Abstract
A recent model for the mechanism of intrinsic transcription termination involves dissociation of the RNA from forward-translocated (hypertranslocated) states of the complex [Yarnell WS, Roberts JW (1999) Science, 284:611–615]. The current study demonstrates that halted elongation complexes of T7 RNA polymerase in the absence of termination signals can also dissociate via a forward-translocation mechanism. Shortening of the downstream DNA or the introduction of a stretch of mismatched DNA immediately downstream of the halt site reduces a barrier to forward translocation and correspondingly reduces the lifetime of halted complexes. Conversely, introduction of a cross-link downstream of the halt site increases the same barrier and leads to an increase in complex lifetime. Introduction of a mismatch within the bubble reduces a driving force for forward translocation and correspondingly increases the lifetime of the complex, but only for mismatches at the upstream edge of the bubble, as predicted by the model. Mismatching only the two most upstream of the eight bases in the bubble provides a maximal increase in complex stability, suggesting that dissociation occurs primarily from early forward-translocated states. Finally, addition in trans of an oligonucleotide complementary to the nascent RNA just beyond the hybrid complements the loss of driving force derived from placement of a mismatch within the bubble, confirming the expected additivity of effects. Thus, forward translocation is likely a general mechanism for dissociation of elongation complexes, both in the presence and absence of intrinsic termination signals.
Keywords: hypertranslocation, hybrid, stability, topological lock
Transcription lies at the heart of cellular gene expression. Unlike replication, transcription is not distributive; that is, any dissociation of an elongation complex is a terminal event. Consequently, elongation complexes must be highly stable while transcribing at up to several hundred bases per second. Recent studies demonstrate, however, that elongation is not a uniform process, with reasonably long-lived, sequence-dependent pauses occurring stochastically (1–4). Indeed, the elongation phase is well known to be subject to regulation through attenuation: sequence-dependent signals leading to pause, arrest, slippage, or termination (5). Elongating RNA polymerases must hold the DNA template and the RNA product tightly enough to be highly stable through nonterminating pauses, yet they must be able to dissociate the complex in response to specific sequences in the DNA. Understanding this interplay requires understanding mechanisms of dissociation.
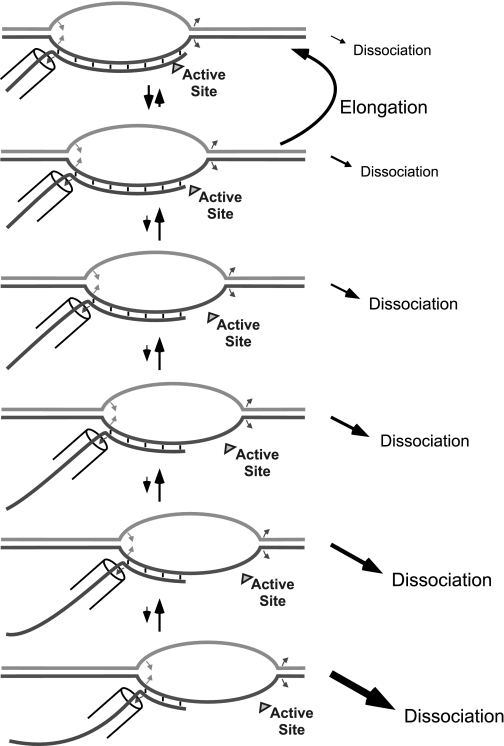
In classic rho-independent or “intrinsic” termination, dissociation is thought to occur as the polymerase slows in response to sequence, and structure begins to form concurrently in the nascent transcript (6–8). A run of encoded U's has been proposed both to slow transcription and to weaken the RNA–DNA hybrid. Recently, it has been proposed that, rather than direct dissociation of the nascent transcript from a complex halted at the primary termination site (with or without allosteric assistance), dissociation occurs primarily from forward-translocated states (9–11). In forward-translocated (hypertranslocated) states, the hybrid is shortened, weakening the binding of the RNA to the complex. Additionally, shortening of the hybrid may also serve to remove a topological locking of the RNA onto the template strand. As illustrated in Fig. 1, dissociation of the RNA transcript would be substantially faster from forward translocated states than from the initially paused state. Although forward-translocated states are expected to be energetically disfavored relative to the initial on-pathway state and so would be expected to exist at lower populations, the difference in the kinetics of dissociation could compensate such that the dominant dissociative pathway proceeds via forward translocated states.
Fig. 1.
Forward translocation as a mechanism of elongation complex dissociation. Halted complexes distribute among various forward-translocated states, with complex dissociation being favored from those states with the smallest hybrids.
The model shown in Fig. 1 predicts certain behaviors. In general, forward translocation requires melting of the DNA duplex downstream and dissociation of the most upstream base(s) in the hybrid. These costs are partially offset by the energy gained by collapse of a base pair in the DNA at the upstream edge of the bubble. Thus, one would expect that altering the balance between these effects could shift the distribution toward more forward-translocated states and therefore toward or away from dissociation. As demonstrated previously, a direct crosslink between the DNA strands downstream of the halt site should prevent forward translocation and is in fact seen to reduce termination from an intrinsic terminator (9).
We propose that this model for dissociation is more general and applies not only to termination mediated by external assistive forces but also to dissociation of halted elongation complexes in the absence of external forces. We also propose that weakening of the complex via forward translocation is a general mechanism for complex dissociation, applicable not only to the multisubunit family of RNA polymerases but also to the evolutionarily unrelated single-subunit RNA polymerases best represented by the RNA polymerase from bacteriophage T7. Weakening of the complex via forward translocation is a general mechanism for complex dissociation. Indeed, footprinting studies have suggested that artificially halted (by nucleotide starvation) T7 RNA polymerase elongation complexes exist in translocational states beyond the simple pre- and posttranslocated “on-pathway” states (12).
In the current study, we use the T7 RNA polymerase system to examine the stability of halted elongation complexes as a function of perturbations to the system that are intended to either favor or disfavor the distribution toward forward-translocated states. Those that favor forward translocation lead to an expected decrease in stability of the complex, whereas those that disfavor forward translocation increase the stability of the elongation complexes.
Results
In the experiments that follow, elongation complex stability is determined by measuring steady-state turnover with transcription halted at a unique site in the DNA. The more unstable complexes dissociate readily and so exhibit higher total turnover. The halted complexes are expected to decay with a first-order rate constant, characterized by an average half-life (lifetime) that can be derived from measurements of turnover. The first-order rate constant is estimated as the velocity of RNA synthesis divided by the concentration of the limiting species, in this case the enzyme [promoter binding and initiation are fast relative to the turnover rates seen here (13, 14)]. We have recently shown that measurements of elongation complex stability can be dominated by “bumping” at 1:1 or higher ratios of enzyme to DNA (15). To provide a true measure of intrinsic complex stability, a low polymerase:DNA ratio was therefore used to eliminate bumping between trailing and halted complexes.
Role of the Downstream DNA in Complex Stability.
Elongation complexes halted internally are substantially more stable than complexes run off to the end of linear DNA. Conventional wisdom would conclude therefore that interactions between the protein and downstream DNA contribute to the stability of internally halted complexes. Alternatively (or in addition), downstream duplex DNA presents a barrier to forward translocation, leading to increased stability in internally halted complexes.
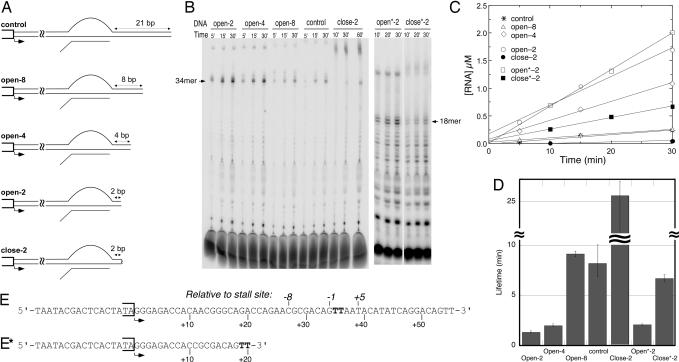
To quantitatively assess the contributions of downstream DNA to halted complex stability, constructs were prepared containing varying lengths of DNA downstream of a halt site positioned 34 bases from the promoter, as shown in Fig. 2A. The results indicate that including eight base pairs downstream of the halt is sufficient to provide maximal stability, whereas reduction of this duplex to four and two base pairs, respectively, progressively destabilizes the complex. This result is consistent with either of the two models for the contributions of downstream DNA to complex stability.
Fig. 2.
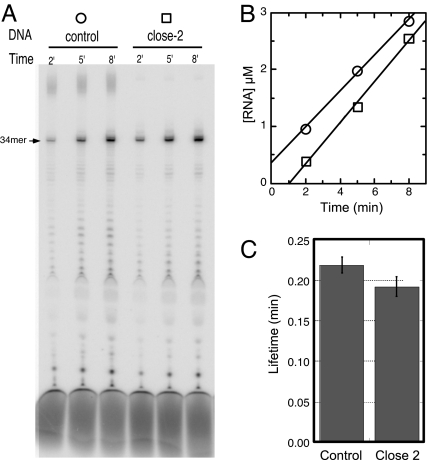
Contributions of downstream DNA to stability. (A) DNA constructs with varying lengths of downstream DNA. Sequences up to the halt site are identical for all constructs and allow halting at position +34 in the presence of GTP, ATP, and CTP. All constructs are fully duplex; the bubble in a halted complex is shown for clarity. In close-2 and close*-2, the end of the DNA is closed by an 18-atom polyethylene glycol spacer that covalently links the template and nontemplate strands, preventing full melting. (B) Denaturing gel electrophoresis of labeled RNA products at increasing time intervals. Reactions at 37°C contained 0.1 μM enzyme and 0.5 μM DNA (0.2/1.0 μM for the open*-2 and close*-2 constructs). (C) Stalled RNA products from B were quantified, and the data were plotted as a function of time. The slopes (velocities) divided by the concentration of enzyme were used as an estimate for the first-order rate constants (koff) for dissociation, the rate limiting step in turnover. (D) The rate constants were then used to calculate the half-lives of the halted elongation complexes by using t½ = ln(2)/koff. (E) The nontemplate strand sequence of the control construct (others have the same sequence truncated appropriately). (E*) The nontemplate strand sequence of the constructs open*-2 and close*-2.
If melting into the downstream DNA is necessary for dissociation, as predicted by forward translocation, then in the construct containing only two base pairs past the halt site, the introduction of a cross-link should restore stability (9). The construct “close-2” does just this, and the results show a large increase in the lifetime of halted complexes. However, this construct leads to apparently larger transcripts that may arise from turn-around synthesis, a phenomenon known to be sequence-dependent and thought to be more prominent with increasingly stable halted complexes (16). To measure complex stability in the absence of this complicating effect, we repeated the cross-linking experiment with a different terminal sequence, shown in constructs close*-2 and open*-2. The results confirm that cross-linking of the end of the duplex yields a 3-fold increase in stability, comparable with that of an internally stalled complex.
This result not only supports the forward-translocation model, but also suggests that interactions between the polymerase and the duplex downstream are not significant in stabilizing elongation complexes. Thus the relative instability of complexes run off to the end of linear DNA arises primarily via the removal of a barrier to forward translocation.
Altering Energy Barriers to and Driving Forces for Forward Translocation.
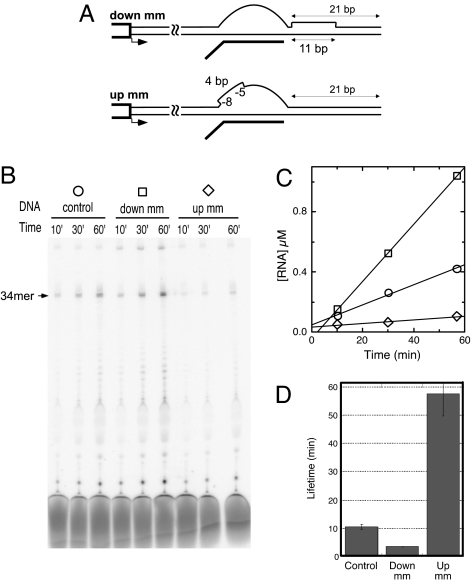
Presumably, the reannealing of the upstream DNA duplex is the major driving force for the forward movement of the transcription bubble, and opening of the downstream DNA duplex is an energy cost for forward translocation. We have designed constructs that introduce local mismatches (mm) in the DNA to either remove the driving force of upstream reannealing (up mm in Fig. 3A) or remove the need to melt downstream DNA (down mm in Fig. 3A) (10). Both constructs are based on the control construct described in Fig. 2. In the first case (up mm), nontemplate strand bases at positions −8 to −5 relative to the halt site were replaced by GCGC. In the second case (down mm), nontemplate strand bases at positions +1 through +11 relative to the halt site were replaced by the sequence AACCGCGGCGC.
Fig. 3.
Reducing the downstream barrier to and the upstream driving force for forward translocation. (A) Constructs used. (B) Denaturing gel electrophoresis of reactions as described in Fig. 2. (C and D) Quantification of stability as described in Fig. 2.
As predicted by the model, decreasing/removing the downstream barrier to melting by the introduction of an extended stretch of mismatched bases just downstream of the bubble (down mm in Fig. 3A) leads to an approximately 3-fold decrease in the stability of the complex. Removal of the energetic barrier to downstream duplex melting presumably shifts the distribution of complexes (see Fig. 1) toward more forward-translocated states, thereby leading to higher overall rates of dissociation.
In contrast, mismatching four bases at the upstream edge of the bubble (up mm in Fig. 3A) removes a driving force for forward translocation: collapse of the upstream edge of the bubble as the fixed length bubble translocates forward (17). The observed result is an ≈5-fold increase (relative to the control) in the lifetime of halted elongation complexes. This result provides complementary support for forward translocation as a mechanism for complex dissociation.
Forward Translocation Is Driven by Collapse of the Upstream Edge of the Bubble.
The observation that the introduction of a mismatch into the bubble region, as shown in Fig. 3A, leads to an increase in stability of the halted elongation complex is consistent with the forward-translocation model but is also consistent with other more simple models. Release of the transcript by an overall and direct collapse of the bubble would also predict the observed behavior. The forward-translocation model requires, specifically, collapse from the upstream end of the bubble. In most of the forward-translocated states of Fig. 1, the downstream bases from the initial bubble remain melted, such that mismatching these should have no effect.
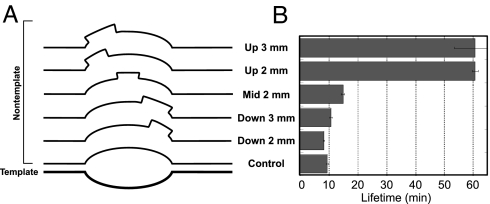
The constructs presented in Fig. 4A are aimed to test these predictions (10). As predicted by the model, those constructs containing a mismatch at the upstream end of the bubble show a large increase in complex lifetime, whereas similar mismatches at the downstream end show no detectable effect. Indeed, a two-base mismatch at the upstream end increases stability ≈5-fold, whereas a three-base mismatch downstream has no significant effect. It is interesting that a two-base mismatch located in the center of the bubble increases complex stability only slightly. This finding suggests that dissociation of the complexes occurs from translocational states (see Fig. 1) not too far removed from the initial pause site (wherein the middle part of the bubble has not yet collapsed).
Fig. 4.
Collapse of the upstream edge of the bubble helps to drive forward translocation. Placement of two- or three-base mismatches at different positions in the bubble demonstrates that stability derives from decreasing reannealing at the upstream edge only. (A) Shown are constructs used; control is fully matched. (B) Lifetimes of halted complexes on the indicated constructs. Reactions and analyses are the same as for Fig. 2.
Complex Dissociation via Bumping Does Not Require Forward Translocation.
We have recently shown that under 1:1 or excess enzyme conditions, the primary mechanism for elongation complex dissociation is displacement by bumping from a second, trailing RNA polymerase (15). Under these conditions, elongation complexes show a much shorter effective lifetime. Because collision by a trailing RNA polymerase would logically “push” the halted polymerase forward, displacement of the halted complex could also occur via a forward-translocation mechanism. To test this hypothesis, we carried out measurements of complex lifetimes on two of the previously characterized constructs: the control DNA with 21 base pairs downstream of the halt and the DNA construct containing a covalent cross-link two bases downstream of the halt site (close-2). As demonstrated above, the latter is substantially more stable, because conventional forward translocation should be impossible.
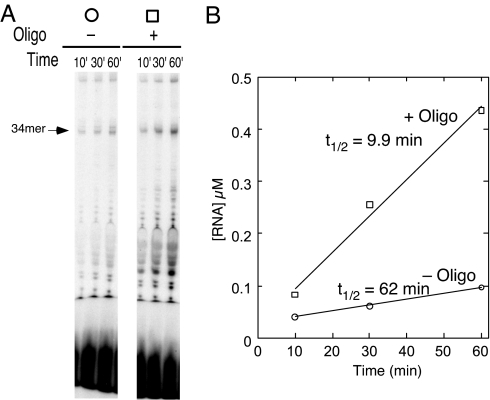
The results presented in Fig. 5show much shorter complex lifetimes under conditions of enzyme in excess, as reported previously (15). However, no difference in complex stability is observed for the two constructs. This suggests that dissociation via bumping does not require forward translocation of the halted complex.
Fig. 5.
Bumping does not require forward translocation. In order make bumping the dominant mechanism of dissociation, experiments were carried out as described in Fig. 2 but with concentrations of enzyme and DNA reversed (enzyme, 0.5 μM; DNA, 0.1 μM). (A) Denaturing gel electrophoresis of reactions. (B and C) Quantification of stability is the same as in Fig. 2. Note the much shorter lifetimes compared with previous figures.
Structure in the Nascent RNA Complements Loss of Driving Force from Weakened Upstream Bubble Collapse.
For intrinsic termination, the complex is not fully halted but is presumably transiently halted and/or slowed substantially in its normal elongation. As recently proposed, formation of structure in the nascent transcript, pulling the RNA through the exit channel, provides a driving force for forward translocation that is not present in normal paused or halted complexes (9, 10). Formation of structure in the transcript has also been modeled by the addition in trans of an oligonucleotide complementary to the initially exiting RNA (11, 18).
To add this additional driving force to our study, we have carried out a similar experiment in which we added an excess of an oligonucleotide complementary to transcript positions −24 to −9 (relative to the halt site). In this case, for the transcribed DNA, we used a construct (up 2) that is mismatched from positions −8 to −7 relative to the halt site and so is unusually stable. The results presented in Fig. 6demonstrate that annealing of the complementary oligonucleotide to the exiting RNA approximately replaces the driving force lost by introduction of the mismatch. The lifetime of the complex is reduced ≈6-fold, restoring it to the original control level. As proposed for termination, formation of structure in the nascent RNA effectively “pulls” the RNA from the complex, driving the complex forward.
Fig. 6.
Introduction of a new driving force complements the loss of upstream bubble collapse. Transcribing from template (up 2), which contains a mismatch at positions −8 and −7 in the bubble (see Fig. 4), an oligonucleotide complementary to bases −24 to −9 of the RNA transcript was added in ≈150-fold excess over DNA (to drive binding).
Discussion
Transcription elongation complexes possess a dual need of being extremely stable while transcribing but readily dissociable in response to signals encoded in the DNA. It has been proposed that sequence-dependent termination proceeds via forward translocation (in the absence of synthesis) of the complex, resulting in a progressive decrease in the size of the RNA–DNA hybrid (9, 10). Thus, although complexes with an eight-base pair RNA–DNA hybrid are extremely stable against dissociation (via both the intrinsic stability of the protein-bound hybrid and a topological “locking” of the transcript around the template strand), as forward progression proceeds, this stability decreases. As shown in Fig. 1, dissociation of the RNA would then occur primarily from a forward-translocated state. At termination, this process is proposed to be driven by formation of structure in the nascent RNA at the protein surface. The primary driving force is then the pulling of the 5′ end of the RNA from the RNA–DNA hybrid.
RNA polymerases are known to pause, perhaps stochastically, along the DNA in the absence of traditional terminating sequences (2). Halted complexes can be created and characterized in vitro at precise positions in the DNA by the simple absence of one or more nucleoside triphosphates from the medium. Ultimately, such complexes will dissociate with a characteristic average lifetime. The current study asks whether these complexes also dissociate primarily via a forward-translocation mechanism.
Runoff Complexes Lack a Barrier to Forward Translocation.
As illustrated in Fig. 2, complexes run off to the end (or halted very near the end) of the DNA duplex are significantly less stable than those halted internally. The simplest interpretation of this result is that the polymerase possesses stabilizing interactions with the downstream DNA duplex, and analysis of the crystal structures of elongation complex models would appear to support this proposal (19, 20). Indeed, although the presence of only eight base pairs downstream of the halt site provides maximal stability, progressive shortening of the DNA results in a corresponding decrease in the stability of the complex.
This behavior is also predicted, however, by the forward-translocation model for dissociation. To forward translocate, the complex must melt DNA downstream of the halt site. Removal of downstream DNA reduces an energetic barrier to forward translocation, such that the distribution of complexes will be shifted toward the more forward-translocated states, from which RNA release occurs more rapidly. For the (unstable) complex halted two bases short of the end of the duplex, we can restore this energetic barrier by the introduction of a polyethylene glycol linker connecting the 5′ and 3′ ends of the downstream duplex DNA. As predicted by the forward-translocation model, the stability of the complex is restored. This return to stability occurs in the absence of potential downstream interactions, arguing either that downstream interactions contribute to stability but that the instability generated by their deletion is offset by the increase in stability afforded by the cross-link or more simply that downstream duplex interactions are of significantly less importance in their contribution to elongation complex stability.
Removal of the Downstream Barrier in Internally Halted Complexes.
For internally halted complexes, we can remove the downstream barrier to forward translocation by simply replacing a stretch of duplex DNA with intentionally mismatched DNA. The results summarized in Fig. 3 (down mm) demonstrate an effect analogous to removal of the downstream DNA, with essentially the same results. Complexes halted at a site with mismatched DNA downstream of the halt site are substantially less stable than control complexes. The downstream barrier has been removed, allowing a distributional shift toward forward-translocated complexes with shorter hybrids.
Removal of an Upstream Driving Force.
Although the need to melt DNA downstream of the transcription bubble presents a barrier to forward translocation, collapse of the open bubble at its upstream end should drive complexes toward forward-translocated states. Hence, mismatching of bases known to lie at the upstream end of the open bubble should remove that driving force, leading to more stable elongation complexes. The T7 RNA polymerase elongation complex has been shown to possess a melted bubble extending approximately eight base pairs upstream of the halt site (12, 21). We have now demonstrated (Fig. 3, up mm) that mismatching of the four bases inside the upstream edge of the bubble is observed to dramatically increase the stability of halted complexes.
Any model for dissociation of a halted elongation complex would predict that mismatches within the bubble will lead to increased stability, and this has been observed (22). The forward-translocation model uniquely predicts that only mismatches at the upstream edge will lead to such stability, because collapse proceeds sequentially from the upstream edge (10). Mismatching inside the downstream edge of the bubble should have no effect on stability, because all but the most extreme forward-translocated states remain melted in those bases.
This predicted behavior is demonstrated nicely by the data summarized in Fig. 4. Indeed, an increase in stability is observed only for mismatches of the two or three most upstream bases. Even a two-base mismatch in the center of the bubble has little effect on stability. This latter result suggests that dissociation occurs from complexes that have forward translocated by only a few bases (the uppermost states in Fig. 1). If eight base pairs is the minimal length required to achieve a topological lock, then shortening of the hybrid by two or three bases would be sufficient to release that lock.
An eight-base hybrid duplex would not be expected to be stable in solution (23, 24). Clearly, to the extent that hybrid stability contributes to retention of the transcript in an elongation complex, protein–duplex interactions must contribute to this stability. In any case, shortening of the hybrid by only two bases represents a significant fractional decrease in stability. These results would also argue for dissociation from complexes that have forward translocated by only a few bases, as observed here.
Introduction of a Termination-Like Driving Force.
In order to forward translocate, the enzyme must melt downstream DNA and must dissociate RNA from the hybrid duplex, but this is assisted by collapse of the upstream edge of the bubble. In class I (rho-independent) termination, the latter is facilitated by formation of a hairpin in the RNA. Completion of the hairpin requires that RNA be extruded from the RNA exit channel, at the expense of dissociating RNA from the hybrid. Hairpin formation has been simulated by the annealing, in trans, of an oligonucleotide complementary to the nascent RNA just 5′ of the heteroduplex. To test this model in general and to test the complementarity of the various forces, we have carried out a similar experiment on the DNA construct containing a two-base mismatch just within the upstream edge of the bubble. As predicted by the model, addition of a short piece of DNA complementary to bases −9 to −24, relative to the halt site, decreases the lifetime of the complex 10-fold.
Bumping Does Not Require Forward Translocation.
In a recent study, we showed that for T7 RNA polymerase, halted elongation complexes are subject to dissociation via bumping from a trailing RNA polymerase, even at 1:1 ratios of enzyme to DNA (15). To characterize the intrinsic dissociation of halted elongation complexes, the current work avoids this by carrying out reactions with a 5-fold molar excess of DNA.
In complementary work presented here, in Fig. 5, we have shown that bumping in the T7 RNA polymerase system need not proceed via a forward-translocation mechanism. Reversing the ratio of enzyme to DNA leads to conditions in which bumping should dominate and, indeed, the average lifetime of the complexes is reduced dramatically. However, introduction of the polyethylene glycol cross-link just downstream of the halt site, although physically limiting forward translocation, yields no stabilization of the complexes. The force of the trailing RNA polymerase can apparently displace the halted complex via other mechanisms.
General Predictions of the Model.
The observation that reduction of the driving force derived from collapse of the upstream edge of the bubble can be offset by addition of a driving force from an oligonucleotide complementary to the nascent transcript confirms that one must consider carefully the balance of effects introduced by other factors that might influence these processes. For example, it has been demonstrated that elongation complexes of T7 RNA polymerase are less stable on supercoiled than on relaxed DNA (22). Supercoiling would be expected to reduce the energy required to melt downstream DNA and so should reduce the barrier to forward translocation, yielding a less stably halted complex. However, supercoiling would also reduce the driving force derived from collapse of the upstream edge of the bubble and so should yield a more stably halted complex. The observation that complexes on supercoiled DNA are less stable suggests that the reduction in the driving force (rewinding) upstream may be less important than the reduction in the energetic barrier to melting downstream.
It is intriguing to speculate that these effects may be reflected in sequence-dependent variations in elongation complex stability (25) (all of the current studies were carried out on DNA with identical template strand sequences). Thus sequences that weaken upstream bubble collapse may lead to longer lifetimes, whereas sequences that favor melting downstream of the bubble may lead to shorter lifetimes. In predicting such effects, one will have to include not only dinucleotide step thermodynamics but also the energetic sequence-dependence of the edges of the bubbles, perhaps approximated by thermodynamic studies of dangling ends (23, 24, 26, 27). In addition, these duplexes, whether at the upstream or downstream edges of the bubble or in the RNA–DNA hybrid, are not free in solution but may interact with the protein in ways not accounted for by solution–phase model duplexes.
To date, there is no direct evidence for backtracking in T7 RNA polymerase complexes. In contrast to forward tracking, backtracking of a halted complex does not lead to a reduction in the size of the hybrid and so should yield relatively stable complexes in all backtracked states. Failure to partition substantially into these competing states may, in part, explain the relative instability of halted T7 RNA polymerase complexes as compared with those of the multisubunit enzymes, even at low ratios of enzyme to DNA.
Finally, these data point to the relative ease with which RNA polymerase can adopt off-pathway translocated states, consistent with the notion that the action of exonuclease, for example, in exonuclease footprinting can “push” halted complexes from their targeted halt site (12). Indeed, other progressively translocating enzymes may not only perturb the halted complex but may contribute to its instability.
The forward-translocation model for intrinsic termination has now been logically extended to complexes halted in the absence of terminator sequences. The current study argues that in the T7 RNA polymerase system, and likely in the multisubunit RNA polymerases, forward translocation is the primary mechanism for general dissociation of halted elongation complexes. This model also explains the decreased stability of complexes run off to the end of linear DNA, without the need to invoke the loss of stabilizing interactions with DNA downstream of the active site.
Methods
Expression and Purification of RNA Polymerase.
N-terminal His-tagged T7 RNA polymerase was prepared from Escherichia coli strain BL21 carrying the plasmid pBH161, kindly supplied by William T. McAllister (University of Medicine and Dentistry of New Jersey, Stratford, NJ). The enzyme was then purified by an affinity column with nickel nitrilotriacetic acid (NTA) agarose beads (Qiagen, Valencia, CA). The purity of the enzyme was verified by SDS/PAGE analysis to be >95%. The concentration of the enzyme was determined by its absorbance at 280 nm by using ε280 = 1.4 × 105 M−1·cm−1. The enzyme was stored in 20 mM potassium phosphate, 50% glycerol, 1 mM EDTA, 1 mM DTT, and 100 mM sodium chloride at −20°C.
Preparation of DNA Templates.
Oligonucleotides were synthesized by the phosphoramidite method using an Applied Biosystems (Foster City, CA) Expedite 8909 DNA synthesizer. All reagents were purchased from Transgenomic (Omaha, NE) and Glen Research (Sterling, VA). The synthesized ssDNAs were purified by denaturing polyacrylamide gel electrophoresis. DNA from an excised gel slice was electro-eluted using an Elu-Trap apparatus (Schleicher & Schüll, Dassel, Germany), followed by ethanol precipitation. The concentrations of purified oligonucleotide were determined as described in ref. 28. The concentrations of purified oligonucleotide containing the polyethylene glycol spacer (Spacer 18; Glen Research) were determined by Quant-it PicoGreen dsDNA Assay kit (Invitrogen, Carlsbad, CA). The purity of the oligonucleotides was confirmed by denaturing gel electrophoresis of end-labeled ssDNA. Double strand DNA templates were prepared by mixing a 1:1 molar ratio of two complementary single strands in TE buffer (10 mM Tris, I mM EDTA, pH 7.8) and heating to 90°C with slow cooling to room temperature (the same annealing process was also applied to the single oligonucleotide containing the Spacer 18 linker).
Transcription Assays.
Unless otherwise noted, transcription reactions were carried out at 37°C in a total volume of 10 μl. T7 RNA polymerase was present at 0.1 μM, with DNA template at 0.5 μM (for the reactions described in Fig. 5, the concentrations were reversed). After incubating at 37°C for 1 min, NTPs were added to a final concentration of 400 μM each to initiate the reaction, and RNA transcripts were labeled with [α-32P]GTP (3,000 Ci/mmol) (1 Ci = 37 GBq) (PerkinElmer Life Sciences, Boston, MA) for quantification. The final reaction buffer contained 30 mM Hepes (pH 7.8), 0.4 mM EDTA, 15 mM magnesium acetate, 25 mM potassium glutamate, and 0.05% Tween 20. Reactions were quenched with an equal volume of transcription stop solution (8 M urea/50 mM EDTA/0.01% each of bromophenol blue and xylene cyanol) at different time intervals (ranging from 5 to 60 min). The samples were then heated to 90°C for 5 min, cooled rapidly, and then loaded onto a 20% polyacrylamide, 7 M urea gel. After electrophoresis, the gels were dried, and RNA transcripts were quantified with a Typhoon 9210 imager (Amersham Biosciences, Piscataway, NJ).
In these steady-state reactions, dissociation is rate limiting, such that the dissociation rate koff is approximated by the turnover rate. In the assays presented here, the slope from a plot of RNA produced vs. time yields Vmax. Division of this maximal velocity by the concentration of the limiting species (0.1 μM, the enzyme concentration in all but the data of Fig. 5, the DNA concentration in those data) estimates the turnover rate for the reaction, which in turn approximates the complex dissociation rate, koff. The half-life (lifetime) of a halted complex is then related to the dissociation rate by the equation: t½ = ln(2)/koff (22, 29).
Acknowledgments
This work was supported by National Institutes of Health Grant 1R01 GM55002. Y. Z. was supported by National Research Service Award T32 GM08515 from the National Institutes of Health.
Abbreviation
- mm
local mismatches.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 3.Davenport RJ, Wuite GJ, Landick R, Bustamante C. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 4.Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Proc Natl Acad Sci USA. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winkler ME, Yanofsky C. Biochemistry. 1981;20:3738–3744. doi: 10.1021/bi00516a011. [DOI] [PubMed] [Google Scholar]
- 6.von Hippel PH, Yager TD. Science. 1992;255:809–812. doi: 10.1126/science.1536005. [DOI] [PubMed] [Google Scholar]
- 7.Mooney RA, Artsimovitch I, Landick R. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uptain SM, Kane CM, Chamberlin MJ. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 9.Santangelo TJ, Roberts JW. Mol Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 10.Park JS, Roberts JW. Proc Natl Acad Sci USA. 2006;103:4870–4875. doi: 10.1073/pnas.0600145103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yarnell WS, Roberts JW. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Sousa R. J Mol Biol. 2000;303:347–358. doi: 10.1006/jmbi.2000.4150. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmine I, Martin CT. J Mol Biol. 2001;305:559–566. doi: 10.1006/jmbi.2000.4316. [DOI] [PubMed] [Google Scholar]
- 14.Patel SS, Bandwar RP. Methods Enzymol. 2003;370:668–686. doi: 10.1016/S0076-6879(03)70055-X. [DOI] [PubMed] [Google Scholar]
- 15.Zhou Y, Martin CT. J Biol Chem. 2006;281:24441–24448. doi: 10.1074/jbc.M604369200. [DOI] [PubMed] [Google Scholar]
- 16.Rong M, Durbin RK, McAllister WT. J Biol Chem. 1998;273:10253–10260. doi: 10.1074/jbc.273.17.10253. [DOI] [PubMed] [Google Scholar]
- 17.Ryder AM, Roberts JW. J Mol Biol. 2003;334:205–213. doi: 10.1016/j.jmb.2003.09.039. [DOI] [PubMed] [Google Scholar]
- 18.Artsimovitch I, Landick R. Genes Dev. 1998;12:3110–3122. doi: 10.1101/gad.12.19.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 20.Yin YW, Steitz TA. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 21.Liu C, Martin CT. J Mol Biol. 2001;308:465–475. doi: 10.1006/jmbi.2001.4601. [DOI] [PubMed] [Google Scholar]
- 22.Mentesana PE, Chin-Bow ST, Sousa R, McAllister WT. J Mol Biol. 2000;302:1049–1062. doi: 10.1006/jmbi.2000.4114. [DOI] [PubMed] [Google Scholar]
- 23.SantaLucia J., Jr Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.SantaLucia J, Jr, Hicks D. Annu Rev Biophys Biomol Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 25.Lee DN, Phung L, Stewart J, Landick R. J Biol Chem. 1990;265:15145–15153. [PubMed] [Google Scholar]
- 26.Bommarito S, Peyret N, SantaLucia J., Jr Nucleic Acids Res. 2000;28:1929–1934. doi: 10.1093/nar/28.9.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tadigotla VR, O Maoileidigh D, Sengupta AM, Epshtein V, Ebright RH, Nudler E, Ruckenstein AE. Proc Natl Acad Sci USA. 2006;103:4439–4444. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schick C, Martin CT. Biochemistry. 1993;32:4275–4280. doi: 10.1021/bi00067a016. [DOI] [PubMed] [Google Scholar]
- 29.Diaz GA, Rong M, McAllister WT, Durbin RK. Biochemistry. 1996;35:10837–10843. doi: 10.1021/bi960488+. [DOI] [PubMed] [Google Scholar]