technical support. This work was supported by a grant from the National Cancer Institute (CA47296) to G. L and core Grant CA16672.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: CK, casein kinase; PKC, protein kinase C; PMSF, phenylmethylsulfonyl fluoride; CIP, calf intestinal alkaline phosphatase.
References
- 1.Hainaut P, Soussi T, Shomer B, Hollstein M, Grenblatt M, Hovig E, Harris C C, Montesano R. Nucleic Acids Res. 1997;25:151–157. doi: 10.1093/nar/25.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finlay C A, Hinds P W, Levine A J. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 3.Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Proc Natl Acad Sci USA. 1989;86:8763–8767. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen P-L, Chen Y M, Bookstein R, Lee W-H. Science. 1990;250:1576–1580. doi: 10.1126/science.2274789. [DOI] [PubMed] [Google Scholar]
- 5.Baker S J, Markowitz S, Fearon E R, Willson J K, Vogelstein B. Science. 1990;249:912–915. doi: 10.1126/science.2144057. [DOI] [PubMed] [Google Scholar]
- 6.Kastan M B, Onyekwere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 7.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 8.Lees-Miller S P, Sakaguchi K, Ullrich S J, Appella E, Anderson C W. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence J J. Proc Natl Acad Sci USA. 1992;89:11627–11647. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne D M, Campbell D G, Caudwell F B, Meek D W. J Biol Chem. 1994;269:9253–9260. [PubMed] [Google Scholar]
- 11.Milne D M, Palmer R H, Campbell D G, Meek D W. Oncogene. 1992;7:1361–1369. [PubMed] [Google Scholar]
- 12.Meek D W, Simon S, Kikkawa U, Eckhart W. EMBO J. 1990;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hupp T R, Meek D W, Midgley C A, Lane D P. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- 14.Hao M, Lowy A M, Kapoor M, Deffie A, Liu G, Lozano G. J Biol Chem. 1996;271:29380–29385. doi: 10.1074/jbc.271.46.29380. [DOI] [PubMed] [Google Scholar]
- 15.Milne D M, Palmer R H, Meek D W. Nucleic Acids Res. 1992;20:5565–5570. doi: 10.1093/nar/20.21.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hupp T R, Sparks A, Lane D P. Cell. 1995;83:237–245. doi: 10.1016/0092-8674(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 17.Rolley N, Milner J. Oncogene. 1994;9:3067–3070. [PubMed] [Google Scholar]
- 18.Fuchs B, O’Connor D, Fallis L, Scheidtmann K H, Lu X. Oncogene. 1995;10:789–793. [PubMed] [Google Scholar]
- 19.Fiscella M, Zambrano N, Ullrich S J, Unger T, Lin D, Cho B, Mercer W E, Anderson C W, Appella E. Oncogene. 1994;9:3249–3257. [PubMed] [Google Scholar]
- 20.Reinke V, Lozano G. Radiat Res. 1997;148:115–122. [PubMed] [Google Scholar]
- 21.Michalovitz D, Halevy O, Oren M. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 22.Takenaka I, Morin F, Seizinger B R, Kley N. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- 23.Kastan M B, Zhan Q, El-Diery W S, Carrier F, Jacks T, Walsh W V, Plunkett B S, Vogelstein B, Fornace A J., Jr Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 24.Zhan Q, Carrier F, Fornace A J., Jr Mol Cell Biol. 1993;13:4242–4250. doi: 10.1128/mcb.13.7.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.El-Diery W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 26.Shieh S-Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 27.Milne D M, Campbell L E, Campbell D G, Meek D W. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- 28.Shaw P, Freeman J, Bovey R, Iggo R. Oncogene. 1996;12:921–930. [PubMed] [Google Scholar]
- 29.Hansen S, Hupp T R, Lane D P. J Biol Chem. 1996;271:3917–3924. doi: 10.1074/jbc.271.7.3917. [DOI] [PubMed] [Google Scholar]
- 30.Wu Y, Liu Y, Lee L, Milner Z, Kulesz-Martin M. EMBO J. 1994;13:4823–4830. doi: 10.1002/j.1460-2075.1994.tb06808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaziri H, West M D, Allsopp R C, Davison T S, Wu Y-S, Arrowsmith C H, Poirier G G, Benchimol S. EMBO J. 1997;16:6018–6033. doi: 10.1093/emboj/16.19.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu Y, Huang H, Miner Z, Kulesz-Martin M. Proc Natl Acad Sci USA. 1997;94:8982–8987. doi: 10.1073/pnas.94.17.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okorokov A L, Ponchel F, Milner J. EMBO J. 1997;16:6008–6017. doi: 10.1093/emboj/16.19.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hainaut P, Milner J. Cancer Res. 1993;53:1739–1742. [PubMed] [Google Scholar]