Abstract
Positive-strand RNA viruses direct different virus-specific processes during their infection of host cells. Fundamental events such as viral RNA genome replication are controlled by viral regulatory RNA elements (REs). Here, we have investigated the possibility of specifically modulating the action of a viral RE using RNA aptamer technology. Through rational design, a tombusvirus RE, which has the structure of a perfect RNA stem loop in the plus-strand RNA genome, was replaced with a theophylline-binding RNA aptamer sequence, an imperfect stem loop. The aptamer–RE hybrid was designed so that, upon binding theophylline, it would become more stable and structurally mimic the functional RE (i.e., represent a ligand-inducible RE riboswitch). Initial experiments were conducted with a small noncoding virus genome-derived RNA replicon, and the results showed that replication was inducible, up to ≈10-fold, in a theophylline-specific and dose-dependent manner. A similar level of theophylline-dependent induction was also observed when a full-length viral genome containing an RE riboswitch was tested. Analysis of this engineered viral genome revealed that this RE, located in the 5′ untranslated region, specifically mediates efficient accumulation of plus-strands of the virus genome. Therefore, in addition to allowing for modulation of virus reproduction, the RE riboswitch system also provided insight into RE function. The ability to chemically induce a viral process via modulation of virus genome structure could be useful for basic and applied aspects of research.
Keywords: aptamer, gene regulation, riboswitch, RNA structure, tombusvirus
RNA aptamers are RNA molecules that are capable of specifically binding to cognate ligands. Small molecule-binding RNA aptamers were first identified through artificial selection by using systematic evolution of ligands by exponential enrichment (i.e., SELEX) (1). Surprisingly, some of these RNA aptamers showed favorable binding affinities and high levels of specificity (1). These observations suggested that natural RNA aptamers might exist and function in nature. Indeed, naturally occurring RNA aptamers, present as components of riboswitches, were subsequently identified in bacteria and shown to be involved in regulating translation or transcription of cellular mRNA (2). These riboswitches contain two mutually exclusive structures, one functional and the other nonfunctional, and ligand binding to the aptamer domain within the riboswitch acts to stabilize one of the two structures (2). In this way, the presence or absence of the ligand dictates which RNA structure dominates and, thus, the activity of the functional RNA module.
Engineered RNA-based regulation of gene expression in living cells is now a rapidly expanding area of biotechnology. RNA aptamers that bind to small-molecule ligands (e.g., theophylline) have been instrumental in the development of novel systems that allow for ligand-dependent regulation of mRNA translational activity (3–10), ribozyme activity (10, 11), or other cellular processes (3, 9, 12, 13). In these systems, the aptamers are able to directly or indirectly influence their targets through ligand-binding-induced changes in RNA structure (3–13). This ability to modulate RNA conformation makes aptamers amenable for use in the development of other biologically based regulatory systems. In this respect, an unexplored application for small molecule-binding RNA aptamers is in the modulation of viral processes.
Viruses that possess RNA genomes represent obvious candidates for the development of this type of regulatory system. During RNA virus infections, various processes are controlled by the action of viral genomic RNA sequences and/or higher-order RNA structures, herein termed regulatory RNA elements (REs). Accordingly, the ability to modulate the activity of an RE would provide a means to control the viral process that it directs, and this could potentially be accomplished through rational design with RNA aptamer technology. This goal could be achieved by integrating an RNA aptamer into the viral RNA genome in such a manner that binding of its cognate ligand affects the structure and function of an RE, thereby creating an RE riboswitch. Here, we have explored the use of RNA aptamer technology for activating viral RNA replication in cells. The tombusvirus tomato bushy stunt virus (TBSV) is particularly well suited for this purpose as the structures of the REs involved in regulating the replication of its 4.8-kb-long single-stranded, positive-sensed RNA genome are among the best characterized (14) (Fig. 1A). Using TBSV, we show that an RE-riboswitch strategy can be applied successfully to modulate the activity of viral RNA replication and can provide useful information on RE function. The utility of such systems for basic and applied studies is discussed.
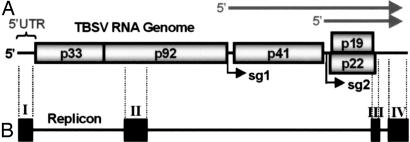
Fig. 1.
Schematic structures of the TBSV genome and a viral replicon. (A) Linear representation of the TBSV RNA genome. Encoded viral proteins are depicted as boxes with their molecular masses (in thousands) prefixed by “p.” The 5′ UTR is delineated. Initiation sites for sg mRNA transcription are labeled sg1 and sg2, and corresponding structures of the two sg mRNAs are represented by arrows above the genome. (B) The TBSV-derived replicon shown is composed of four noncontiguous regions (I–IV) that correspond to different segments of the viral genome (delineated by vertical dotted lines). Note that region I in the replicon is the 5′ UTR from the viral genome. The horizontal line joining the four regions represents genomic segments that are not present in the replicon.
Results and Discussion
Designing a Theophylline-Inducible Viral RNA Replicon.
A replication-specific RE in the 5′ untranslated region (UTR) of the TBSV genome was selected to assess the possibility of controlling virus RNA accumulation with a small molecule-binding RNA aptamer. The 166-nt-long 5′ UTR folds into a complex higher-order RNA structure that includes a small stem loop (SL), termed SL5, which is an RE that is necessary for efficient viral RNA replication (15) (Fig. 2A). SL5 separates two RNA domains, the T-shaped domain and the downstream domain, which interact via a functionally relevant pseudoknot, PK-TD1 (15) (Fig. 2A). The importance of the SL5 RE for viral RNA replication was demonstrated previously by using a small, replicable, noncoding TBSV genome-derived RNA, herein termed replicon (Fig. 1B). Although this viral RNA encodes no proteins, it maintains replication REs and is efficiently amplified when cotransfected into host cells along with a full-length “helper” TBSV genome, which provides essential viral replication proteins (p33 and p92) in trans (14). Previous studies using this helper/replicon system showed a direct correlation between the thermodynamic stability of the SL5 and the efficiency of replicon amplification in vivo (15). Specifically, the introduction of mismatches into the lower stem portion of this RE inhibited viral RNA replication, whereas a compensatory mutant containing transposed base pairs was able to replicate to WT levels (15). Importantly, the same study also showed that the identity of the residues in the stem and terminal loop of SL5 was not relevant to its activity and that SL5 operates as a plus-strand structure (i.e., it functions as part of the 5′ UTR) (15).
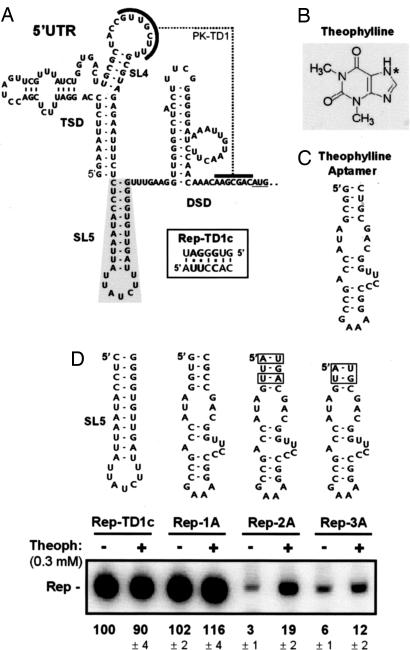
Fig. 2.
Designing and testing a ligand-inducible viral replicon. (A) RNA secondary structure of the 5′ UTR of the TBSV RNA genome (which also corresponds to region I of the replicon). The 5′-proximal T-shaped domain (TSD) and 3′-proximal downstream domain (DSD) are indicated. These domains are separated by SL5, which is outlined by shading. A tertiary interaction, PK-TD1, is indicted, and substitutions in Rep-TD1c are shown in the box. (B) Structure of theophylline (*, N7). (C) A theophylline-binding RNA aptamer (1). (D) The structures of putative RE riboswitches that were used to replace SL5 in Rep-TD1c are shown at the top. Sequence differences in the closing stems are boxed. Replicons were cotransfected with helper TBSV genome sg1T100 into plant-cell protoplasts, incubated in the absence (−) or presence (+) of 0.3 mM theophylline, and their accumulation levels quantified by Northern blot analysis at 22 h after cotransfection. The values at the bottom represent means that were normalized to that for uninduced Rep-TD1c (set at 100%) with standard deviations from three separate experiments.
The defined structural requirement and critical role of the SL5 RE made it an ideal experimental candidate for conversion into an RE riboswitch. The strategy devised to accomplish this involved replacing SL5 with an RNA aptamer. The rationale for a SL5-to-aptamer substitution was that an unbound aptamer would mimic a destabilized nonfunctional SL5 RE, whereas a ligand-bound aptamer would emulate a stable functional SL5, thus potentially allowing for ligand-mediated control of viral RNA replication (i.e., an inducible “on” riboswitch). A theophylline-binding RNA aptamer was selected for the replacement because its size and shape are similar to those of SL5 (1) (Fig. 2 A and C). Additionally, this aptamer–ligand pair displays a high degree of specificity and affinity (Kd = 0.2–0.4 μM), and theophylline (Fig. 2B) is membrane-permeable and relatively noncytotoxic (5). The initial screening for an appropriate RE riboswitch involved replacing the SL5 RE in the replicon with a typical aptamer (Fig. 2C) (1) or modified versions that contained different closing base pairs (Fig. 2D). However, mfold secondary structure analysis of the RNA sequences in these replicons revealed that a section of the ligand-binding pocket in the aptamer (5′-GGCAGC) could potentially base pair to a nearby complementary sequence in the loop of SL4 within the T-shaped domain (5′-GUUGCU). The latter sequence participates in a pseudoknot interaction, PK-TD1, that is essential for viral RNA replication (15) (Fig. 2A), whereas the former sequence is critical for ligand binding (16). Therefore, to preclude this possible dual interference, the L4 sequence and its complementary pseudoknot partner sequence in the downstream domain were modified to create Rep-TD1c (Fig. 2A Inset); thereby providing a context that was predicted to be free of such steric hindrance. Importantly, previous studies showed that replicons containing this modified pseudoknot are fully functional (15). Consequently, Rep-TD1c was used as the base replicon for testing potential RE riboswitches (Fig. 2D). Also, to avoid possible RNA recombination between the helper virus genome and the riboswitch-containing replicons (17) (that could restore a WT SL5 in the latter), the helper TBSV genome used in cellular cotransfection studies, sg1T100, contained a modified functional 5′ UTR that did not contain SL5. Rep-TD1c derivatives containing different potential RE riboswitches in place of the SL5 RE were analyzed in vivo for replicon responsiveness to theophylline.
Analysis of Theophylline-Induced Replication of a Viral RNA Replicon.
Cells were cotransfected with in vitro-generated RNA transcripts of helper sg1T100 viral genome and different riboswitch-containing replicons and then incubated in liquid medium in the presence or absence of theophylline. Three representative examples from a larger pool of candidates that were screened (data not shown) are presented in Fig. 2D. Results from the replicon analysis were divided into three categories based on theophylline responsiveness and replicon accumulation: (i) non- or weakly responsive with constitutively high levels of replicon accumulation, (ii) non- or weakly responsive with constitutively low levels of replicon accumulation, or (iii) responsive with inducible replicon accumulation. Rep-2A (iii) showed a notable increase in accumulation in the presence of 0.3 mM theophylline, whereas Rep-1A (i) and Rep-3A (ii), which contained aptamers with closing stems that were predicted to be either more or less stable, respectively, were far less responsive (Fig. 2D). The notable induction observed for Rep-2A indicated that an appropriate balance of stability for the unbound aptamer was achieved; it was unstable enough to inhibit viral RNA replication, yet ordered enough to bind to its ligand.
Next, a ligand dose–response assay was carried out to assess the effectiveness of induction. With increasing concentrations of theophylline, Rep-2A accumulation was enhanced up to ≈10-fold, or to ≈25% of the level of the Rep-TD1c control (Fig. 3A). Maximal induction was achieved with 0.75 mM ligand, whereas further increases in concentration led to reduced levels of accumulation. To determine whether the observed induction required a functional aptamer, a replicon containing a mutated aptamer was tested (Fig. 3B). Rep-2Am, in which an essential nucleotide in the theophylline-binding pocket was substituted (16), was not responsive to theophylline (Fig. 3A). In addition, the necessity and specificity of theophylline for induction was evaluated by using caffeine as a ligand. Caffeine is a molecular analogue of theophylline that differs in that it contains a methyl group at N7 (Fig. 2B). This structurally similar ligand was not able to activate replication of Rep-2A (Fig. 3A). Collectively, these analyses indicate that the induction of viral RNA replication is aptamer- and ligand-dependent, as well as theophylline-specific and ligand-dose-dependent.
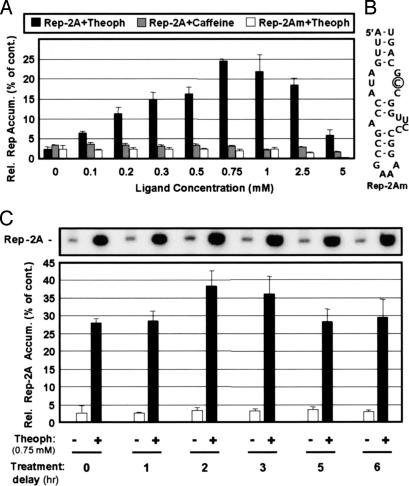
Fig. 3.
Analysis of an RE riboswitch in a viral replicon. (A) The effect of increasing concentrations of ligand (theophylline or caffeine) on the relative levels of Rep-2A or Rep-2Am accumulation in plant-cell protoplasts at 22 h after cotransfection. (B) Modified RE riboswitch in Rep-2Am (the substitution is circled). (C) Induction of Rep-2A accumulation at various times after cotransfection. For both A and C, the graphed values represent means that were normalized to that for uninduced Rep-TD1c control (set at 100%) with standard deviations from three separate experiments. cont., control.
Effective induction was also observed when theophylline treatment was delayed for up to 6 h after cotransfection of viral RNAs (note that Rep molecules are very stable within cells) (15) (Fig. 3C). This result demonstrates that ligand treatment does not have to immediately follow transfection of its target RNA and suggests that riboswitch-containing viral RNAs that are expressed constitutively in cells at low levels from either a DNA vector or a genome-incorporated transgene would also be inducible by using this approach. Indeed, the ability to control both the timing and level of activation provides a powerful combination for modulating a viral process.
Design and Analysis of Theophylline-Induced Replication of a Full-Length TBSV Genome.
Next, theophylline-based induction of replication was tested in the more complex and biologically relevant context of the TBSV genome. Gen-60A through Gen-66A were constructed from PK-TD1cT, which is a TBSV genome that contains the same pseudoknot substitutions present in Rep-TD1c (Fig. 2A Inset). Interestingly, the RE riboswitch that functioned best in the replicon (Rep-2A) was not nearly as responsive when tested in the full-length TBSV genome, Gen-60A (Fig. 4A). This finding was not surprising because the genome contains an additional ≈4 kb of sequence that could affect the structure and/or stability of the aptamer. Additional screening of a small group of viral genomes that contained aptamers with different closing stems in place of SL5 allowed for identification of one, Gen-65A, that was suitably responsive (Fig. 4A). Gen-65A accumulation was inducible (≈10-fold) in a dose-dependent manner to a level ≈50% that of the control PK-TD1cT genome (Fig. 4B), thus verifying that this regulatory strategy is also amenable to a complete virus genome.
Fig. 4.
Screening and identification of an RE riboswitch for induction of TBSV genome (Gen) accumulation. (A) Structures of putative RE riboswitches that were introduced in place of SL5 in the TBSV genome PK-TD1cT. Sequence differences in the closing stems are boxed. RE-riboswitch-containing viral genomes were transfected into plant-cell protoplasts, incubated in the absence or presence of theophylline (0.5 mM), and accumulation levels were monitored 22 h after transfection by Northern blotting. Gen-65A (shaded) was identified as the most responsive to theophylline. (B) Dose-dependent theophylline induction of Gen-65A. Protoplasts were transfected with Gen-65A and incubated in the presence of different concentrations of theophylline. Gen-65A accumulation levels were quantified by Northern blot analysis at 22 h after transfection. The graphed values represent means that were normalized to that for the uninduced PK-TD1cT genome control (set at 100%) with standard deviations from three separate experiments. cont., control.
To gain possible insight into the function of the SL5 RE in genome replication, the accumulation levels of both plus and minus strands of viral RNAs, including subgenomic (sg) mRNAs, were analyzed. In the absence of theophylline, Gen-65A genome levels were comparatively low, whereas its associated sg-mRNA levels were similar to those for the control genome, PK-TD1cT (Fig. 5A, compare lanes 2 and 3). The corresponding levels of minus-strand sg mRNAs for Gen-65A were also similar to those of the control genome, but the minus-strand genome level was greater than that of the control genome (Fig. 5B, compare lanes 2 and 3). With increasing concentrations of theophylline, the relative plus-strand genome levels were markedly restored, with sg mRNA levels increasing moderately (Fig. 5A, lanes 4–8). In contrast, the corresponding minus-strand genome and sg-mRNA levels were much less affected by changes in theophylline concentration (Fig. 5B, lanes 4–8).
Fig. 5.
Analysis of viral RNA accumulation in Gen-65A infections. (A) A representative Northern blot showing accumulation of plus strands for TBSV viral RNAs. The concentration of theophylline that transfected plant-cell protoplasts were incubated in posttransfection is indicated at the top, and the positions of the genome (g) and sg mRNAs (sg1 and sg2) are indicated to the left. (B) A representative Northern blot showing accumulation of minus strands for TBSV viral RNAs, as described in A. (C) Schematic representation of TBSV genome replication and sg-mRNA transcription and the effect of disrupting SL5 RE activity. The TBSV genome is shown in the middle. Synthesis of progeny genomes, i.e., replication, proceeds via the synthesis of a full-length complementary minus-strand RNA of the genome that then serves as the template for progeny genome production (as shown in the upper half). In contrast, transcription of sg mRNAs involves premature termination of minus-strand synthesis followed by use of the 3′-truncated minus strand generated as a template for sg-mRNA transcription (as shown in the lower half). A defective SL5 RE (depicted by “X”) specifically inhibits the accumulation of progeny genomes, suggesting a defect in plus-strand genome synthesis.
These findings are significant in several respects. First, they indicate that SL5, which functions as a plus-strand RE in the genome (15), is required for efficient accumulation of progeny viral genomes. This concept is supported by the fact that, in the absence of theophylline (which corresponds to an inactive or “off” mode for the RE riboswitch), the levels of progeny genomes from Gen-65A were dramatically decreased, whereas activation of the RE riboswitch by the addition of ligand alleviated this defect (Fig. 5A). Second, these data show that this RE is not required for efficient synthesis of genome minus strands, as minus strands accumulated efficiently when the RE function was not induced (Fig. 5B). Third, the results show that this RE is not necessary for the synthesis of sg mRNAs, as they accumulated well, regardless of RE activity (Fig. 5A). Importantly, when a WT SL5 was destabilized (by the introduction of mismatches in its stem) in the context of a WT TBSV genome, the same profile of plus- and minus-strand viral RNAs as described above for the uninduced RE riboswitch was observed (data not shown). This result suggests that the preferential defect in plus-strand viral genome accumulation seen in Fig. 5 is authentic and not specifically related to the presence of the RE riboswitch. Instead, the effect correlates well with the lower thermodynamic stability of the unbound aptamer structure, as compared with its ligand-bound counterpart (ΔGbind ≈ −8.9 kcal/mol) (18).
Collectively, this RE riboswitch analysis has (i) confirmed that the RE functions in the plus strand, given that the stereospecificity of the RNA aptamer restricts its activity to the plus strand, and (ii) defined the RE as a major determinant of viral plus strand genome accumulation, suggesting that it may function specifically in promoting synthesis of progeny genomes (Fig. 5C). Thus, although the main objective of this work was to provide a proof-of-concept for a chemically-inducible viral RE-riboswitch system, further facile analysis of the associated viral phenotypes revealed important insights into the structure and function of the RE and, moreover, demonstrated that the system can be useful for addressing fundamental questions about molecular mechanisms.
Utility of RNA Aptamer-Based Control of Viral Processes.
Through rational design, we have succeeded in generating an in vivo viral system based on TBSV that allows for the dose-dependent control of virus genome accumulation via the administration of a small molecule ligand. The mechanism of control is unique in that it relies on direct allosteric regulation of a genomic RNA element that functions in a virus-specific process. Additionally, the system allows for systematic assessment of the dose-dependent effect of a specific RNA element within an invariant viral context. In contrast, in traditional approaches, where a series of different mutant REs are analyzed, each of the RE variants may have a different undesirable and/or unanticipated collateral effect, which could lead to misinterpretations. By using a common viral context, as with the RE riboswitch system, the chances of such complications are diminished. Another advantage of this system is that the activity of the RE can be modulated systematically, which allows for assessment of its function over a defined and broad range. This approach could potentially uncover dose-dependent effects that would be missed if the assessment were performed with more traditional all-or-nothing-type mutational approaches. Moreover, by combining the dosage control with temporal modulation, multidimensional analysis of a viral RNA element is also possible.
An obvious question about this regulatory system is: How stable is the engineered RE riboswitch in the viral genome? Maintenance will depend on many factors, including the RE that is targeted, the structure of the riboswitch, the length of the in vivo experiment, and the propensity of the particular virus to evolve. In our study, the Rep-2A and Gen-65A riboswitches were stably maintained in their viral contexts, irrespective of theophylline concentration, over the entire course of the experiments (data not shown). This 22-h period provided us with ample time to induce RE activity and to observe associated phenotypic effects.
A second logical question is how applicable is this approach to other RNA virus systems. The particular design strategy that we used involved the bound aptamer mimicking a functional RE. However, other schemes using a different RNA aptamer or designing the bound aptamer to interfere with the formation of a functional RE (i.e., an inducible “off” RE riboswitch) are also viable alternatives. Of course, some basic knowledge of the target RE structure and/or sequence in a virus is necessary; however, its specific function need not be known, and RE riboswitch analysis could help to uncover this role (as shown in this study). Importantly, the candidates for this strategy extend beyond plus-strand RNA viruses and include those with negative, ambisense, and retroviral RNA genomes, as well as DNA viruses (see below).
Another question is whether this system could be useful for applied technologies (e.g., virus vector systems). In this respect, the RNA-based regulatory approach adds another technique to the existing tool kit for controlling and using viruses for beneficial purposes. Challenges associated with this scheme include the requirement for custom design of the RE riboswitch and its possible instability over extended infections. Custom designing is an unavoidable aspect of this strategy; however, this requirement is common to other regulatory approaches, such as RNAi-mediated targeting of viral RNAs (19). Similarly, the potential instability of the RE riboswitch also applies to virtually all foreign sequences that are inserted into an RNA virus. With respect to these two parameters, a design and test approach will be required. However, one way of getting away from the instability of RNA virus genomes is to use DNA viruses. In this case, the RE riboswitch could be designed to function in one or more of the viral mRNAs transcribed by the DNA virus. For example, it could be devised to control the translational activity of the viral mRNA (and thus expression of its encoded protein), as has been achieved artificially with eukaryotic cellular mRNAs (4). There is obvious potential for this regulatory strategy to be used for (i) modulating virus-vector replication in cases where temporal considerations and/or viral loads are important (e.g., plus-strand RNA vectors and retroviral vectors) and (ii) controlling the amount and timing of production of foreign proteins, potentially at either the level of sg-mRNA transcription (e.g., plus-strand RNA vectors) or viral mRNA translation (e.g., adeno-, baculo-, retrovirus vectors). Moreover, other RNA-based processes such as RNA virus encapsidation, where RNA origins of assembly control assembly, also represent viable targets for regulation. In conclusion, it is hoped that the proof-of-concept provided herein provides incentive for others to pursue the development of novel and useful viral RE riboswitch control systems.
Materials and Methods
Plasmid Construction.
The majority of the clones used in this study were generated from previously described constructs. These include: T100, the WT-TBSV genome construct (20); Rep, a prototypical TBSV defective-interfering RNA (21); and Rep-TD1c, a modified replicon clone (15). Rep-1A, Rep-2A, and Rep-3A were generated from Rep-TD1c by replacing their WT SL5s with modified theophylline-binding aptamers (1). Rep-2Am is a derivative of Rep-2A in which the adenylate, in the CAG portion of the internal loop in the aptamer, was substituted with a cytidylate. sg1T100 is a modified TBSV genome clone generated by replacing its 5′ UTR with the 5′ UTR of sg mRNA1 (which does not contain SL5). Gen-60A through Gen-66A were constructed from PK-TD1cT, which contained the pseudoknot substitutions present in Rep-TD1c. PK-TD1cT replicates its genome and transcribes its sg mRNAs at WT levels (data not shown). Where relevant, detailed structures of the modifications introduced into these different base constructs are presented in the respective figures. All modifications were introduced by using PCR-based mutagenesis and standard cloning techniques. The PCR-derived regions introduced into constructs were sequenced completely to ensure that only the intended modifications were present.
In Vitro Transcription and Protoplast Inoculation.
In vitro RNA transcripts of TBSV replicons and genomes were generated with T7 RNA polymerase as described (22). Preparation and transfection of cucumber protoplasts were carried out as described (22). Briefly, isolated protoplasts (≈300,000) were prepared from cucumber cotyledons and inoculated with in vitro-generated viral transcripts. For all genomes, 3 μg of transcript was used, and for replicons, 1 μg of transcript was used. For replicon experiments, transfected protoplasts were incubated at 22°C for 22 h with or without the indicated concentrations of theophylline or caffeine. Genome analysis was performed in a similar manner except that infected protoplasts were incubated at 27°C.
Viral RNA Analysis.
Total nucleic acid preparations isolated from virus-inoculated protoplasts were subjected to Northern blot analysis for detection of plus- and minus-strand viral RNAs as described (22). Nucleic acids were either treated with glyoxal and separated in 1.4% agarose gels (for genome and subgenomic RNA analysis) or denatured in formamide-containing buffer and separated in 4.5% polyacrylamide/8 M urea gels (for replicon analysis). Equal loading of lanes was confirmed before transfer via staining the gels with ethidium bromide. After electrophoretic transfer to nylon membranes, viral RNAs were detected with 32P-labeled DNA oligonucleotide probes or RNA riboprobe (for plus- and minus-strand detection, respectively) and their relative levels were determined by using a phosphoimager (Bio-Rad, Hercules, CA). RNA secondary structures were predicted with the mfold computer program (23, 24).
Acknowledgments
We thank members of our laboratory for reviewing the manuscript and Marc Fabian for constructing sg1T100. This work was supported by a Natural Sciences and Engineering Research Council of Canada Steacie Fellowship and Discovery grant, and a Canada Research Chair.
Abbreviations
- RE
regulatory RNA element
- SL5
Stem loop5
- TBSV
tomato bushy stunt virus
- sg
subgenomic.
Footnotes
The authors declare no conflict of interest.
References
- 1.Jenison RD, Gill SC, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Breaker RR. ChemBioChem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 3.Isaacs FJ, Dwyer DJ, Collins JJ. Nat Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 4.Werstuck G, Green MR. Science. 1998;282:296–298. doi: 10.1126/science.282.5387.296. [DOI] [PubMed] [Google Scholar]
- 5.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Nat Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 6.Bayer TS, Smolke CD. Nat Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 7.Suess B, Fink B, Berens C, Stentz R, Hillen W. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harvey I, Garneau P, Pelletier J. RNA. 2002;8:452–463. doi: 10.1017/s135583820202633x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An CI, Trinh VB, Yokobayashi Y. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yen L, Svendsen J, Lee JS, Gray JT, Magnier M, Baba T, D'Amato RJ, Mulligan RC. Nature. 2004;431:471–476. doi: 10.1038/nature02844. [DOI] [PubMed] [Google Scholar]
- 11.Soukup GA, Breaker RR. Proc Natl Acad Sci USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim DS, Gusti V, Pillai SG, Gaur RK. RNA. 2005;11:1667–1677. doi: 10.1261/rna.2162205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buskirk AR, Landrigan A, Liu DR. Chem Biol. 2004;11:1157–1163. doi: 10.1016/j.chembiol.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 14.White KA, Nagy PD. Prog Nucleic Acid Res Mol Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- 15.Ray D, Wu B, White KA. RNA. 2003;9:1232–1245. doi: 10.1261/rna.5630203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A. RNA. 2000;6:659–667. doi: 10.1017/s1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White KA, Morris TJ. Proc Natl Acad Sci USA. 1994;91:3642–3646. doi: 10.1073/pnas.91.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouda H, Kuntz ID, Case DA, Kollman PA. Biopolymers. 2003;68:16–34. doi: 10.1002/bip.10270. [DOI] [PubMed] [Google Scholar]
- 19.Ketzinel-Gilad M, Shaul Y, Galun E. J Gene Med. 2006;8:933–950. doi: 10.1002/jgm.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hearne PQ, Knorr DA, Hillman BI, Morris TJ. Virology. 1990;177:141–151. doi: 10.1016/0042-6822(90)90468-7. [DOI] [PubMed] [Google Scholar]
- 21.Wu B, Vanti WB, White KA. J Mol Biol. 2001;305:741–756. doi: 10.1006/jmbi.2000.4298. [DOI] [PubMed] [Google Scholar]
- 22.White KA, Morris TJ. J Virol. 1994;68:14–24. doi: 10.1128/jvi.68.1.14-24.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews DH, Sabina J, Zuker M, Turner DH. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M, Mathews DH, Turner DH. In: RNA Biochemistry and Bio/Technology. Barciszewski J, Clark BFC, editors. Dordrecht, The Netherlands: Kluwer; 1999. pp. 11–43. [Google Scholar]