Abstract
Splicing provides an additional level in the regulation of gene expression and contributes to proteome diversity. Herein, we report the functional characterization of a recently described plant-specific protein, atRSZ33, which has characteristic features of a serine/arginine-rich protein and the ability to interact with other splicing factors, implying that this protein might be involved in constitutive and/or alternative splicing. Overexpression of atRSZ33 leads to alteration of splicing patterns of atSRp30 and atSRp34/SR1, indicating that atRSZ33 is indeed a splicing factor. Moreover, atRSZ33 is a regulator of its own expression, as splicing of its pre-mRNA is changed in transgenic plants. Investigations by promoter-β-glucuronidase (GUS) fusion and in situ hybridization revealed that atRSZ33 is expressed during embryogenesis and early stages of seedling formation, as well as in flower and root development. Ectopic expression of atRSZ33 caused pleiotropic changes in plant development resulting in increased cell expansion and changed polarization of cell elongation and division. In addition, changes in activity of an auxin-responsive promoter suggest that auxin signaling is disturbed in these transgenic plants.
INTRODUCTION
Splicing is an important step in the expression of many eukaryotic genes. It generates mature transcripts by excision of introns and ligation of exons. It also provides an additional level of regulation of gene expression by means of selection of alternative splice sites and subsequent generation of differential transcripts, in many cases encoding functional protein isoforms, thus increasing the coding potential of a genome (Smith and Valcarcel, 2000). Splicing occurs on a large RNP particle, termed spliceosome, which consists of five small nuclear ribonucleoprotein particles (snRNPs) and a large number of non-snRNP proteins (reviewed by Kramer, 1996; Rappsilber et al., 2002; Zhou et al., 2002). The latter include a family of proteins termed serine/arginine-rich (SR) proteins with a common structure, represented by one or two RNA recognition motifs (RRMs), followed by an Arg/Ser-rich (RS-rich) domain involved in protein–protein interactions. These proteins are conserved throughout the metazoa and are represented by two SR proteins in Schizosaccharomyces pombe and by 10 known human proteins (reviewed by Graveley, 2000). Recent analysis of the Arabidopsis genome has revealed at least 18 SR proteins (Lorkovic and Barta, 2002), some of which have been characterized (Lazar et al., 1995; Lopato et al., 1996a,b, 1999a,b; Golovkin and Reddy, 1998, 1999).
SR proteins have multiple functions in the splicing process and are essential for constitutive splicing. Their dual ability to bind to specific RNA sequences and to interact with other proteins makes them essential proteins for splice site selection and assembly of active spliceosomes. By binding to splicing enhancer sequences, they modulate splice site selection in a concentration-dependent manner in vitro and in vivo and can therefore be important regulators of alternative splicing as well (reviewed by Smith and Valcarcel, 2000).
Expression of SR proteins is subjected to tight tissue-specific and developmental regulation. The activity of some SR proteins can be antagonized by members of the heterogenous nuclear ribonucleoproteins (hnRNPs) A/B family and by other SR proteins, and the ratio of the antagonists varies in different tissues. Interestingly, genes encoding SR proteins are often expressed as a set of alternative mRNA species, the ratio of which is also under tissue-specific and developmental control (reviewed by Caceres and Krainer, 1997; Smith and Valcarcel, 2000; see also Lazar et al., 1995; Lazar and Goodman, 2000; Lopato et al., 1996a,b, 1999b; Golovkin and Reddy, 1998). The biological significance of such regulation is not clear.
The ability of most SR proteins to individually complement splicing-deficient HeLa cytosolic extract (which is devoid of SR proteins) suggested that SR proteins have redundant functions in constitutive splicing (reviewed by Manley and Tacke, 1996). Functional redundancy of SR proteins has been shown in Caenorhabditis elegans by RNA interference experiments. Only CeSF2/ASF seemed to be an essential factor because its inactivation led to late embryonic lethality, whereas RNA interference with other CeSR proteins caused no obvious phenotype; however, simultaneous suppression of two or more proteins showed strong and specific phenotypes (Longman et al., 2000). Similarly, deletion of SF2/ASF in chicken B cell line resulted in cell death (Wang et al., 1996), and depletion of B52/SRp55 in Drosophila melanogaster led to lethal defects during development (Ring and Lis, 1994). In both cases, splicing of specific transcripts was affected (Wang et al., 1998; Hoffman and Lis, 2000). Knockout of SRp20 has been shown to block mouse development, indicating an essential function of this protein in mouse (Jumaa et al., 1999), but not in C. elegans (Longman et al., 2000). The different importance of SR proteins in various organisms is also evident for SC35. Inactivation of neither of the two C. elegans homologs showed a phenotype (Longman et al., 2000), whereas, in mouse, conditional depletion of SC35 caused a defect in T cell maturation and modulated alternative splicing of the receptor tyrosine phosphatase CD45, isoforms of which have been shown to differentially regulate T cell activation (Wang et al., 2001).
Overexpression of SR proteins has also been demonstrated to interfere with splicing of particular transcripts and to influence development. Increasing levels of B52/SRp55 in transgenic Drosophila led to defects in development of many cell types (Kraus and Lis, 1994), which could be partially rescued by overproduction of splicing repressor RSF1 (Labourier et al., 1999). Overexpression of atSRp30, one of Arabidopsis homologs of SF2/ASF, resulted in morphological and developmental changes in transgenic plants. Alternative splicing of several endogenous genes was affected, including atSRp30 itself and atSRp34/SR1 (Lopato et al., 1999b). Increasing expression of SC35 in HeLa cells has been shown to decrease endogenous levels of its own mRNA along with changes in its alternative splicing pattern (Sureau et al., 2001). Therefore, the functional significance of a particular SR protein during development seems to vary in different organisms.
As mentioned above, the Arabidopsis SR protein family consists of 18 members (Lorkovic and Barta, 2002), which are arranged in subfamilies of several close homologs. Some are orthologs of vertebrate proteins, but some seem to be plant specific (Lopato et al., 1996a, 2002). Because animal introns cannot be processed in plants (Barta et al., 1986), plant-specific factors are of particular interest to study. Recently, we have identified such a novel plant-specific Arg/Ser-rich protein with unique features in its domain organization (Lopato et al., 2002). This protein, named atRSZ33, has an N-terminal RRM, two zinc knuckles embedded in a basic RS region, and an acidic C-terminal domain. It is a phosphoprotein and is localized in the nucleus in a speckled manner. We have demonstrated that this protein interacts with several known splicing factors, including atSRp34/SR1 (Lazar et al., 1995), atRSZp21 and atRSZp22, homologs of the human 9G8 (Golovkin and Reddy, 1998; Lopato et al., 1999a), and with a novel family of SC35-like splicing factors (Lopato et al., 2002). Together, these data have suggested a role for atRSZ33 in pre-RNA splicing. To gain more insight into possible functions of atRSZ33 in splicing, we applied an overexpression approach, which proved to be successful in studies of other splicing factors (Kraus and Lis, 1994; Labourier et al., 1999; Lopato et al., 1999b; Sureau et al., 2001). This approach is of special interest when studying plant-splicing factors, because plant in vitro splicing assays are not available. Additionally, although there is a growing amount of biochemical and molecular data, very little is known about physiological roles of individual splicing factors in plants. In general, gain-of-function, or overexpression, approach has provided valuable information about many plant genes (Mizukami and Ma, 1992; Kirik et al., 2001; Chen and Chen, 2002; Jasinski et al., 2002; Kandasamy et al., 2002).
In this study, we show that atRSZ33 is indeed a splicing factor because overexpression of the protein causes changes in the splicing pattern of its own and of some other mRNAs. AtRSZ33 is expressed in specific cell types during root and flower development, and throughout all embryogenesis and seed development, thus implicating its possible role in these processes. Ectopic expression of atRSZ33 resulted in severe abnormalities in the phenotype of transgenic plants affecting mostly cell expansion and cell fate, suggesting that proper regulation of atRSZ33 is crucial for plant development.
MATERIALS AND METHODS
Constructs for Promoter Analysis and Overexpression of atRSZ33
The atRSZ33 promoter-GUS fusion construct was described previously (Lopato et al., 2002). atRSZ33 cDNA and genomic clone were amplified by polymerase chain reaction (PCR) by using primers 5′-AACTGGATCCACGCGTCCGCCGAAATTAG-3′ and 5′-AAATGAGCTCGAAACTTTTATAAATCC-3′, containing BamHI and SacI restriction sites, respectively. The PCR conditions were 95°C 1 min, once; 95°C 30 s, 50°C 1 min, 72°C 1 min, 35 times; and 72°C 5 min, once. DNA fragments were verified by sequencing and placed under the control of the 35S RNA promoter from Cauliflower Mosaic Virus (35S CaMV) by replacing the β-glucuronidase (GUS) gene at the BamHI and SacI restriction sites of pBI121 (BD Biosciences Clontech, Palo Alto, CA). This yielded the constructs 35S:catRSZ33 and 35S:gatRSZ33.
Plant Transformation
The constructs described above were introduced into either AGL1 (Lazo et al., 1991) or LBA4404 (Hoekema et al., 1983) Agrobacterium tumefaciens strains. Arabidopsis thaliana (ecotype Columbia, Col-O) plants were used for transformations. Plants carrying the atRSZ33 promoter-GUS fusion were obtained by the floral dip method (Clough and Bent, 1998). Plants carrying 35S:catRSZ33 and 35S: gatRSZ33 were produced by root transformation by using the method described by Valvekens et al. (1988). Selection of transgenic plants was done in the presence of 50 μg/ml kanamycin. Plants containing one copy of transgene were used for further studies. Plants were grown under 16-h light/8-h dark conditions at 23°C.
Analyses of Expression Patterns of atRSZ33
Histochemical GUS expression assays were performed on intact seedlings or excised organs of mature plants as described by Jefferson et al. (1987). Localization of GUS activity in lines obtained from crosses of DR5:GUS plants (Ulmasov et al., 1997) with 35S:gatRSZ33 plants was done according to Malamy and Benfey (1997). For in situ hybridization, seedlings, flowers, and siliques of A. thaliana were fixed in 4% paraformaldehyde and embedded in butyl methyl methacrylate according to Baskin et al. (1992). Sections were 4 μm in thickness. Hybridization was done as described by Dornelas et al. (1999) with a 456-nucleotide fragment of coding sequence spanning the C-terminal SR and serine/proline-rich regions of atRSZ33 protein, amplified with primers 5′-AACGAATTCATGCCCAAGAAGCTTAGGCGCAG-3′ and 5′-ATATAGGATCCTTAAGGAGACTCACTTCC-3′. Probes were obtained according to manufacturer's instructions by using DIG RNA labeling kit (Roche Diagnostics, Mannheim, Germany).
Reverse-Transcription-Polymerase Chain Reaction (RT-PCR) and Western Blot Analyses of Transgenic Plants Carrying 35S:catRSZ33 and 35S:gatRSZ33
Seedlings were germinated and grown on plates with germination medium (Valvekens et al., 1988) under 16-h light/8-h dark conditions at 23°C. Wild-type plants and transgenic line containing the original plasmid pBI121 (BD Biosciences Clontech) served as controls. Isolation of total RNA from 16-d-old plants was performed using RNeasy plant kit (QIAGEN, Hilden, Germany). Reverse transcription was done with a primer derived from the 3′-untranslated region (3′-UTR) of atRSZ33: 5′-AAATGAGCTCGAAACTTTTATAAATCC-3′. Primers for PCR were designed for the beginning and the end of coding region: 5′-ATATACCATGGCTCGCTATGATGATC-3′ and 5′-ATATAGGATCCTTAAGGAGACTCACTTCC-3′. PCR was performed with 25 cycles as described above. All RT-PCR products were sequenced.
Total protein extracts from 16-d-old wild-type and transgenic plants were prepared according to Zahler et al. (1992). SR proteins were purified from 3-wk-old wild-type plants as described in Lopato et al. (1996a). Proteins were separated on a 12% SDS-PAGE gel. Detection was performed using either chicken preimmune serum or polyclonal antibodies raised against atRSZ33 (Lopato et al., 2002).
Analyses of the Phenotype of 35S:atRSZ33 Plants
For analyses, plants were grown in soil in a growth chamber under 16-h light/8-h dark conditions at 23°C. Examination of early seedling development was carried on seedlings germinated and grown on GM medium (Valvekens et al., 1988) under the same conditions. The development of embryos, wherever necessary, was monitored in heterozygous plants to correlate stages of development of transgenic and wild-type seeds within the same silique. Tissues for microscopic examination were fixed in ethanol/acetic acid (9:1) from 2 h to overnight, washed twice in 90% ethanol and cleared in chloral hydrate/glycerol (2.5 g of chloral hydrate in 1 ml of 30% glycerol) solution. Samples were viewed using Nomarski optics.
To monitor auxin levels, DR5:GUS plants (Ulmasov et al., 1997) were used to pollinate plants carrying 35S:gatRSZ33.
For regeneration assay, 50-leaf explants of 10-d-old control transgenic seedlings, carrying pBI101, and of 35S:gatRSZ33 seedlings were incubated for 1 wk on callus induction medium, containing 1.0 mg/l α-naphthalene acetic acid and 0.1 mg/l 6-benzylaminopurine and then transferred to the medium for shoot induction, containing 0.2 mg/l α-naphthalene acetic acid and 1.0 mg/l 6-benzylaminopurine (van der Graaff and Hooykaas, 1998). After 5 d, leaf explants were examined and photographed.
RT-PCR Analyses of Particular Genes in 35S:gatRSZ33 Plants
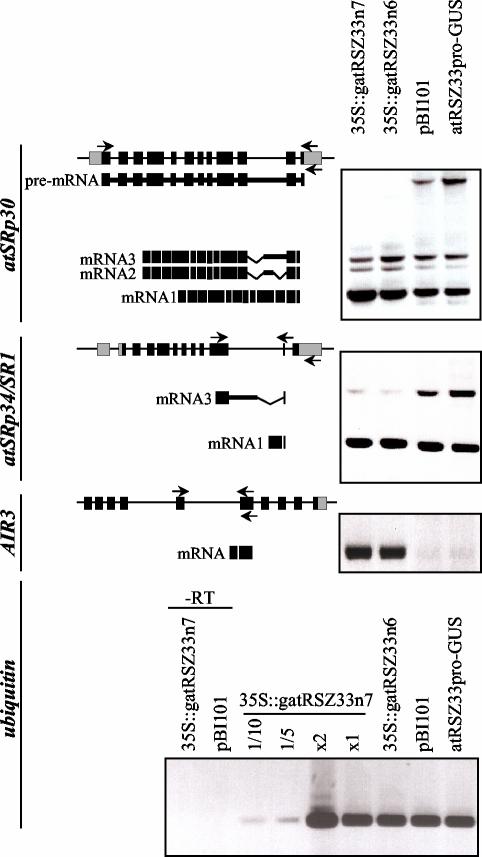
Seedlings were germinated and grown for 16 d on vertically oriented plates with GM medium (Valvekens et al., 1988) under 16-h light/8-h dark conditions at 23°C. Isolation of total RNA from roots of control or transgenic Arabidopsis plants was done using RNeasy plant kit (QIAGEN). Primers for reverse transcription were as follows: 5′-AAATGAGCTCAAATGTATATGTATGAAAAACC-3′ (atSRp30), 5′-AATGAGCTCGAAACGATATCTTCAAAAAAAAAC-3′ (atSRp34/SR1), 5′-TTGTAATGTTTTCCATTACCG-3′ (AIR3; Neuteboom et al., 1999), and 5′-CTCCTTCTTTCTGGTAAACGT-3′ (ubiquitin; Sablowski and Meyerowitz, 1998). Primers for PCR were as follows: 5′-ATATACCATGGGTAGCCGATGGAATCGTAC-3′ and 5′-ATATAGGATCCTATCTTGATCTTGATCTTG-3′ (atSRp30), 5′-ATAGGATCCAGGAGCAGAAGTCCCAAGGCAAAG-3′ and 5′-ATATAGGATCCCATTTTACCTCGATGGAC-3′ (atSRp34/SR1), 5′-TGTGTGGCCAGAATCAAAGAGC-3′ and 5′-AGAACAAGATTGCTAGCAAAC-3′ (AIR3), and 5′-GGTGCTAAGAAGAGGAAGAAT-3′ and 5′-CTCCTTCTTTCTGGTAAACGT-3′ (ubiquitin). RT-PCR of ubiquitin was used to control loading. PCR was performed with 25 cycles as described previously. The number of PCR cycles necessary for semiquantitative representation was determined by taking RNA samples for reverse transcription reaction in 1/10, 1/5, 2×, and 1× consecutive dilutions. To exclude the possibility that amplification products arise from DNA contamination of RNA samples control RT-PCRs were performed where reverse transcriptase was excluded from RT reaction. All RT-PCR products were sequenced to verify their origin.
Accession Numbers for Nucleotide Sequences
Accession numbers for nucleotide sequences are as follows: atRSZ33, AJ293801; atSRp30, AJ131214; atSRp34/SR1, AF173640; and AIR3, AF098632.
RESULTS
Transgenic Plants Overexpressing atRSZ33 Have Reduced Viability
To investigate whether different levels of atRSZ33 would affect splicing, we used an overexpression approach. We established transgenic Arabidopsis plants transformed either with the cDNA (35S:catRSZ33) or with the genomic sequence (35S:gatRSZ33) of atRSZ33 fused to the 35S CaMV promoter, which is constitutively active in most plant tissues. Complete development of plants was hindered by strong abnormalities in cell and organ shape. The majority of T0 35S:catRSZ33 lines had a stronger phenotype than 35S: gatRSZ33 plants and were sterile. Only three 35S:catRSZ33 lines showing no obvious morphological changes gave T1 progeny with wild-type phenotype. 35S:gatRSZ33 transgenic T0 plants had reduced fertility due to abnormal flower development; however, several T1 lines with phenotypic changes were obtained. Flower abnormalities in T0 and subsequent generations included changes in organ identity and multiplication of organs in all four whorls. Examination of pollen development revealed heterogeneous population, which contained a lot of dead pollen. Pollen germination test in vitro showed that germination ability was impaired; pollen tubes were shorter, thicker, and often branched. Siliques, which were formed, had a reduced number of seeds (our unpublished data).
Segregation analysis of 35S:gatRSZ33 T2 progenies showed that about one-third of the seedlings were kanamycin sensitive. Ratios for lines used in further analyses were as follows: line 35S:gatRSZ33 n4–2,4:1; line 35S:gatRSZ33 n6–2,1:1; and line 35S:gatRSZ33 n7–1,8:1. Analysis of T3 progenies of kanamycin-resistant T2 plants revealed that the population of homozygous plants is reduced, suggesting a lower viability. In contrast, 35S:catRSZ33 T2 progenies segregated as 3:1, and again no changes in the phenotype of these plants were observed.
The low proportion of 35S:gatRSZ33 homozygous plants together with observations made on the regeneration process and flower development suggested that overexpression of atRSZ33 can reduce viability of plants, especially during fertilization and/or seed development.
Because the surviving 35S:catRSZ33 plants did not show any phenotype, it was important to check the amount of atRSZ33 protein in transgenic plants. Western blot analysis, performed with total protein extracts from 16-d-old wild-type (Figure 1A, lane 1) and T3 homozygous transgenic plants (three 35S:catRSZ33 lines, lanes 3–5; one weaker 35S: gatRSZ33 line, lane 6; and two 35S:gatRSZ33 lines with stronger phenotype, lanes 7 and 8), detected elevated levels of atRSZ33 protein only in plants transformed with the genomic construct (Figure 1A, lanes 6–8) but not in plants containing the cDNA construct (lanes 3–5). This implies that the latter ones do not, or only in small amounts, overexpress atRSZ33, which is consistent with the observation that these plants did not show any phenotype. In 35S:gatRSZ33 lines the amount of overproduced protein correlated with the severity of the phenotype. The overproduced protein comigrated with atRSZ33 detected in an SR protein preparation from wild-type plants (Figure 1A, lane 9), which has been previously shown to be enriched in atRSZ33 (Lopato et al., 2002). Thus, only 35S:gatRSZ33 transgenic lines had elevated levels of atRSZ33 protein, which correlated with the severity of phenotype (see below), and these lines were used for further analyses.
Figure 1.
Protein and RNA analyses of 35S:atRSZ33 transgenic Arabidopsis plants. (A) Western blot analysis of total protein extracts from control and T3 homozygous 35S:atRSZ33 lines (lanes 1–8) and SR proteins preparation from wild-type plants (lane 9). Lanes 1 and 2, wild-type plants and plants containing pBI121, respectively; 35S: catRSZ33 lines, lanes 3–5; one weaker 35S:gatRSZ33 line, lane 6; and two 35S:gatRSZ33 lines with stronger phenotype, lanes 7 and 8. Asterisks indicate unspecific bands. Arrow indicates position of atRSZ33 protein. (B) RT-PCR analysis of atRSZ33 transcripts in T0 generation. Lane 1, control plants containing pBI121; plants transformed with cDNA (lanes 2 and 3) and with genomic construct (lanes 4 and 5). (C) RT-PCR analysis of atRSZ33 transcripts in T3 generation (top). Lanes 1–8, same plants as indicated in A. Asterisk indicates unspecific band. Schematic representation of atRSZ33 gene structure, its mRNA isoforms detected in transgenic 35S:gatRSZ33 plants, and deduced proteins (bottom). Exons are shown as black boxes, 5′ and 3′-UTRs are gray boxes. 3′alt ss-3′ alternative splice site in the second intron. Bold lines represent included sequences of the second and third introns. Stars indicate premature stop codons. Deduced protein structures are shown as black, RRM; gray, RS-rich region; white, two zinc knuckles; light gray, SP region; and striped boxes, sequences included due to alternative splicing. Positions of primers used for RT-PCR analyses are shown as arrows. Schematic drawings are not to scale.
AtRSZ33 Overexpression Influences Splicing of Its Own mRNA
Surprisingly, analyses using RT-PCR (Figure 1B) and Northern blotting (our unpublished data) revealed that plants showing phenotype in T0 generation of 35S:catRSZ33 transgenic lines had altered splicing pattern of atRSZ33 (Figure 1B, lanes 2 and 3). Because the cDNA construct does not contain introns, alternative mRNAs in this case can only originate from the endogenous atRSZ33 gene. Interestingly, when the genomic clone of atRSZ33 is used for transformation the amount of alternative transcripts is higher than in 35S:catRSZ33 T0 lines (Figure 1B, lanes 4 and 5), indicating that they originate also from the transgenic copy of atRSZ33.
Because Western blot analysis showed that 35S:gatRSZ33 plants had elevated amounts of atRSZ33, and protein levels were not changed in phenotypically normal 35S:catRSZ33 lines, we analyzed the transcript patterns of atRSZ33 in homozygous transgenic lines of the same T3 generation.
While in the 35S:catRSZ33 lines, only correctly spliced mRNA1 encoding the full-length protein was present (Figure 1C, top, lanes 3–5), making them undistinguishable from wild-type (lane 1) and from the pBI121 control line (lane 2), the lines containing the genomic construct showed several transcript bands (Figure 1C, lanes 6–8). Sequencing revealed that mRNA4 contained an alternatively spliced second intron and an unspliced third intron. mRNA3 included the same part of the second intron, but the third intron was spliced normally. In contrast, splicing of the second intron was not affected in mRNA2, but the third intron was retained similarly to mRNA4. Only one of these transcripts, namely, mRNA3, has been detected in wild-type plants and only at the first days after germination (Lopato et al., 2002). The alternative splicing events generate premature stop codons in the included part of the second intron in mRNA4 and mRNA3 and in the unspliced third intron of mRNA2 potentially leading to proteins carrying one RRM (mRNA2) or only part of it (mRNA3 and mRNA4) (Figure 1C, scheme). The analyses of T0 and further generations of transgenic lines carrying the cDNA construct show that overexpression of atRSZ33 regulates splicing of the endogenous gene. However, this regulation is insufficient because no overexpressing cDNA line was present in the next generation. In contrast, the same type of regulation seems to down-regulate the overexpression of the genomic construct, so further generations of overexpressing lines could be recovered in this case. Together, these results suggest that atRSZ33 regulates its own expression on the posttranscriptional level.
Overexpression of atRSZ33 Influences the Expression of Particular Genes
Because atRSZ33 has structural features of splicing factors and influences processing of its own mRNA, we decided to check whether overexpression of atRSZ33 affects splicing of other genes in vivo.
Out of several genes tested, which were known to be regulated by alternative splicing, two showed altered splicing patterns in atRSZ33-overexpressing lines (Figure 2). AtSRp30 is expressed as a set of at least three different transcripts, the ratio of which is under temporal and spatial control (Lopato et al., 1999b). RT-PCR analysis showed that, in addition to the different splice forms, unspliced RNA of atSRp30 is present in control lines. In the lines overexpressing atRSZ33, no unspliced RNA was detected, but the level of mRNA1 was increased (Figure 2). The enhancement of atSRp30 expression by improving splicing efficiency correlates well with partially overlapping phenotypes of atRSZ33 and atSRp30 overexpressing plants.
Figure 2.
Analysis of transcript patterns of atSRp30, atSRp34/SR1, and AIR3 in plants overexpressing atRSZ33. Primers used for RT-PCR are shown by arrows on the schemes of corresponding genes. Exons are shown as boxes and introns as lines (bold lines are introns included in the mRNAs). 5′- and 3′-UTRs are gray, and coding regions are black. 35S:gatRSZ33n6 and 35S:gatRSZ33n7 are two independent transgenic lines overexpressing atRSZ33. The following control lines were used: transgenic line carrying pBI101, and transgenic line containing atRSZ33 promoter-GUS fusion. RT-PCR of ubiquitin was used to control loading. 1/10, 1/5, x2 and x1 are consecutive dilutions of RNA sample from 35S:gatRSZ33n7 line. Controls for DNA contamination of all RNA preparations were done by omitting reverse transcriptase, and controls for ubiquitin only are shown (–RT).
Splicing of atSRp34/SR1 (Lazar et al., 1995; Lopato et al., 1999b) was also modulated by overexpression of atRSZ33. Control plants showed the presence of two main products, one corresponding to mRNA1 with a correctly processed intron 10 and another corresponding to mRNA3 retaining part of this intron due to usage of an alternative 5′ splice site (Figure 2; Lopato et al., 1999b). In transgenic lines, production of mRNA3 was inhibited and correct splicing of the tenth intron of atSRp34/SR1 was promoted (Figure 2). No unspliced product of atSRp34/SR1 was detected in any lines.
Because analysis of the phenotype of 35S:gatRSZ33 plants (see below) revealed changes in the cell shape and auxin signaling, the expression of additional genes, which are involved in these processes, was tested. AIR3 encodes an Arabidopsis subtilisin-like serine protease, which accumulates during auxin-induced lateral root formation and is proposed to be localized outside the cell or at the plasma membrane and to be involved in changing cell wall structure (Neuteboom et al., 1999). RT-PCR analysis did not detect any alternative splicing products of AIR3. However, we observed a drastic increase of AIR3 mRNA in the transgenic lines (Figure 2). Up-regulation of the steady-state level of AIR3 mRNA in lines overexpressing atRSZ33 was also detected by Northern blot analysis (our unpublished data).
Expression Patterns of atRSZ33 in Plant Development
Preliminary observations showed that expression of atRSZ33 under 35S CaMV promoter led to strong changes in the phenotype. Our previous data have shown that atRSZ33 is expressed only in roots and flowers of Arabidopsis (Lopato et al., 2002). To better interpret this phenotype, we performed a detailed analysis of the spatial and temporal expression of atRSZ33 by using a promoter-reporter gene fusion and in situ hybridization.
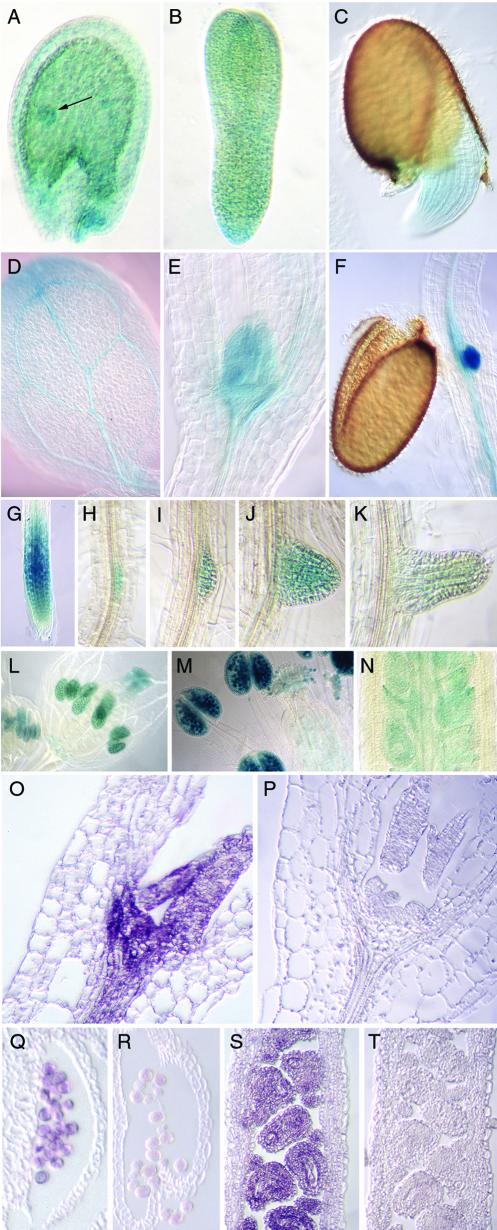
Arabidopsis plants, transformed with a construct containing a 949-base pair promoter and the 5′-UTR of atRSZ33 fused to the reporter GUS gene, were used for analysis of the expression patterns. The atRSZ33 promoter seemed to be active during embryogenesis and seed development. Figure 3A shows its expression in the Arabidopsis seed with the embryo at globular stage. The promoter remains active throughout embryogenesis until embryo and seed maturity, and it does not show any cell-type specificity at these stages (Figure 3, A and B).
Figure 3.
Expression patterns of atRSZ33. Histochemical localization of atRSZ33 promoter-GUS activity (A–N) and in situ hybridization of atRSZ33 transcripts (O–T). (A) Immature seed with embryo at the globular stage of development (arrow). (B) Torpedo stage embryo. (C) Germinating seed. (D–F) Arabidopsis seedling, showing expression in the tip and in the vasculature of cotyledon (D), in the shoot apical meristem (E) and at the site of secondary root formation at the hypocotyl-root junction (F). (G) Expression in the elongation zone of primary root. (H–K) Different stages of the lateral root formation. (L) Young flowers with expression in the tapetum within anthers and in stigma and style. (M) Flower after opening with expression in mature and germinating pollen, in stigma and ovules. (N) Part of immature silique with staining in developing seeds, funiculi and septum. (O) and (P) Shoot apical meristem of 5-d-old seedling. (Q and R) Mature pollen within anther. (S and T) Young silique with developing seeds. (O, Q, and S) Antisense probe. (P, R, and T) Sense probe.
During seed germination, atRSZ33 is expressed in the radicle (Figure 3C). In young seedlings, GUS activity was detected in cotyledons with the highest levels in the tips and vascular system (Figure 3D), in the shoot apical meristem (Figure 3E), and in the hypocotyl/root junction with a maximum at the site of secondary root formation (Figure 3F). Examination of the primary root revealed that the atRSZ33 promoter was mostly active in the elongation zone (Figure 3G); however, the activity was also detected in the very first stages of lateral root formation (Figure 3H). Further development of lateral roots is accompanied by a continuous expression of the atRSZ33 promoter (Figure 3, I–K). This particular expression pattern of atRSZ33 in roots suggests a role in lateral root formation and cell elongation.
During flower development, staining was detected in the tapetal layer within anthers of very young flower buds, but it also occurred later in the style and stigma (Figure 3L). After tapetum degradation, the atRSZ33 promoter becomes active in mature pollen (Figure 3M), which was also confirmed in mature pollen isolated from the anthers and assayed separately (our unpublished data). The activity was also shown during pollen germination both in vivo as seen in pollen attached to stigma (Figure 3M). After fertilization, the activity occurs within ovules (Figure 3M). In the immature silique, GUS expression has been found in a septum, funiculi, and developing seeds (Figure 3N).
To confirm the expression pattern of the atRSZ33 promoter-GUS construct, in situ hybridization analysis has been performed on selected tissues. Sense and antisense probes corresponding to the C-terminal part of atRSZ33 have been selected to prevent cross-hybridization with highly homologous RRMs. AtRSZ33 transcripts have been detected in the shoot apical meristem and in the developing leaf primordia (Figure 3O), in the mature pollen within anther (Figure 3Q) and in the developing seeds within the silique (Figure 3S). No signal could be detected in control experiments with the sense probe (Figure 3, P, R, and T). The results obtained by in situ hybridization corroborate the expression patterns observed in transgenic plants carrying atRSZ33 promoter-GUS fusion (Figure 3, E, M, and N).
As predicted from Northern blot analysis (Lopato et al., 2002), no atRSZ33 expression could be detected in stems or in leaves of Arabidopsis by using either atRSZ33 promoter-GUS fusion or in situ hybridization.
Ectopic Expression of atRSZ33 Causes Pleiotropic Changes in the Phenotype of Arabidopsis Plants
Effect of atRSZ33 Overexpression on Embryogenesis and Early Seedling Development Because atRSZ33 has been found to be expressed during embryo and seed development, and we have observed a low seed set and viability while obtaining transgenic plants, possible effects of atRSZ33 overexpression on embryogenesis were examined. The most obvious alteration of embryo development in the overexpressing plants was the formation of twin embryos (Figure 4, B–D). We have never observed this phenomenon in control plants, whereas in homozygous transgenic plants ∼3–5% of the seeds contained two embryos (3.3% [6 of 184 seeds] in the weaker line, 4.4% [4 of 91 seeds] and 5.2% [7 of 135 seeds] in two stronger lines). Seed examination after germination revealed a lower amount of twin seedlings (∼2%), suggesting that not all such altered seeds/embryos are viable. Twin seedlings had either similar or different morphology (Figure 4, K and J, respectively). Formation of the second seedling with a single collar-like cotyledon was the most frequent alteration.
Figure 4.
Influence of atRSZ33 overexpression on the embryo and seedling development. (A) Wild-type seed with embryo at globular stage. (B) 35S:gatRSZ33 seed, containing twin embryos indicated by arrows 1 and 2. (C and D) Close-up of embryos shown in B. (E and F) Close-up of wild-type and 35S:gatRSZ33 embryos at globular stage, respectively, showing abnormal division in suspensor (arrow in F). (G and H) Wild-type and transgenic embryos at heart stage. (I) Eight-day-old wild-type seedling. (J, M, and N) Eight-day-old transgenic seedlings. (K) Three-day-old transgenic seedling. (J) Twin seedlings, one of them with single cotyledon. (K) Equally developed twin seedlings. (L and M) Seedlings with single cotyledon. Arrows in J, L, and M point out shoot apical meristems. Bars, 50 μm (A–H) and 1 mm (I–N).
Closer examination of twin embryos showed that they might share suspensor structures (Figure 4, C and D). We observed multiple suspensor abnormalities in transgenic plants, including additional divisions, changes in the orientation of cell division plane and cell expansion (Figure 4F), which, probably, can lead to the formation of secondary embryos.
In addition, establishment of bilateral symmetry, when the wild-type embryo proceeds from globular to heart stage (Figure 4G), was impaired in some of the transgenic embryos (Figure 4H). Up to ∼10% of seeds contained “monocotyledonous” embryos (6.6% [10 of 151 seeds] in the weaker line, 7.9% [11 of 140 seeds], and 10.8% [16 of 148 seeds] in two stronger lines). Seedlings originating from such single-cotyledon embryos were also detected (Figure 4, J, M, and L).
In general, development of the seedlings was retarded and growth of shoot apical and root meristems was inhibited (Figure 4N). These seedlings had shorter hypocotyls (Figure 4, I–N) in comparison with control plants (2.3 ± 0.4 mm in the weaker line, 1.8 ± 0.5 and 1.5 ± 0.3 mm in two stronger lines, 3.0 ± 0.4 mm in the control line), and both hypocotyls and cotyledons were thicker. Seedlings were compressed in the apical-basal axis and cells seemed to be more expanded.
Analysis of embryo and seedling development in several transgenic lines revealed that the extent of morphological changes in 35S:gatRSZ33 lines correlates with amount of overexpressed protein.
Ectopic Expression of atRSZ33 Alters Cell Expansion and Cell Shape Examination of the cell size in the cleared samples of the same regions of cotyledons and hypocotyls taken from the control (Figure 5, A and B) and the transgenic seedlings of the same age (Figure 5, E and F) revealed that cell size was increased in 35S:gatRSZ33 lines. However, cells in the hypocotyl were shorter than those of control plants, but their number was not changed, which explains the stunted morphology of the transgenic seedlings.
Figure 5.
Ectopic expression of atRSZ33 affects cell size and shape, stomatal development, and formation of ectopic meristems. Plants, containing 35S:gatRSZ33 (E–H, J, and K) were compared with wild-type plants (A–D, and I) of the same age and grown under the same conditions. (A and E) Cotyledons. (B and F) Hypocotyls. (C and G) Root hairs. Arrows in G point out root hairs with abnormal branched shape. (D and H) Trichomes. (I) Stomata in the leaf epidermis of wild-type plants, arrows point out stomata precursors formed due to asymmetric cell divisions, arrows point out areas with multiple misoriented cell divisions. (J) Stomatal development in the leaf epidermis of 35S:gatRSZ33 plants. (K) Formation of stomatal clusters in the leaf epidermis of 35S:gatRSZ33 plants. (L) Ectopic structure (arrow) on the cotyledon of 10-d-old transgenic seedling. Note abnormal shape of the cotyledon surface. (M) Abaxial side of the rosette leaf with ectopic structures formed over midvein and on the edge of the leaf (indicated by arrows). (N) Ectopic meristem developed on the cotyledon surface (arrow points out enlarging trichome precursor). (O) Ectopic meristem formed in the epidermal layer of rosette leaf. Bars, 100 μm (A, B, E, and F), 200 μm (C, D, G, and H) and 25 μm (N and O).
Additionally, root hairs in the transgenic plants had abnormal shapes with bulges and branches (Figure 5G), they could be short and more expanded, or of normal length but wavy (our unpublished data), in contrast to the wild type, where root hairs are tubular cells formed due to polarized cell expansion at the tip region (Figure 5C). Interestingly, the shape of trichomes in leaves was also changed by ectopic expression of atRSZ33 (compare Figure 5, D and H). The trichomes had significantly more branches than the wild-type ones. The spatial distribution of the trichomes was not affected.
In summary, ectopic expression of atRSZ33 led to an enhancement of cell expansion and to change of cell shape by affecting the direction of cell expansion.
Ectopic Expression of atRSZ33 Affects Stomata Development and Formation of Meristems Examination of cotyledon and leaf epidermis revealed that stomata development is affected by ectopic expression of atRSZ33. Transgenic plants have clustered stomata (Figure 5K) in contrast to the wild-type Arabidopsis (Figure 5I), where all stomatal units, consisting of one stomate, are separated by at least one epidermal cell. These clusters were of different shape, and the number of guard cells varied among clusters. Expression of atRSZ33 influenced stomata development more strongly in the cotyledons than in the leaf epidermis. Examination of early stages of stomatal development showed that there are multiple divisions of meristemoids within future clusters and some of those divisions are misoriented (Figure 5J), resulting presumably in the stomatal unit consisting of more than one stomate. Interestingly, we observed also archshaped clusters of actively dividing cells, and development of some of them was accompanied by enormous cell expansion resulting in cell death (our unpublished data). Thus, ectopic expression of atRSZ33 impairs normal stomatal development, which is controlled by frequency and orientation of asymmetric divisions in the epidermis.
Because we observed areas with enhanced cell divisions in the epidermis (Figure 5J), we assume that they could serve as a source for the formation of ectopic structures, which were found on the abaxial and adaxial sides of cotyledons and leaves of transgenic plants (Figure 5, L and M). Some of these structures morphologically resembled shoot apical meristems with leaf primordia, which developed trichome initials (Figure 5, N and O).
Interestingly, overexpression of atRSZ33 also led to excessive cell divisions in the shoot apical meristem and formation of multiple leaf primordia with highly expanded cells. Further development of these primordia was arrested (our unpublished data). In contrast, cell divisions in the root meristem were inhibited. Roots of transgenic plants were significantly shorter than those of controls (Figure 4, I, K–N), and the root meristem itself had a distorted organization (Figure 6, A–C). The number of lateral root primordia was higher in 35S:gatRSZ33 plants, but their further development was in many cases impaired (our unpublished data).
Figure 6.
Changes in the activity of DR5-GUS promoter construct and of the in vitro regeneration process in 35S:gatRSZ33 plants. (A–C) DR5: GUS activity in the root meristems of primary roots of the control DR5: GUS plants and two homozygous DR5:GUS/35S:gatRSZ33 lines (weak and strong lines), respectively. Arrow in C points out the cell with residual GUS activity. (D and G) DR5:GUS activity in the rosette leaves of control and DR5:GUS/35S:gatRSZ33 plants, respectively. (E) Close-up of rosette leaf of control plant. (H) Close-up of the same area of rosette leaf in the DR5:GUS/35S:gatRSZ33 plant, showing enhanced DR5:GUS activity and additional formation of provascular bands. (F) Close-up of control rosette leaf, including midvein. (I) Close-up showing accumulation of DR5:GUS activity near midvein in the rosette leaf of DR5:GUS/35S:gatRSZ33 plant. GUS activity is visible as blue staining on A–C and as a pink staining in dark field on D–I. (J) Control leaf explant with multiple green clusters of shoot meristems on the shoot-induction medium. (K) 35S:gatRSZ33 leaf explant with roots on the same medium. Bars, 50 μm (A–C), 500 μm (D and G), and 100 μm (E, F, H, and I).
Ectopic Expression of atRSZ33 Changes Activity of an Auxin-responsive Promoter and Affects the In Vitro Regeneration Process The dramatic changes in 35S: gatRSZ33 plants implied that hormone levels in these plants might be altered. Many of the observed features are regulated by levels and the exact distribution of the plant growth regulator auxin (Friml and Palme, 2002).
To visualize auxin localization in the atRSZ33 transgenic plants, we took advantage of a synthetic promoter, containing seven tandem repeats of an auxin-responsive element, TGTCTC, fused to a minimal 35S CaMV promoter and reporter gene β-glucuronidase (DR5:GUS; Ulmasov et al., 1997). Two 35S:gatRSZ33 lines, differing in the severity of their phenotypes and in their levels of overexpressed protein, were used for crosses with plants containing DR5:GUS. Roots of plants homozygous for both 35S:gatRSZ33 and DR5:GUS showed a dramatic decrease in the activity of DR5:GUS, as well as mislocalization of its expression (Figure 6, A–C). Changes in the DR5:GUS activity were more prominent in the line with higher overexpression of atRSZ33 (compare Figures 1A, lanes 6 and 8, and 6, B and C). DR5: GUS was reported to be active not only in the roots but also in the tips of cotyledons and of young leaves as detected after a longer staining period (Ulmasov et al., 1997). Examination of the rosette leaves of plants homozygous for 35S: gatRSZ33 and DR5:GUS revealed an unusual pattern of DR5 expression (Figure 6, D–I). These plants showed an accumulation of DR5:GUS activity as clusters of bands of irregular shape (Figure 6G). Closer examination revealed that staining depicted enhanced formation of provascular bands (Figure 6H). This pattern was never observed in control DR5:GUS plants (Figure 6, D and E). Interestingly, examination of the leaf epidermis over these areas showed enhanced rates of irregular cell divisions, similar to ones found in atRSZ33 transgenic plants during stomatal development (our unpublished data; Figure 5J). Unusual accumulation of GUS activity was also detected close to midveins (Figure 6I), coinciding with the formation of ectopic structures over midveins in 35S:gatRSZ33 plants (Figure 5M).
The observed accumulation of DR5:GUS activity in the leaves prompted us to check whether the changed auxin levels would affect the regeneration process in vitro. Leaf explants of ten days old control and 35S:gatRSZ33 seedlings were subjected to treatment that leads to shoot induction (Graaff van der and Hooykaas, 1998). Briefly, seedlings were incubated for 1 wk on callus induction medium and then transferred to the medium for shoot induction. In 5 d, leaf explants of control lines showed formation of the multiple clusters of shoot meristems (Figure 6J), which developed further into shoot-like structures. In contrast, leaf explants of 35S:gatRSZ33 plants produced roots (Figure 6K). Longer incubation of these explants on the shoot induction medium led to the enormous expansion of the cells both in the roots and in the explants with subsequent necrosis (our unpublished data).
Together, these results suggest that plants ectopically expressing atRSZ33 might have reduced levels of auxin in the root tips and higher levels of auxin in leaves.
DISCUSSION
Many splicing factors have been shown to be tissue specific and developmentally regulated both at transcriptional and posttranscriptional levels, thus indicating their crucial role in splicing events at specific stages of organism development (Lazar et al., 1995; Lopato et al., 1996a,b, 1999b; Caceres and Krainer, 1997; Golovkin and Reddy, 1998; Lazar and Goodman, 2000; Smith and Valcarcel, 2000). In this study, we demonstrate that expression of the putative splicing factor atRSZ33 is subjected to tissue-specific and developmental control because it was detected during embryo and seed development, as well as in other rapidly growing zones of the plant, including flowers, shoot apical meristem, and developing lateral root primordia. Expression of atRSZ33 has been also found in the tip of the primary root with maximum in cells of the elongation zone, indicating that it may be essential for cell expansion rather than for cell divisions in the meristems. In line with the studies of atRSZ33 localization, our results demonstrate that ectopic expression of atRSZ33 has dramatic effects on various developmental processes in Arabidopsis. This is consistent with results from different organisms, which show that interference with expression levels of SR proteins affects development (Kraus and Lis, 1994; Wang et al., 1996; Jumaa et al., 1999; Lopato et al., 1999b; Hoffman and Lis, 2000; Longman et al., 2000; Wang et al., 2001).
Interestingly, transformation of plants with genomic or cDNA constructs of atRSZ33 had a different impact on plant viability. The majority of 35S:catRSZ33 lines had a stronger phenotype in T0 generation than 35S:gatRSZ33 lines (i.e., higher regeneration efficiency, more expanded cells, stronger abnormalities in flower development), but no progeny of these plants were obtained, which suggests that strong overexpression of atRSZ33 is lethal. This is further supported by the fact that only phenotypically normal plants with inactive atRSZ33 cDNA transgene, showing no changes either in RNA or in protein levels, could proceed to further generations. Additionally, analysis of 35S:catRSZ33 plants of T0 generation revealed alternative transcripts, which can only originate from endogenous atRSZ33, indicating that atRSZ33 autoregulates splicing of its own pre-mRNA. Negative feedback regulation by alternative splicing of their own pre-mRNAs leading to truncated proteins (Wang et al., 1996; Jumaa and Nielsen, 1997; Lopato et al., 1999b) or to altered mRNA stability (Sureau et al., 2001) had been demonstrated for other splicing factors as well. When the genomic clone of atRSZ33 is used for transformation, the amount of alternative transcripts is higher than in 35S:catRSZ33 T0 lines (Figure 1B). Accumulation of alternative transcripts can be partially explained by overloading the splicing machinery by high amounts of atRSZ33 pre-mRNA. However, it is also likely that overexpressed atRSZ33 protein modulates splicing of the transgene in a similar manner, as described for the endogenous gene in 35S:catRSZ33 T0 lines. Therefore, 35S: gatRSZ33 plants can modulate the overexpression of atRSZ33 much better than 35S:catRSZ33 plants. However, higher amounts of overexpressed protein caused more severe phenotype in 35S:gatRSZ33 T3 lines. Thus, we can assume that atRSZ33 protein levels over a certain threshold are deleterious for plant development.
Interestingly, only one of the alternative transcripts of atRSZ33, namely, mRNA3, using a 3′ alternative splice site in the second intron, has been found in wild-type plants during early stages of development (Lopato et al., 2002), and two other transcripts, differing in processing of the second intron, but both retaining the third intron, have been detected only in transgenic plants, suggesting an influence of atRSZ33 on the inclusion of the third intron.
Although we have found that atRSZ33 can regulate splicing of its own pre-mRNA, thus implicating its function in splicing, finding target genes for atRSZ33 is hindered by the fact that there are very few examples in plants describing tissue-specific or developmentally regulated splicing events in particular genes. Using overexpressing lines, we tested the effect of atRSZ33 on splicing of two genes encoding SR proteins, atSRp30 and atSRp34/SR1, which have been previously shown to be regulated by alternative splicing (Lazar et al., 1995; Lopato et al., 1999b). We have found that pre-mRNA of atSRp30 is spliced more efficiently in transgenic lines overexpressing atRSZ33, and that the amount of transcript encoding full-length atSRp30 protein was increased. This agrees well with some phenotypic changes in plants overexpressing atSRp30 (Lopato et al., 1999b), like increased cell size, trichomes with higher number of branches, shortened and more branched root system. The splicing pattern of atSRp34/SR1 was also modulated by atRSZ33 because almost no alternative transcript of atSRp34/SR1 was detected in 35S:gatRSZ33 root samples, in contrast to controls. Because both atSRp30 and atSRp34/SR1 are expressed during different stages of lateral root development (Lopato et al., 1999b), altered splicing patterns of these genes may explain the distorted root phenotype of 35S:gatRSZ33 plants.
Ectopic expression of atRSZ33 caused pleiotropic changes in the phenotype of transgenic plants. We have detected increase of cell size in various tissues. In hypocotyls, cells acquired more isodiametrical shape, but the number of the cells was not changed, thus explaining the stunted phenotype of the seedlings. The shape of root hairs and trichomes, together with impaired growth of pollen tubes, indicates that these types of cells have lost their ability to direct axis of expansion. Changes in the cell shape, misorientation of cell division plane, and additional divisions have been detected in the suspensor, possibly leading to the formation of twin embryos in the 35S:gatRSZ33 transgenic lines. Several mutants affecting cell divisions in the suspensor were described in Arabidopsis. Interestingly, one of them, sus2, has a T-DNA insertion in a gene with high similarity to the yeast PRP8 gene, which encodes an essential spliceosomal protein (Meinke, 1996). Whether there is a functional relationship of atRSZ33 and SUS2 in the splicing process needs further investigation.
Another interesting feature of plants ectopically expressing atRSZ33 was the formation of clusters with multiple stomata. In wild type, stomatal units consist of one stomate and are separated by at least one epidermal cell. Spatial distribution of stomata and their development are controlled by the frequency and orientation of asymmetric divisions in the epidermis (Berger and Altmann, 2000). Detailed analysis of epidermal tissue in transgenic plants has revealed multiple divisions in the area of future cluster and at least some of those divisions are misoriented. A similar effect on stomatal development can be achieved when wild-type seedlings are grown under high humidity or treated with 1-aminocyclopropane-1-carboxylate, which is a precursor of the plant hormone ethylene (Serna and Fenoll, 1997). Possibly, atRSZ33 ectopic expression can influence one of these processes. Additionally, several mutants, such as flp, tmm, and sdd, with defects at different stages of stomatal development were described, and recently genes affected in two of these mutants were cloned (Berger and Altmann, 2000; Nadeau and Sack, 2002). The SDD gene encodes a subtilisin-like serine protease, which is involved in controlling cell division orientation during stomatal development and is proposed to process a precursor molecule involved in signaling during meristemoid activity (Berger and Altmann, 2000). Interestingly, we have demonstrated that another subtilisin-like serine protease AIR3 is strongly up-regulated in the roots of 35S:gatRSZ33 plants (Figure 2). AIR3, which might be involved in signal transduction by modification of cell wall components during lateral root development is also expressed in wild-type leaf tissues (Neuteboom et al., 1999), but it is not known whether AIR3 plays any role in stomata development.
We assumed that some of the phenotypic features in the 35S:gatRSZ33 plants could be related to changes in plant hormone levels. The plant hormone auxin affects cell divisions, cell elongation, and differentiation, and influences initiation of organ formation (Davies, 1995). In addition, auxin distribution is an important regulator for pattern formation at different stages of plant development (reviewed by Palme and Galweiler, 1999; Sabatini et al., 1999). Crosses of 35S:gatRSZ33 plants with plants carrying an auxin responsive DR5:GUS reporter gene (Ulmasov et al., 1997) have demonstrated that its activity is dramatically reduced within root tips, and the location of its residual activity is shifted in comparison to control plants. However, the reporter gene activity was up-regulated in leaves and pattern of its expression depicted enhanced vascular strand formation. In vitro regeneration assay corroborated the data on elevated amounts of auxin in the leaves of 35S:gatRSZ33 plants, because we observed a shift from shoot to root formation. We have not detected any changes in the expression of several tested genes of the auxin pathway, including AUX1 (Bennett et al., 1996) and PIN1 (Galweiler et al., 1998) (our unpublished data). It is not clear whether redistribution of auxin in tissues of 35S:gatRSZ33 plants results from the changes in the cell shape and polarity, thus changing the localization of auxin influx and/or efflux carriers, or whether some genes of the auxin pathway are affected by atRSZ33 directly. In addition, the possibility of changes downstream of the auxin-signaling pathway should be considered.
The overexpression approach was for us the method of choice, which allowed us to check activity of atRSZ33 in plants as no in vitro splicing system is available in plants. Interestingly, despite the restricted expression pattern of atRSZ33 mainly in the roots and flowers, overexpression of atRSZ33 caused dramatic changes in plant development. We are currently searching for a mutant of atRSZ33 to validate the biological significance of these pleiotropic changes. Although the loss-of-function approach proved to be successful in dissecting the function of many genes, it is worth mentioning that mutants of some splicing factors are lethal (Ring and Lis, 1994; Wang et al., 1996; Jumaa et al., 1999). On other hand, knockouts may have a problem when studying members of multigene families with possible overlapping functions. In this case, simultaneous disruption of several genes is necessary to investigate their function and to reveal phenotypic changes. Indeed, only simultaneous suppression of two or more SR proteins in C. elegans showed strong and specific phenotypes (Longman et al., 2000). Whether atRSZ33 has an essential or redundant function in plant development still remains to be investigated.
In conclusion, our previous results (Lopato et al., 2002) and the ones reported in this article show that atRSZ33 is clearly involved in splicing. However, because of the uniqueness of its domain structure, it is also challenging to determine whether atRSZ33 has activities in other aspects of plant RNA metabolism.
Acknowledgments
We thank Zdravko Lorković, Marie-Theres Hauser, and Christian Luschnig for critical reading of the manuscript and helpful discussions; Viktor Voronin for help with in situ hybridization; and Tom Guilfoyle and Christian Luschnig for seeds of DR5:GUS plants. This work was supported by grants from the Österreichischer Fonds zur Förderung der wissenschaftlichen Forschung to AB (P12364-GEN, SFB-F017/10/11).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–02–0109. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0109.
Abbreviations used: 35S CaMV, 35S RNA promoter from cauliflower mosaic virus; GUS, β-glucuronidase; RNP, ribonucleoprotein; RRM, RNA recognition motif; RS, arginine- and serinerich; snRNP, small nuclear ribonucleoprotein; SR, serine- and arginine-rich; UTR, untranslated region.
References
- Barta, A., Sommergruber, K., Thompson, D., Hartmuth, K., Matzke, M.A., and Matzke, A.J.M. (1986). The expression of a nopaline synthase-human growth hormone chimaeric gene in transformed tobacco and sunflower callus tissue. Plant Mol. Biol. 6, 347–357. [DOI] [PubMed] [Google Scholar]
- Baskin, T.I., Busby, C.H., Fowke, L.C., Sammut, M., and Gubler, E. (1992). Improvements in immunostaining samples embedded in methacrylate: localization of microtubules and other antigens throughout developing organs in plants of diverse taxa. Planta 187, 405–413. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273, 948–950. [DOI] [PubMed] [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Caceres, J.F., and Krainer, A.R. (1997). Mammalian pre-mRNA splicing factors. In: Eukaryotic mRNA Processing: Frontiers in Molecular Biology, ed. A.R. Krainer, Oxford: IRL Press, 174–212.
- Chen, C., and Chen, Z. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). Plant Hormones, Dordrecht, The Netherlands: Kluwer Academic Publishers.
- Dornelas, M.C., Wittich, P., von Recklinghausen, I., van Lammeren, A., and Kreis, M. (1999). Characterization of three novel members of the Arabidopsis SHAGGY-related protein kinase (ASK) multigene family. Plant Mol. Biol. 39, 137–147. [DOI] [PubMed] [Google Scholar]
- Friml, J., and Palme, K. (2002). Polar auxin transport - old questions and new concepts? Plant Mol. Biol. 49, 273–284. [PubMed] [Google Scholar]
- Galweiler, L., Guan, C., Muller, A., Wisman, E., Mendgen, K., Yephremov, A., and Palme, K. (1998). Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282, 2226–2230. [DOI] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S. (1998). The plant U1 small nuclear ribonucleoprotein particle 70K protein interacts with two novel serine/arginine-rich proteins. Plant Cell 10, 1637–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin, M., and Reddy, A.S. (1999). An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1–70K protein. J. Biol. Chem. 274, 36428–36438. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. (2000). Sorting out the complexity of SR protein functions. RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekema, A., Hirsch, P.R., Hooykaas, P.J.J., and Schilperoort, R.A. (1983). A binary plant vector strategy based on separation of vir and T-region of the A. tumefaciens Ti-plasmid. Nature 303, 179–180. [Google Scholar]
- Hoffman, B.E., and Lis, J.T. (2000). Pre-mRNA splicing by the essential Drosophila protein B52: tissue and target specificity. Mol. Cell. Biol. 20, 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski, S., Riou-Khamlichi, C., Roche, O., Perennes, C., Bergounioux, C., and Glab, N. (2002). The CDK inhibitor NtKIS1a is involved in plant development, endoreduplication and restores normal development of cyclin D3; 1-overexpressing plants. J. Cell Sci. 115, 973–982. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa, H., and Nielsen, P.J. (1997). The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16, 5077–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumaa, H., Wei, G., and Nielsen, P.J. (1999). Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9, 899–902. [DOI] [PubMed] [Google Scholar]
- Kandasamy, M.K., McKinney, E.C., and Meagher, R.B. (2002). Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13, 251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik, V., Schnittger, A., Radchuk, V., Adler, K., Hulskamp, M., and Baumlein, H. (2001). Ectopic expression of the Arabidopsis At-MYB23 gene induces differentiation of trichome cells. Dev. Biol. 235, 366–377. [DOI] [PubMed] [Google Scholar]
- Kramer, A. (1996). The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Kraus, M.E., and Lis, J.T. (1994). The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol. Cell. Biol. 14, 5360–5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labourier, E., Bourbon, H.M., Gallouzi, I.E., Fostier, M., Allemand, E., and Tazi, J. (1999). Antagonism between RSF1 and SR proteins for both splice-site recognition in vitro and Drosophila development. Genes Dev. 13, 740–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, G., and Goodman, H.M. (2000). The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 42, 571–581. [DOI] [PubMed] [Google Scholar]
- Lazar, G., Schaal, T., Maniatis, T., and Goodman, H.M. (1995). Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl. Acad. Sci. USA 92, 7672–7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo, G.R., Stein, P.A., and Ludwig, R.A. (1991). A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967. [DOI] [PubMed] [Google Scholar]
- Longman, D., Johnstone, I.L., and Caceres, J.F. (2000). Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Forstner, C., Kalyna, M., Hilscher, J., Langhammer, U., Indrapichate, K., Lorkovic, Z.J., and Barta, A. (2002). Network of interactions of a novel plant-specific Arg/Ser-rich protein, atRSZ33, with atSC35-like splicing factors. J. Biol. Chem. 277, 39989–39998. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Gattoni, R., Fabini, G., Stevenin, J., and Barta, A. (1999a). A novel family of plant splicing factors with a Zn knuckle motif: examination of RNA binding and splicing activities. Plant Mol. Biol. 39, 761–773. [DOI] [PubMed] [Google Scholar]
- Lopato, S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A.R., and Barta, A. (1999b). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Mayeda, A., Krainer, A.R., and Barta, A. (1996a). Pre-mRNA splicing in plants: characterization of Ser/Arg splicing factors. Proc. Natl. Acad. Sci. USA 93, 3074–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopato, S., Waigmann, E., and Barta, A. (1996b). Characterization of a novel arginine/serine-rich splicing factor in Arabidopsis. Plant Cell 8, 2255–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., and Barta, A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30, 623–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44. [DOI] [PubMed] [Google Scholar]
- Manley, J.L., and Tacke, R. (1996). SR proteins and splicing control. Genes Dev. 10, 1569–1579. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W. (1996). Embryo-defective mutants of Arabidopsis: cellular functions of disrupted genes and developmental significance of mutant phenotype. In: Embryogenesis: The Generation of a Plant, ed. T.L. Wang and A. Cuming, Oxford: Bios Scientific Publishers Limited, 35–50.
- Mizukami, Y., and Ma, H. (1992). Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. [DOI] [PubMed] [Google Scholar]
- Nadeau, J.A., and Sack, F.D. (2002). Control of stomatal distribution on the Arabidopsis leaf surface. Science 296, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Neuteboom, L.W., Ng, J.M., Kuyper, M., Clijdesdale, O.R., Hooykaas, P.J., and van der Zaal, B.J. (1999). Isolation and characterization of cDNA clones corresponding with mRNAs that accumulate during auxin-induced lateral root formation. Plant Mol. Biol. 39, 273–287. [DOI] [PubMed] [Google Scholar]
- Palme, K., and Galweiler, L. (1999). PIN-pointing the molecular basis of auxin transport. Curr. Opin. Plant Biol. 2, 375–381. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., and Mann, M. (2002). Largescale proteomic analysis of the human spliceosome. Genome Res 12, 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring, H.Z., and Lis, J.T. (1994). The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol. 14, 7499–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini, S., et al. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Sablowski, R.W., and Meyerowitz, E.M. (1998). A homolog of no apical meristem is an immediate target of the floral homeotic genes apetala3/pistillata. Cell 92, 93–103. [DOI] [PubMed] [Google Scholar]
- Serna, L., and Fenoll, C. (1997). Tracing the ontogeny of stomatal clusters in Arabidopsis with molecular markers. Plant J. 12, 747–755. [DOI] [PubMed] [Google Scholar]
- Smith, C.W., and Valcarcel, J. (2000). Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Sureau, A., Gattoni, R., Dooghe, Y., Stevenin, J., and Soret, J. (2001). SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 20, 1785–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens, D., Van Montague, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants using kanamycin selection. Proc. Natl. Acad. Sci. USA 85, 5536–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff, E., and Hooykaas, P.J.J. (1998). Transformation of Arabidopsis thaliana C24 leaf discs by Agrobacterium tumefaciens. In: Arabidopsis Protocols, vol. 82, ed. J. Martinez-Zapater and J. Salinas, Totowa, NY: Humana Press, 245–258. [DOI] [PubMed] [Google Scholar]
- Wang, H.Y., Xu, X., Ding, J.H., Bermingham, J.R., Jr., and Fu, X.D. (2001). SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell 7, 331–342. [DOI] [PubMed] [Google Scholar]
- Wang, J., Takagaki, Y., and Manley, J.L. (1996). Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 10, 2588–2599. [DOI] [PubMed] [Google Scholar]
- Wang, J., Xiao, S.H., and Manley, J.L. (1998). Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 12, 2222–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler, A.M., Lane, W.S., Stolk, J.A., and Roth, M.B. (1992). SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6, 837–847. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., and Reed, R. (2002). Comprehensive proteomic analysis of the human spliceosome. Nature 419, 182–185. [DOI] [PubMed] [Google Scholar]