Abstract
We examine here the mechanisms ensuring the fidelity of RNA synthesis by RNA polymerase III (Pol III). Misincorporation could only be observed by using variants of Pol III deficient in the intrinsic RNA cleavage activity. Determination of relative rates of the reactions producing correct and erroneous transcripts at a specific position on a tRNA gene, combined with computational methods, demonstrated that Pol III has a highly efficient proofreading activity increasing its transcriptional fidelity by a factor of 103 over the error rate determined solely by selectivity (1.8 × 10−4). We show that Pol III slows down synthesis past a misincorporation to achieve efficient proofreading. We discuss our findings in the context of transcriptional fidelity studies performed on RNA Pols, proposing that the fidelity of transcription is more crucial for Pol III than Pol II.
Keywords: nucleotide selectivity, transcriptional fidelity
Both the replication and expression of genetic material occur with extremely high fidelity. During genome replication in Escherichia coli, one error is created per 109nucleotides incorporated (1, 2). This high fidelity results in part from the high accuracy of DNA synthesis by DNA-dependent DNA polymerases (Pols). The mechanisms whereby these enzymes ensure the fidelity of DNA synthesis are well characterized. DNA Pols are highly selective for the correct nucleotide, using it 103–106 times more efficiently than an incorrect one. Furthermore, any errors created can be immediately corrected by an intrinsic proofreading nuclease activity (2). Importantly, different DNA Pols may have a different level of fidelity, being optimized for the particular context in which they perform DNA synthesis (2–4).
The expression of genetic material is also highly accurate. In E. coli cells under standard growth conditions, the error rate of transcription is 10−5. Theoretical considerations argue in favor of the importance of transcriptional fidelity for the cell (5). Several experimental approaches have confirmed this importance in vivo and especially under conditions favoring nucleotide triphosphate (NTP) misincorporation (6–8). However, only a few studies have investigated the molecular mechanisms that underlie the fidelity of RNA synthesis by DNA-dependent RNA Pols.
Presteady-state kinetic analyses of nucleotide addition by the E. coli RNA Pol indicated that synthesis occurs through a branched kinetic mechanism with the enzyme switching between an activated state and a state termed “unactivated” at each template position. This unactivated state, absent from DNA Pols, is characterized by an extremely slow phosphotransfer reaction and can be reversed to the active state only in the presence of the correct NTP (9, 10). Fidelity of transcription is ensured by the selectivity of the Pol for the correct NTP and by kinetic trapping of the ternary complexes in the unactivated state in the absence of the correct NTP or after a misincorporation (9). Furthermore, recent studies have demonstrated that transcriptional fidelity is intimately linked to the mechanism of elongation, be it through the flexibility of the F bridge domain (11), the preloading of NTPs and dynamic error correction (12), or the association of the trigger loop with the matched NTP in the A site, thus coupling NTP recognition and phosphodiester bond catalysis (13).
Initially, unlike DNA Pols, RNA Pols had been thought to contain no proofreading activity. RNA-cleavage within ternary complexes was subsequently noted for RNA Pols from all domains of life (14–17). Unlike DNA Pols, this activity resides in the Pol active site and for it to be efficient the E. coli RNA Pol requires accessory proteins, GreA or GreB (18, 19). GreA/B can improve the overall fidelity of the E. coli RNA Pol (9). Similarly, TFS, the archeal functional equivalent of GreA/B, reduces the steady-state error rate of the archaeal RNA Pol (20).
In eukaryotes, three similar but distinct forms of RNA Pol transcribe each a different set of genes in the nuclear genome. Only the fidelity of the mRNA-synthesizing Pol II has been investigated to date. The selectivity of Pol II for the correct NTP was measured in vitro (21), and, paralleling the situation in E. coli, the role of TFIIS, the Pol II cleavage-stimulatory factor, in permitting efficient proofreading was determined (21, 22). Interestingly, at least in the yeast Saccharomyces cerevisiae, the errors committed by Pol II during the synthesis of mRNAs may be masked by the higher error-rate of mRNA translation (23). In E. coli during translation one error occurs per 104 residues (5).
The two other nuclear Pols, Pol I and Pol III, synthesize only untranslated RNAs. Thus, the fidelity of their transcription may be of even greater importance for the cell; however, their fidelity has not been examined. Elongation complexes of Pol I appear to contain a dissociable TFIIS-like activity (24). On the other hand, Pol III appears to be unique, because, unlike Pol I and II, it has a high intrinsic RNA-cleavage activity that does not require an accessory factor (16).
In this study, we examined nucleotide misincorporation by cleavage-competent and cleavage-deficient S. cerevisiae Pol III ternary complexes at a specific position on a tRNA gene. The results define the strategies used by a eukaryotic RNA Pol with high intrinsic cleavage activity to ensure the accuracy of its transcription. We used computational methods to combine the experimental observations and predict the steady-state error rate of Pol III and to demonstrate the equally high contribution of proofreading and nucleotide selectivity to the overall fidelity of Pol III.
Results
Pol IIIΔ, an Incomplete Form of Pol III Lacking RNA-Cleavage Activity, Can Quantitatively Incorporate an Incorrect Nucleotide.
To gain an insight into the mechanisms ensuring the fidelity of Pol III transcription, we initially examined the ability of the wild-type Pol III to incorporate an incorrect base. Transcription by Pol III of the SUP4 template preassembled with transcription factors TFIIIC and TFIIIB was initiated by the addition of ATP, CTP, and radiolabeled UTP. Pol III formed stable ternary complexes containing a 17-mer RNA, due to the absence of GTP required for synthesis at positions +18 and +19 of the template (Fig. 1 A and B). These were purified from free NTPs and further incubated with each of the four NTPs. As expected, the 17-mer was correctly extended to a 19-mer in the presence of GTP (Fig. 1B, lane 2) whereas no synthesis was observed with the other NTPs (Fig. 1B, lanes 3–5). Instead, the intrinsic RNA cleavage activity of Pol III led to the disappearance of the 17-mer generating shorter RNAs (Fig. 1B, lanes 3–5). In contrast to this apparent inability of Pol III to incorporate a mispaired nucleotide, Chédin et al. (25) have reported that transcription of SUP4 in the absence of GTP by Pol IIIΔ, an incomplete form of Pol III lacking the C11, C37 and C53 subunits, resulted in formation of an 18-mer transcript differing at the 3′ end from the 17-mer produced by Pol III (25, 26). The authors suggested that Pol IIIΔ incorporated a mispaired nucleotide at position +18, which failed to be subsequently removed because of the lack of RNA-cleavage activity in Pol IIIΔ resulting from the absence of the TFIIS-like C11 subunit (25). We wanted to test this hypothesis and determine which base was misincorporated at position +18. The SUP4 gene was transcribed in the absence of GTP by Pol IIIΔ preincubated with the recombinant C11 subunit (rC11). The resulting halted ternary complexes contained a 17-mer RNA and were purified to remove rC11 and the NTPs (25, 26). Traces of the 18-mer transcript remained, which could be attributed to a somewhat reduced cleavage efficiency of rC11 compared with the endogenous C11, and a negligible amount of a slippage product (27) was observed (Fig. 1B, lane 6). The purified complexes were assayed for their capacity to incorporate a single nucleotide at position +18. In the presence of ATP, Pol IIIΔ quantitatively synthesized an 18-mer (Fig. 1B, lane 8). This 18-mer could not have resulted from incorporation of ITP that might have contaminated our ATP stocks, because in the presence of ITP, Pol IIIΔ produces a 19-mer (data not shown) and a 19-mer was not observed after 5 min (Fig. 2B) nor after 60 min of incubation with ATP (data not shown). Thus, the appearance of an 18-mer revealed that the enzyme incorrectly incorporates an A instead of a G. Misincorporation of UTP occurred less efficiently (Fig. 1B, lane 10), whereas no incorporation of CTP was observed (Fig. 1B, lane 9). As for the wild-type enzyme, a 19-mer transcript was synthesized in the presence of GTP (Fig. 1B, lane 7). However, on longer incubation in the presence of GTP, Pol IIIΔ also synthesized a 20-mer transcript, indicating that it misincorporated a G instead of a U at this position (data not shown). The results demonstrated that Pol IIIΔ is capable of misincorporation.
Fig. 1.
A Pol III form devoid of RNA cleavage activity incorporates a mispaired nucleotide. (A) Template-strand sequence in the beginning of SUP4 (gray) and the sequence of the 17-mer RNA (black). (B) (Upper) Shows the relevant sequence of SUP4 (gray) and the 3′-terminal sequence of the 17-mer (black), with X denoting the position at which the NTP incorporation was tested. (Lower) Purified ternary complexes containing radiolabeled 17-mer SUP4, Pol III, or Pol IIIΔ were analyzed either directly (lanes 1 and 6) or after a 5-min incubation with 600 μM of the noted NTP. Arrow, residual 18-mer; asterisks, slippage product. (C) SUP4 was transcribed for 5 min in the presence of ATP, CTP and radiolabeled UTP by Pol III, Pol IIIΔ alone, or Pol IIIΔ preincubated for 10 min with the indicated purified recombinant subunits.
Fig. 2.
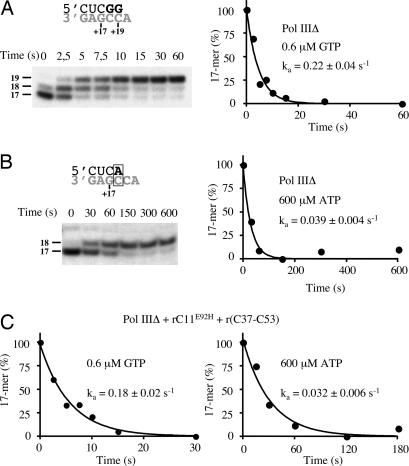
Pol III selectivity determined in the absence of RNA-cleavage activity. (A) (Upper) Shows the relevant sequence of SUP4 (gray) and the 3′-terminal sequence of the 17-mer (black) with the newly added bases in bold. (Lower) Time-course analysis of incorporation of GTP (0.6 μM) by ternary complexes containing SUP4, Pol IIIΔ and the radiolabeled 17-mer. For each reaction time, the amount of the residual 17-mer was quantified and expressed as percentage of the initial amount (t = 0) after subtraction of the lowest quantity of 17-mer remaining unreacted. Data were fitted to a single-exponential function and the apparent rate constant (ka) calculated. (B) Same as in A, except that the ternary complexes were incubated with ATP (600 μM) instead of GTP. The mismatch formed is framed on the sequence representation. (C) Same as in A except that the ternary complexes were preincubated for 10 min with the cleavage-incompetent rC11E92H and r(C37-C53), before being allowed to react with either GTP (0.6 μM) (Left) or ATP (600 μM) (Right).
RNA Cleavage Activity of Pol III Is Sufficient to Prevent Observable Misincorporations.
From the above observations, we could not conclude whether Pol IIIΔ produced quantitatively a mispaired transcript solely because it lacked RNA-cleavage activity or because the missing C11, C37, and/or C53 subunits were also important in preventing misincorporation. Therefore, it remained unclear whether Pol III did not incorporate an incorrect base to an observable degree, or whether misincorporation occurred but was masked by the efficient subsequent cleavage of the RNA. To address this issue, we used a Pol III form containing a complete set of subunits but lacking the intrinsic RNA-cleavage activity. We have recently shown that Pol IIIΔ reconstituted with rC37, rC53, and the mutant rC11E92H subunit displays the same transcriptional properties in all of the in vitro tests examined (initiation, elongation rate, termination, and reinitiation efficiency) as the wild-type enzyme except that it lacks RNA-cleavage activity (26). When transcription of SUP4 was carried out in the absence of GTP, Pol IIIΔ alone synthesized an 18-mer (Fig. 1C, lane 2), whereas Pol IIIΔ preincubated with the three wild-type recombinant subunits, and the wild-type enzyme synthesized a 17-mer RNA (Fig. 1C, lanes 3 and 1, respectively). However, when we used the rC11E92H mutant subunit instead of the wild-type rC11, an 18-mer was synthesized (Fig. 1C, lane 4). These observations strongly suggest that the synthesis of a 17-mer by the wild-type Pol III reflects a steady state in which misincorporations at position +18 occur but are rapidly corrected by the efficient RNA cleavage activity intrinsic to Pol III.
Pol III Incorporates a Correct Base 103 Times More Efficiently Than an Incorrect One.
The errors committed by Pol IIIΔ allowed us to characterize the specificity of nucleotide incorporation by Pol III. We determined the incorporation rates for the correct and an incorrect nucleotide at position +18 of the SUP4 gene in a time-course analysis of single nucleotide addition to a 17-mer within Pol IIIΔ ternary complexes. The incorporation of the correct nucleotide (GTP) proceeding extremely rapidly, the reaction was slowed by reducing the GTP concentration to 0.6 μM. Under these conditions, the reaction was virtually complete by 15 s (Fig. 2A). Because the quantity of GTP greatly exceeds that of the ternary complexes in the reaction, the concentration of GTP can be considered to remain constant and the incorporation of G to occur with pseudo-first-order kinetics. By fitting a single exponential equation to the time-dependent loss of the 17-mer, the pseudo-rate constant (ka) for the incorporation of the correct nucleotide (GTP) was estimated to be 0.22 s−1 with a standard error of 0.04 s−1 (Fig. 2A). The incorporation of the preferred incorrect nucleotide (ATP) was much slower, taking 2.5 min for the reaction to go to completion in the presence of 600 μM ATP (Fig. 2B). The pseudo-rate constant for this reaction was 0.039 ± 0.004 s−1 (Fig. 2B). When the concentration of ATP was reduced to 60 μM, the pseudo-rate constant was equivalently reduced by an order of magnitude [supporting information (SI) Fig. 6], indicating that ATP was not saturating at 600 μM, and allowing us to extrapolate the rate of ATP incorporation at 0.6 μM to 3.9 × 10−5 s−1. Hence, the correct base was incorporated ≈6 × 103 times faster than the preferred incorrect one at nucleotide concentrations below the effective Km for both the correct and incorrect nucleotide.
Because our measurements were performed with Pol IIIΔ, it was important to determine whether the selectivity observed reflected that of the wild-type Pol III deprived only of RNA cleavage activity. We used the rC11E92H mutant subunit, together with the r(C37-C53) complex, to reconstitute a Pol III lacking only its intrinsic RNA cleavage activity (26). Examination of the misincorporation rate for this enzyme form revealed it to have the same rate of addition of the correct and incorrect nucleotide as Pol IIIΔ (Fig. 2C). Thus, the selectivity measured for Pol IIIΔ also reflects the selectivity of the wild-type Pol III.
RNAs Containing a 3′-End Mismatch Are Cleaved More Rapidly Than Those Containing a Correctly Paired End.
To clarify the importance of the RNA cleavage activity versus selectivity in ensuring the fidelity of Pol III transcription, we sought to quantify the rate of the former reaction. Pol III ternary complexes containing a 17-mer were purified and allowed to cleave the RNA in transcription buffer in the absence of NTPs (Fig. 3A). We determined the apparent rate constant for the cleavage of an RNA with a correctly paired 3′ end from the time-dependent loss of the 17-mer (Fig. 3A) to be 0.10 ± 0.01 s−1. However, the important parameter in proofreading by Pol III is the rate of cleavage of an RNA containing a mispaired nucleotide at its 3′-end. This rate cannot be directly measured, because the mispaired 18-mer is undetectable and supposedly short-lived within Pol III ternary complexes. Therefore, purified ternary complexes containing Pol IIIΔ and the 3′ end-mispaired 18-mer were incubated with rC11 and r(C37–C53). We compared the rate of cleavage of the 18-mer in such reconstituted wild-type-like ternary complexes to the rate of cleavage of the correct 17-mer transcript in similarly obtained complexes. Because the cleavage of the 18-mer was too rapid at 25°C, the reactions were performed in cleavage buffer at 16°C. Despite the reconstituted Pols cleaving only up to 40% of the RNA initially present (SI Fig. 7), we clearly observed that the 18-mer transcript was cleaved 8 times more rapidly than the 17-mer (Fig. 3B). This higher rate of the 18-mer cleavage was not due to a different sequence context (position 18 versus 17), because an 18-mer transcript ending with a 3′-OMeG was cleaved at a rate very similar to that of the 17-mer (SI Fig. 8). Thus, the cleavage of a 3′-end mispaired transcript by Pol III is more efficient than the cleavage of a correctly paired one, indicating that the enzyme discriminates between correct and incorrect bases during both synthesis and cleavage reactions. We extrapolated from the rate of 18-mer cleavage relative to that of the 17-mer by the reconstituted Pol III to the wild-type Pol III, estimating the rate of cleavage of the 3′-end mispaired 18-mer transcript at 25°C by Pol III to be 0.83 s−1 (for details, see “Note 1” in SI Text).
Fig. 3.
Time-course analysis of RNA cleavage by Pol III. (A) Time-course of RNA cleavage in transcription buffer in the absence of NTPs by purified ternary complexes containing SUP4, Pol III, and the radiolabeled 17-mer. Data were analyzed as described for Fig. 2A. (B) Purified ternary complexes containing SUP4, PolIIIΔ and either the (radiolabeled) correctly 3′-end-paired 17-mer or 3′-end mispaired 18-mer transcript were preincubated for 10 min with wild-type rC11 and r(C37–C53) and the RNA-cleavage reaction was initiated by starting incubation in cleavage buffer (at 16°C). The data were analyzed as described for Fig. 2A.
Misincorporation Slows the Incorporation of the Next Nucleotide.
To appreciate the importance of the cleavage reaction in the fidelity of Pol III, we had to determine the rate of the competing step, the addition of the correct nucleotide after a misincorporation. In the presence of 600 μM GTP, purified Pol IIIΔ ternary complexes extended the 3′-end mispaired 18-mer to a 19-mer that subsequently disappeared giving a 20-mer transcript (Fig. 4). Essentially all of the initially present 18-mer was lost by 60 min (Fig. 4). The simplest explanation for the transient appearance of a 19-mer followed by formation of a 20-mer is that the 20-mer arises from a misincorporation of GTP instead of UTP at position +20 and that the rate-limiting step in the formation of the 20-mer is the passage from +18 to +19, i.e., the incorporation of a correct base after a mispaired base. The apparent rate constant for disappearance of the 18-mer was 94 × 10−5 ± 4 × 10−5 s−1 (Fig. 4). Thus, the incorporation of a correct base after a mispaired base is ≈105 times slower than after a correctly paired one. Furthermore, our analyses demonstrate that the addition of a ribonucleotide after a misincorporation is the slowest step in the synthesis of error-containing transcripts by Pol III. In contrast, this step is not slower than the actual misincorporation in the case of E. coli RNA Pol and Pol II (9, 21).
Fig. 4.
Addition of a correct nucleotide after misincorporation is extremely inefficient. (A) (Upper) The relevant sequence of SUP4 (gray) and the 3′-terminal sequence of the 18-mer (black) with the newly added bases in bold and the mismatches framed. (Lower) The time-course analysis of GTP (600 μM) incorporation by purified ternary complexes containing SUP4, Pol IIIΔ, and the radiolabeled 3′-end mispaired 18-mer transcript. (B) The data were analyzed as described for Fig. 2A.
Selectivity and Proofreading Contribute Equally to the Fidelity of Pol III Transcription.
We sought a computational approach to integrate our experimental results and estimate the contributions of proofreading and selectivity to the fidelity of Pol III transcription at a particular position of an RNA chain. Our observations were consistent with the existence of two competing pathways leading to an insertion followed by a fixation of either a correct or an incorrect base (Fig. 5). The system of reactions we propose to be important in determining the fidelity of Pol III is characterized by six kinetic parameters, the first-order rate constants k1–k6 represented in Fig. 5. A ternary complex containing a transcript of n − 1 nucleotides can incorporate at the nth position either a correct nucleotide [giving the product (n) with the first-order rate constant k1] or an incorrect nucleotide [product (n*), k2]. Each product can either undergo the cleavage reaction (k4 and k5, respectively), regenerating an RNA of n − 1 nucleotides, or be extended by the incorporation of the correct nucleotide to give the (n + 1) or (n* + 1) species, respectively (k6 and k3) (Fig. 5).
Fig. 5.
Modeling the reactions determining Pol III fidelity. Diagram describing the reactions determining the fidelity of Pol III transcription, where: (n − 1) − RNA species of length (n − 1) within a Pol III ternary complex, (n) − product of the correct incorporation at the template position n, (n*) − product of misincorporation at position n, (n + 1) − product of the extension of the correctly paired species (n), and (n* + 1) − product of the extension of the incorrectly paired species (n*).
Two simplifications were made in this model: (i) The cleavage of the (n) or (n*) species will give a product of length n − x, with x ≥ 1, but because the extension from position n − x to n − 1 (for x > 1) is rapid, we assumed that the cleavage of the (n) or (n*) RNAs would directly generate the (n − 1) species. (ii) We did not include the cleavage of the (n + 1) or (n* + 1) transcripts in the model because, once a correct nucleotide is incorporated at position n + 1, the forward synthesis rates may be overwhelming and thus no longer have a bearing on the misincorporation at position n. We discuss later a more complex situation created when this second simplification is altered (see Discussion).
One measure of transcriptional fidelity of Pol III at a particular position in an RNA chain is the ratio of the concentrations of the correct to the incorrect product [n + 1]/[n* + 1], which is the inverse of the error rate. We can calculate that, after an infinite time, the [n + 1]/[n* + 1] ratio reaches:
Assuming that the incorporations of correct ribonucleotides onto a correctly paired 3′-end will occur with similar rates relative to the rate of misincorporation or extension past a misincorporation, we can state that k1 = k6, giving:
 |
In essence, the fidelity of Pol III transcription is insured by two processes: selectivity and proofreading. Selectivity is given by the ratio of the rates of incorporation of correct to incorrect nucleotide (k1/k2). Proofreading is represented by the ratio of the rate of cleavage of an incorrectly paired nucleotide relative to the rate of its extension (k5/k3). Indeed, visual inspection of formula (Eq. 2) and computer simulations (SI Fig. 9), confirmed that it is not the absolute rate of cleavage that determines fidelity of Pol III transcription but the k5/k3 ratio.
It is obvious from formula (Eq. 2) that if k1 ≫ k4 and k5/k3 ≫ 1, as is the case observed for Pol III (Table 1), fidelity is given by
i.e., it is the product of “selectivity” by “proofreading”. The rate constants measured in this study for the incorporation of GTP and misincorporation of ATP at position +18 of SUP4 indicate that k1/k2 = 5.6 × 103 at GTP and ATP concentrations below the effective Km of the enzyme for these nucleotides. Measurements of cleavage and extension past the misincorporation indicate that k5/k3 = 0.9 × 103, at NTP = 600 μM. A reduction in [NTP] would result in lower k3 and higher k5/k3 (see SI Fig. 9). Hence, at nonsaturating concentrations of nucleotides,
meaning that Pol III would incorporate one error per 5 × 106 bases synthesized. Computer simulations revealed that the limit conditions are reached rapidly (SI Fig. 9).
Table 1.
Summary of the rate constants determined in this study
| Complex* | ka (s−1) | Ref. in Fig. 5 | |
|---|---|---|---|
| Incorporation† | 5′…CG | 2.2 10−1‡ | k1 |
| 3′…GCC…5′ | |||
| 5′…CA | 3.9 × 10−2 | k2 | |
| 3′…GCC…5′ | |||
| 5′…CAG | 9.4 × 10−4 | k3 | |
| 3′…GCC…5′ | |||
| Cleavage | 5′…C | 1 × 10−1 | k4 |
| 3′…GCC…5′ | |||
| 5′…CA | 8.3 × 10−1§§ | k5 | |
| 3′…GCC…5′ | |||
*The upper sequence shows the 3′-end of the RNA, the lower sequence shows the template DNA strand, and mispaired RNA bases are italicized.
†The incorporation of the underlined base is shown. The corresponding NTP is at 600 μM unless noted.
‡The measurement was made at 0.6 μM GTP.
§Determined from the relative rate of cleavage of 18-mer vs. 17-mer and the measured rate constant for the 17-mer cleavage (see Note 1 in SI Text).
Therefore, the efficiency of proofreading by Pol III appears to be within the order of magnitude of the selectivity ratio, indicating that both processes have a significant contribution to Pol III fidelity.
Discussion
In this study, we show that proofreading is a major determinant of the fidelity of Pol III transcription. The measurements of relative reaction rates combined with computer analyses demonstrate that nucleotide selectivity and proofreading contribute equally to Pol III fidelity at the examined position on the SUP4 gene. Further studies may reveal how these contributions may vary in different sequence contexts or even in vivo and whether our conclusions are generally applicable.
Mechanics of Error Avoidance and Error Correction by Pol III.
As noted in the Introduction, few studies have examined the fidelity of RNA Pols. Still, we find it important to make some comparisons between the functioning of different RNA Pols, although the comparisons remain speculative, because they are based on a limited number of ternary complexes.
The selectivity measured for Pol III (5.6 × 103) is similar to that reported for the E. coli RNA Pol and Pol II (103 and >5 × 102, respectively) (9, 21), indicating that the selectivity of RNA Pols may be in the lower end of the range (103–106) reported for DNA Pols (2). We have measured the rate of the most-likely misincorporation, which may have led to an underestimate of selectivity. At the same time, maximal error rates may have also been measured for other RNA Pols, because preloading of NTPs may reduce their error incorporation (12).
The proportion of E. coli RNA Pol or Pol II ternary complexes able to incorporate a correct nucleotide was higher than that capable of misincorporation (9, 21), which Erie and coworkers interpreted as indicating that the polymerase could assume an unproductive conformation (i.e., become “unactivated”) at each synthesis step, thus preventing misincorporation. Single-molecule analyses on bacterial enzyme confirmed recently this hypothesis (28). The existence of such an unactivated state was also inferred for Pol III from detailed analyses of RNA chain elongation (29). We did not evidence, however, any difference in the proportion of ternary complexes incorporating the correct or an incorrect nucleotide. This observation reflects that the temporal resolution of our experiments may not be fine enough to allow detection of such a state. Alternatively, although highly speculative, one cannot formally exclude that Pol III requires intrinsic cleavage activity to enter an unactivated state, because unlike our study, that of Matsuzaki and coworkers used a cleavage-competent enzyme (29).
Pol III appears to have an exceptionally high contribution of proofreading to its fidelity; the magnitude of this contribution, as determined in this study (103), is high even when compared with the range reported for the proofreading activities of DNA Pols (few- to >100-fold) (2). Interestingly, both DNA Pols and Pol III rely strongly on kinetic partitioning between cleavage and extension reactions for efficient proofreading despite crucial differences in the mechanism and site of the cleavage activity.
We demonstrated that the rate of cleavage of nascent transcripts was stimulated by a mismatched base at the 3′ end of the RNA. This may be the extreme case of the preference to cut the nascent RNA just 5′ of an unstable RNA-DNA base pair, demonstrated for Pol III (30). This extends observations made on RNA-DNA “dumbbell” templates, using Pol II+TFIIS (22). Importantly, in our study, the rates of both cleavage and extension are measured under physiologically more relevant conditions, within authentic ternary complexes halted on a class III gene.
C11, the protein essential for the cleavage and thus the proofreading activity of the polymerase, is a Pol III subunit, making it very likely that the role of proofreading in the fidelity of the Pol will be important in different sequence contexts and in vivo. Furthermore, our calculations may have underestimated the contribution of cleavage to Pol III fidelity. In the model presented (Fig. 5), the assumption was made that the rate of extension of the (n* + 1) species by a correct base is equal to the rate of extension of (n) to (n + 1) and cancels out the effects of (n* + 1) cleavage. However, precedents exist in DNA Pols for a reduction in the synthesis rates several bases past the misincorporation, occurring because of distortions in the active site (31). If we include the extension/cleavage of the (n* + 1) species in our model, we find that the fidelity of transcription, as measured in this case by the (n + 2):1/m (n + 2)/(n* + 2). (n* + 2) ratio, increases substantially if the rate of conversion of (n* + 1) to (n* + 2) drops below ≈2 s−1 to reach 4.4 × 109when this rate is as low as that of (n*) → (n* + 1) (SI Fig. 10 and SI Text, “Note 2”). This increase in fidelity occurs through an increase in proofreading. The fidelity of Pol III probably lies between these two extremes: the rate of (n* + 1) → (n* + 2) is not as low as that of (n*) → (n* + 1), because on addition of all four NTPs, the 3′-end mispaired 18-mer can be extended to the complete SUP4 transcript in a time-course similar to that in Fig. 4 without the appearance of a 19-mer (data not shown). A further determinant of the in vivo fidelity is the local NTP concentration, and computer simulations show that fidelity is linearely proportional to the synthesis rates over at least two orders of magnitude (data not shown).
RNA Cleavage Activity and in Vivo Properties of Pol III.
Even if all RNA Pols use generally similar mechanisms to achieve high fidelity, Pol III appears to display specific properties. Why, for example, does Pol III have a high intrinsic cleavage activity, whereas other nuclear RNA Pols do not? This property may be due to the differences in their mechanism of transcription, and reflect their respective biological roles. With respect to the mechanism of transcription, the simplest explanation would be that Pol III is less apt at avoiding transcriptional errors than Pol II. However, the selectivity of the two polymerases appears comparable.
Currently, the role of the Pol II cleavage factor, TFIIS, in transcriptional fidelity is unclear. It is not a constitutive partner of Pol II (32, 33), and there is no evidence for in vivo TFIIS-mediated Pol II proofreading under standard growth conditions, i.e., in absence of stress (34). However, conditions favoring transcriptional errors stimulate the association of TFIIS with elongating Pol II (33), and the deletion the TFIIS-encoding gene in yeast (PPR2) causes sensitivity to the same conditions (35). TFIIS may also have other functions, such as in initiation of transcription (36) or in elongation (37).
The in vivo nonessential or essential character of the cleavage activity is another striking difference between the Pol II and the Pol III systems. PPR2 is not essential for yeast viability (38). Rpb9 subunit of Pol II, a C11 paralogue recently identified as important for the in vivo fidelity of Pol II and suggested to act in proofreading (34), is also encoded by a nonessential gene. These findings strongly suggest that Pol II RNA cleavage activity is not essential in vivo. In stark contrast, the point mutations in the C11 subunit that abolish RNA cleavage activity are lethal [double D91A E92A mutant (25); or E92H mutant, data not shown]. Because the only known transcriptional defect of Pol III-containing rC11E92H in vitro is the absence of cleavage activity (26), this activity may be essential for Pol III in vivo. A simple explanation for this difference would be that transcriptional fidelity is more crucial for Pol III than for Pol II, because the former's untranslated RNA products are direct players in crucial cellular processes, notably translation. The idea that eukaryotic RNA Pols may have different accuracies depending on the class of transcripts they produce was also proposed based on observations made in E. coli (39). Indeed, with respect to protein-coding genes, it appears that translation rather than Pol II transcription is the error-prone process (23), although this may depend on the gene examined (40).
The essential character of Pol III cleavage activity may also reflect more profound differences in the in vivo mechanics of Pol III and II transcription. The efficient transcription of class III genes proceeds through facilitated reinitiation by the enzyme during which Pol III has been suggested to remain on the template (41, 42). The movement of the enzyme on a gene would thus be extremely reduced, making it difficult, if at all possible, for the cell to distinguish between truly elongating Pol III and blocked Pols, thus precluding the possibility of the latter's degradation to save the transcription of the gene. In contrast to Pol II, which has been observed to become ubiquitylated and degraded under the conditions favoring elongation blocs in vivo (43), RNA cleavage and resynthesis may be the only means for the cell to deal with elongation-blocked Pol III.
Materials and Methods
Purification of RNA Pol III, Class III Transcription Factors and Expression and Purification of Recombinant Pol III Subunits.
Purification of Pol III and class III transcription factors was described in ref. 44. Pol IIIwt was purified from the yMR4 strain, whereas Pol IIIΔ was purified from either C37HA-Ct or yEL7 strain (26). Regardless of the source, Pol IIIΔ behaved the same in all of the tests performed. Purified recombinant (His7-C37–C53) complex, C11 and C11E92H were obtained as described (25, 26). rC11, rC11E92H, and the r(C37–C53) complex were purified to homogeneity (26), as monitored by Coomassie blue staining.
Promoter-Directed in Vitro Transcription, Ternary Complex Purification, Single-Nucleotide Addition and Cleavage Assays.
NTPs and [α-32P]UTP were from Amersham Biosciences (Little Chalfont, U.K.). Promoter-directed in vitro transcription reactions were performed in 50 μl of transcription buffer (20 mM Hepes KOH, pH 7.5/0.1 mM EDTA/5 mM MgCl2/5 mM DTT/5% glycerol/90 mM KCl). pRS316-SUP4 plasmid (250 ng) was incubated in this buffer with 50 ng of each rTFIIIC, rTBP, rTFIIIB70, and 0.4 μg of B″ fraction for 20 min at 25°C, followed by addition of native or reconstituted Pol III (50–100 ng), and a further 20-min incubation. Transcription was started by addition of 600 μM ATP and CTP and 3 μM [α-32P]UTP (400 Ci/mmol) and allowed to proceed for 5 min. For add-back experiments, recombinant proteins were added to Pol IIIΔ in PBS, or to ternary complexes, at a molar ratio shown to be efficient in studies of termination, elongation, cleavage, and reinitiation (25, 26) and incubated for 10 min. Unless otherwise noted, all reactions were performed at 25°C. Ternary complexes were purified from 100 μl of transcription reaction by gel-permeation chromatography at 4°C [1 ml of CL2B resin (GE Healthcare Europe, Saclay, France) equilibrated in 40 mM Tris·HCl (pH 8)/90 mM KCl/0.5 mM EDTA/5% glycerol in a 1-ml syringe]. Single-nucleotide addition and cleavage reactions were performed on 20-μl aliquots of purified ternary complexes and initiated by the addition of 20 μl of the reaction mix to give the final concentration of 1× transcription or cleavage buffer with the noted concentration of an NTP. The reactions were stopped and processed as described in ref. 26. Transcripts were analyzed by electrophoresis on denaturing polyacrylamide gels (15%, 20:1 acrylamide:bis-acrylamide ratio). Gels were autoradiographed by using MR film with an intensifying screen (Kodak, Tokyo, Japan). Quantifications were performed with Image Quant software, Version 1.2. The lowest quantity of the 17-mer or 18-mer observed in the reactions was subtracted from all of the values and the data obtained were normalized so that the amount of the oligomer at time 0 is 100%. The normalized values were fitted to a single exponential equation (y = Ae− kx, where A is the initial quantity of oligomer), and the rate constant (k) was calculated by using Prism 4 software (GraphPad, San Diego, CA). R2 values were generally >0.98 (all were >0.94).
Computational Analyses.
Computational analyses were performed with MatLab software (MathWorks, Natick, MA). For details, see SI Text.
Supplementary Material
Acknowledgments
We thank Valérie Goguel for a critical reading of the manuscript, André Sentenac for his constant support, Cécile Ducrot and Joël Acker for providing recombinant proteins. Emilie Landrieux was supported by a short-term grant from the Fondation pour la Recherche Médicale.
Abbreviations
- Pol
polymerase
- NTP
nucleotide triphosphate.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704116104/DC1.
References
- 1.Echols H, Goodman MF. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel TA, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Beard WA, Wilson SH. Structure (London) 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- 4.Tippin B, Pham P, Goodman MF. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Ninio J. Biochimie. 1991;73:1517–1523. doi: 10.1016/0300-9084(91)90186-5. [DOI] [PubMed] [Google Scholar]
- 6.Dukan S, Farewell A, Ballesteros M, Taddei F, Radman M, Nystrom T. Proc Natl Acad Sci USA. 2000;97:5746–5749. doi: 10.1073/pnas.100422497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taddei F, Hayakawa H, Bouton M, Cirinesi A, Matic I, Sekiguchi M, Radman M. Science. 1997;278:128–130. doi: 10.1126/science.278.5335.128. [DOI] [PubMed] [Google Scholar]
- 8.Bregeon D, Doddridge ZA, You HJ, Weiss B, Doetsch PW. Mol Cell. 2003;12:959–970. doi: 10.1016/s1097-2765(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 9.Erie DA, Hajiseyedjavadi O, Young MC, Von Hippel PH. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 10.Foster JE, Holmes SF, Erie DA. Cell. 2001;106:243–252. doi: 10.1016/s0092-8674(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 11.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 12.Gong XQ, Zhang C, Feig M, Burton ZF. Mol Cell. 2005;18:461–470. doi: 10.1016/j.molcel.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surratt CK, Milan SC, Chamberlin MJ. Proc Natl Acad Sci USA. 1991;88:7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reines D. J Biol Chem. 1992;267:3795–3800. [PMC free article] [PubMed] [Google Scholar]
- 16.Whitehall SK, Bardeleben C, Kassavetis GA. J Biol Chem. 1994;269:2299–2306. [PubMed] [Google Scholar]
- 17.Schnapp G, Graveley BR, Grummt I. Mol Gen Genet. 1996;252:412–419. doi: 10.1007/BF02173006. [DOI] [PubMed] [Google Scholar]
- 18.Fish RN, Kane CM. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 19.Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lange U, Hausner W. Mol Microbiol. 2004;52:1133–1143. doi: 10.1111/j.1365-2958.2004.04039.x. [DOI] [PubMed] [Google Scholar]
- 21.Thomas MJ, Platas AA, Hawley DK. Cell. 1998;93:627–637. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 22.Jeon C, Agarwal K. Proc Natl Acad Sci. 1996;93:13677–13682. doi: 10.1073/pnas.93.24.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw RJ, Bonawitz ND, Reines D. J Biol Chem. 2002;277:24420–24426. doi: 10.1074/jbc.M202059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tschochner H. Proc Natl Acad Sci. 1996;93:12914–12919. doi: 10.1073/pnas.93.23.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chédin S, Riva M, Schultz P, Sentenac A, Carles C. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dieci G, Hermann-Le Denmat S, Lukhtanov E, Thuriaux P, Werner M, Sentenac A. EMBO J. 1995;14:3766–3776. doi: 10.1002/j.1460-2075.1995.tb00046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuzaki H, Kassavetis GA, Geiduschek EP. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 30.Bobkova E, Hall B. J Biol Chem. 1997;272:22832–22839. doi: 10.1074/jbc.272.36.22832. [DOI] [PubMed] [Google Scholar]
- 31.Johnson SJ, Beese LS. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 32.Reinberg D, Roeder RG. J Biol Chem. 1987;262:3331–3337. [PubMed] [Google Scholar]
- 33.Pokholok DK, Hannett NM, Young RA. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- 34.Nesser NK, Peterson DO, Hawley DK. Proc Natl Acad Sci USA. 2006;103:3268–3273. doi: 10.1073/pnas.0511330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Exinger F, Lacroute F. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 36.Prather DM, Larschan E, Winston F. Mol Cell Biol. 2005;25:2650–2659. doi: 10.1128/MCB.25.7.2650-2659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Mol Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- 38.Archambault J, Lacroute F, Ruet A, Friesen JD. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blank A, Gallant JA, Burgess RR, Loeb LA. Biochemistry. 1986;25:5920–5928. doi: 10.1021/bi00368a013. [DOI] [PubMed] [Google Scholar]
- 40.Koyama H, Ito T, Nakanishi T, Kawamura N, Sekimizu K. Genes Cells. 2003;8:779–788. doi: 10.1046/j.1365-2443.2003.00677.x. [DOI] [PubMed] [Google Scholar]
- 41.Dieci G, Sentenac A. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 42.Ferrari R, Rivetti C, Acker J, Dieci G. Proc Natl Acad Sci USA. 2004;101:13442–13447. doi: 10.1073/pnas.0403851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Somesh BP, Reid J, Liu WF, Sogaard TM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Cell. 2005;121:913–923. doi: 10.1016/j.cell.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Huet J, Manaud N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. Methods Enzymol. 1999;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.