Abstract
Androgen receptor (AR) is essential for the growth and progression of prostate cancer in both hormone-sensitive and hormone-refractory disease. A DNA-binding polyamide that targets the consensus androgen response element binds the prostate-specific antigen (PSA) promoter androgen response element, inhibits androgen-induced expression of PSA and several other AR-regulated genes in cultured prostate cancer cells, and reduces AR occupancy at the PSA promoter and enhancer. Down-regulation of PSA by this polyamide was comparable to that produced by the synthetic antiandrogen bicalutamide (Casodex) at the same concentration. Genome-wide expression analysis reveals that a similar number of transcripts are affected by treatment with the polyamide and with bicalutamide. Direct inhibition of the AR-DNA interface by sequence-specific DNA binding small molecules could offer an alternative approach to antagonizing AR activity.
Keywords: prostate cancer, bicalutamide, small molecule, transcription
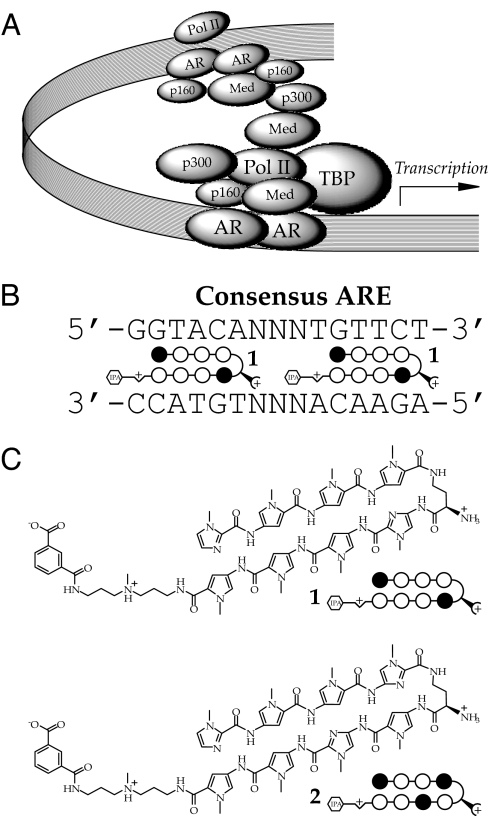
The androgen receptor (AR) is a member of the ligand-activated nuclear receptor family of transcription factors (1). Ligand binding to AR initiates release from the cytoplasm, dimerization, binding to the androgen response elements (ARE) of target genes, and gene activation through interaction with coactivators and the general transcription machinery (2). Functional AREs, consensus sequence 5′-GGTACAnnnTGTTCT-3′ (Fig. 1A) (3) can occur in proximal promoter sequences or in enhancers located up to several thousand base pairs upstream or downstream of the transcription start site.
Fig. 1.
Targeting the ARE with DNA-binding polyamides. (A) Model of the AR transcription complex. (B) Consensus ARE. (C) Structures and ball-and-stick models of polyamides 1, designed to bind the consensus ARE, and 2, a 2-bp mismatch. Imidazole and pyrrole monomer units are represented by filled and open circles, respectively. The isophthalic acid tail moiety is represented by an hexagon.
The regulation of prostate-specific antigen (PSA) (KLK3) expression by AR has been extensively studied as a model for AR-mediated gene activation (4–7). Androgenic induction of PSA is mediated by AR binding to the proximal promoter ≈170 bp from the transcription start site and to several low-affinity AREs in an enhancer ≈4,000 bp upstream (4–6). AREs in both the promoter and enhancer are important for induction after androgen stimulation. AR occupies both the promoter and enhancer regions and recruits transcriptional coactivators including p160 and p300, TATA-binding protein, mediator, and RNA polymerase II to form the AR transcription complex (7, 8). Chromatin-capture assays suggest that the PSA enhancer is located near the promoter in this complex (8).
AR signaling regulates normal prostate development and contributes to the progression of prostate cancer (9). Surgical or drug therapies that act to limit circulating androgen levels or directly antagonize ligand binding to AR initially slow prostate cancer growth (10, 11). However, nearly all patients treated with antiandrogen therapies will eventually develop hormone-refractory disease (12). Dysregulation of AR activity, together with activation of the PTEN/AKT pathway, is thought to contribute to this transition (13). Up-regulation of AR mRNA was found to occur in all transitions from hormone-sensitive to hormone-refractory disease in a mouse tumor-xenograft model of prostate cancer (14). Additionally, a transgenic mouse with a mutated AR that inappropriately interacts with transcriptional coregulators developed metastatic neoplastic disease (15). Mutations in the AR ligand-binding domain can render antagonists such as bicalutamide or flutamide ineffective or, in some models of hormone-refractory disease, convert them to agonists (14, 16). Given that genotropic AR activity is thought to be necessary throughout prostate cancer progression, direct antagonism of AR–DNA binding could inhibit androgen receptor activity in hormone-refractory conditions where androgen antagonists that target the ligand-binding pocket are ineffective (9).
DNA-binding polyamides represent one approach to inhibiting protein–DNA interactions. Polyamides containing N-methylimidazole (Im) and N-methylpyrrole (Py) comprise a class of programmable DNA-binding ligands capable of binding to a broad repertoire of DNA sequences with affinities and specificities comparable to those of natural DNA-binding proteins (17, 18). Sequence specificity is programmed by side-by-side pairings of the heterocyclic amino acids in the minor groove of DNA: Im/Py distinguishes G·C from C·G; Py/Py binds both A·T and T·A (19, 20). Previously, a hairpin polyamide targeted to the hypoxia response element (HRE) inhibited hypoxia-induced expression of several HIF-1-regulated genes, including VEGF, in cultured cells (21, 22).
In this study, we have designed a cell-permeable polyamide to target the sequence 5′-WGWWCW-3′, found in the consensus ARE, with the goal of disrupting AR-mediated gene expression (Fig. 1). We show that this polyamide binds the ARE found in the PSA promoter, inhibits expression of PSA as well as ≈35% of the transcripts that were induced by dihydrotestosterone (DHT) in cultured prostate cancer cells, and reduces AR occupancy at the PSA promoter and enhancer. Down-regulation of PSA by this polyamide was comparable to the effects of the synthetic antiandrogen bicalutamide (Casodex) at the same concentration. A control polyamide targeted to a different sequence had less effect.
Results
Binding Affinities of Polyamides to the ARE of the PSA Promoter.
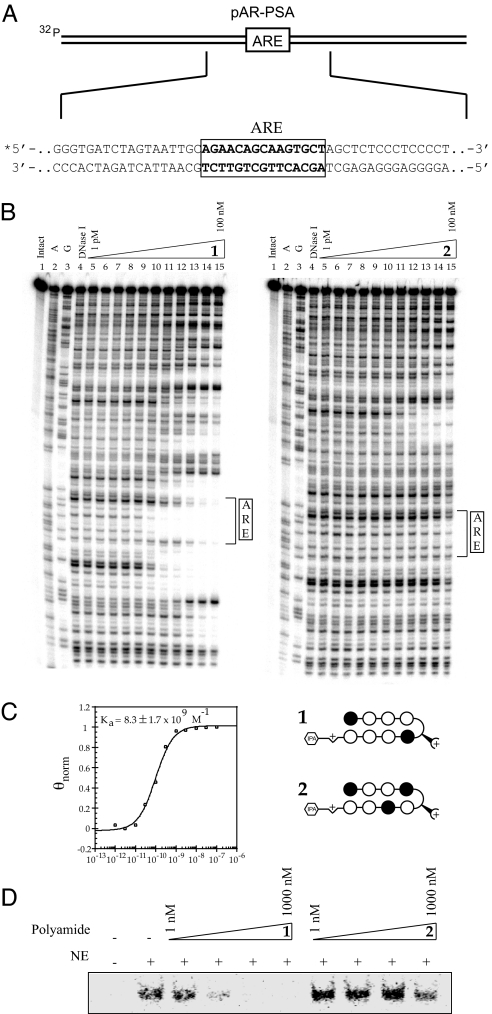
The proximal PSA promoter contains the ARE 5′-AGAACAGCAAGTGCT-3′ (Fig. 2A). The DNA binding of polyamides 1 and 2 on this sequence was measured by quantitative DNase I footprint titrations using a 5′-32P-labeled PCR fragment of pAR-PSA, which contains the PSA ARE. Polyamide 1 has a Ka = 8.3 ± 1.7 × 109 M−1 for the ARE consensus half-site 5′-AGAACA-3′ (Fig. 2B). Binding of polyamide 2, which targets the sequence 5′-WGWCGW-3′, to the ARE is not measurable by these methods (Ka < 1 × 107) (Fig. 2C). Minimal binding of polyamide 1 is observed at the other half-site of the ARE: 5′-AGTGCT-3′, which is formally a single base pair mismatch site for 1. However, 1 is observed to bind the sequence 5′-AGATCA-3′ ≈12 bp 5′ to the ARE, which is an expected binding site for this molecule.
Fig. 2.
Binding of 1 and 2 to the ARE in the PSA promoter. (A) Illustration of pAR and partial sequence of the PSA promoter. (B) Quantitative DNase I footprint titration experiments for polyamides 1 and 2 on the 5′-end-labeled PCR product of plasmid pAR-PSA: lane 1, intact DNA; lane 2, A reaction; lane 3, G reaction; lane 4, DNase I standard; lanes 5–15, 1 pM, 3 pM, 10 pM, 30 pM, 100 pM, 300 pM, 1 nM, 3 nM, 10 nM, 30 nM, 100 nM polyamide, respectively. (C) Isotherm for 1 binding to the ARE half-site 5′-AGAACA-3′. Polyamide 1 has a Ka = 8.3 ± 1.7 × 109 for this site. Polyamide 2 shows no measurable binding in the footprinted region. (D) EMSA of DHT-stimulated LNCaP cell nuclear extract (NE) binding to a 31-bp oligonucleotide duplex containing the PSA promoter ARE in the presence of 1 and 2.
Electrophoretic Mobility Shift Assay.
The effects of polyamides 1 and 2 on the binding of factors present in the nuclear extract isolated from DHT-stimulated LNCaP cells to the ARE site in the PSA promoter was measured by an electrophoretic mobility shift assay (Fig. 2D). Polyamide 1 inhibits binding to the 5′-32P-labeled duplex at concentrations as low as 10 nM. Polyamide 2 has minimal effect at the same concentrations.
Inhibition of Androgen-Inducted PSA Expression.
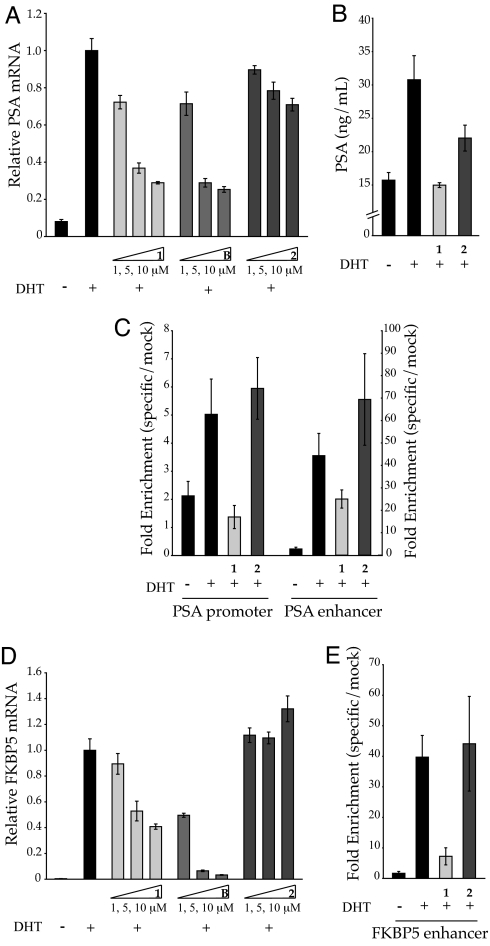
Induction of PSA mRNA by DHT in the presence of polyamides 1 and 2 and bicalutamide in LNCaP cells was measured by quantitative real-time RT-PCR. Bicalutamide and polyamide 1 inhibit the expression of DHT-induced PSA in a dose-dependent manner up to ≈70% at 10 μM, as measured in this assay (Fig. 3A). Polyamide 2 has a more modest effect. Secretion of PSA protein after DHT stimulation of LNCaP cells in the presence of 1 and 2 was measured by ELISA (Fig. 3B). Supernatant concentrations of PSA protein are reduced in cells pretreated with 1 as compared with 2 or an untreated control. AR occupancy at the PSA promoter and enhancer was assessed by chromatin immunoprecipitation (Fig. 3C). Chromatin immunoprecipitation assays with anti-AR antibody treatment indicate decreased occupancy of AR at the PSA promoter and enhancer in the presence of 10 μM 1. Polyamide 2 has minimal effect. Polyamides 1 and 2 display no obvious detrimental effects on cell growth over the course of the experiment. AR mRNA is minimally affected by 1 [see supporting information (SI) Fig. 5].
Fig. 3.
Inhibition of DHT-induced PSA and FKBP5 expression by 1 and 2. (A) Induction of PSA mRNA in the presence of 1 and 2 and bicalutamide, B, measured by quantitative real-time PCR. 1 and bicalutamide inhibit expression of PSA in a dose-dependent manner up to ≈70% at 10 μM. 2 has a more modest effect. (B) Secreted PSA protein measured by ELISA. (C) Chromatin immunoprecipitation assays with anti-AR or mock antibody treatment expressed as fold-enrichment (specific/mock) of DNA sequences at the PSA promoter and enhancer. AR occupancy at the PSA promoter and enhancer is decreased in the presence of 1 (10 μM) but not 2. (D) Induction of FKBP5 mRNA in the presence of 1 and 2 and bicalutamide, B. (E) Chromatin immunoprecipitation assays with anti-AR at the FKBP5 fifth intron enhancer. Polyamide concentrations are 10 μM.
Inhibition of Androgen-Induced FKBP5 Expression.
Recent studies have identified FKBP5 as one of the most strongly induced genes in androgen-stimulated prostate cancer cells (23). Two functional AREs with the sequences 5′-AGCACATCGAGTTCA-3′ and 5′-AGAACAGGGTGTTCT-3′ have been mapped to an enhancer within the fifth intron (24). Polyamide 1 inhibits DHT-induced expression of FKBP5 by ≈60% (Fig. 3D). Bicalutamide was more potent, however, inhibiting expression by almost 95%. Polyamide 2 has minimal effect on FKBP5 expression. Chromatin immunoprecipitation assays indicate decreased occupancy of AR at the FKBP5 intronic enhancer in the presence of 10 μM 1 (Fig. 3E), whereas polyamide 2 has no measurable effect.
Global Effects on Androgen-Induced Gene Expression.
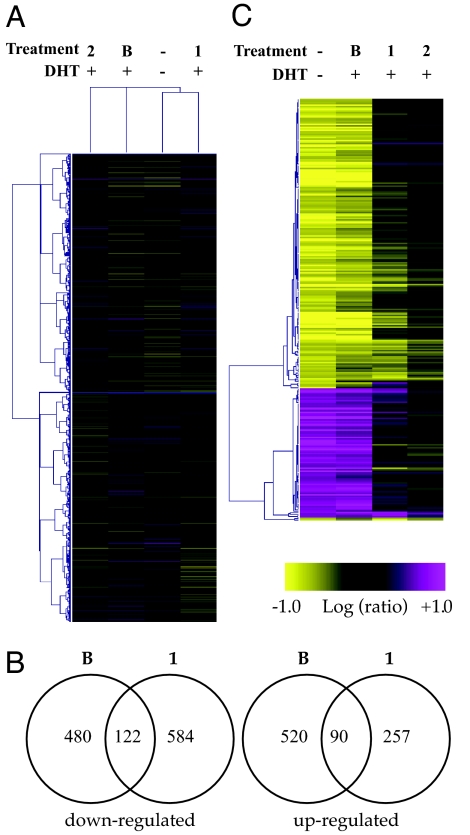
Global effects of polyamides 1 and 2 and bicalutamide on gene expression in DHT-stimulated LNCaP cells were monitored with Affymetrix (Santa Clara, CA) high-density Human Genome U133 Plus 2.0 arrays, which interrogate >50,000 transcripts. As compared with DHT-induced controls, polyamide 1 (10 μM) affected the expression of 1,053 transcripts by at least 2-fold (P ≤ 0.01) (Table 1), which represents less than 2% of interrogated transcripts. Of this total, 706 were down-regulated. At the same threshold, bicalutamide (10 μM) affected the expression of 1,213 transcripts, with 602 of these being down-regulated. Polyamide 2 (10 μM) affected the expression of 379 transcripts, which represents <1% of interrogated transcripts. A divisive clustering analysis over all interrogated transcripts suggests that the expression profiles of cells treated with bicalutamide, 1, and 2 are largely distinct (Fig. 4A). Analysis of transcripts affected by both bicalutamide and 1 shows that 122 and 90 transcripts are commonly down- and up-regulated, respectively, at least 2-fold (P ≤ 0.01) (Fig. 4B). Of the 122 transcripts down-regulated by both bicalutamide and 1, 117 are also observed to be induced by DHT at the same thresholds. Of the 90 up-regulated transcripts, 59 are observed to be repressed by DHT.
Table 1.
Number of transcripts affected relative to DHT-induced controls (P ≤ 0.01)
| Treatment | DHT | Up-regulated (fold change ≥ 2.0) | Down-regulated (fold change ≤ −2.0) | Up-regulated (fold change ≥ 4.0) | Down-regulated (fold change ≤ −4.0) |
|---|---|---|---|---|---|
| — | — | 486 | 782 | 88 | 199 |
| B | + | 611 | 602 | 96 | 133 |
| 1 | + | 347 | 706 | 42 | 126 |
| 2 | + | 95 | 284 | 11 | 32 |
—, No treatment.
Fig. 4.
Global effects on transcripts interrogated by using Affymetrix high-density Human Genome U133 Plus 2.0 arrays. (A) Divisive clustering of all measured transcripts under the four specified conditions: no treatment control, bicalutamide (B, 10 μM), 1 (10 μM), and 2 (10 μM). Clustering was based on an error-weighted Pearson correlation of intensity ratios for each treatment as compared with DHT-induced controls. (B) Ven diagrams representing transcripts down- and up-regulated (|fold change| ≥ 2.0, P ≤ 0.01) by bicalutamide and 1. Numbers inside the intersections represent transcripts affected by both treatments. Of the 122 transcripts down-regulated by both bicalutamide and 1, 117 are also observed to be induced by DHT at the same thresholds. (C) Agglomerative clustering of expression changes of the 199 transcripts induced or repressed 4-fold (P ≤ 0.01) or more by 1 nM DHT under the designated treatment conditions. Of the DHT-induced set, 70 were inhibited by polyamide 1, 20 were inhibited by 2, and 186 were inhibited by bicalutamide (|fold change| ≥ 2.0, P ≤ 0.01). Clustering parameters were the same as in A. Treatments reported are an error-weighted average from three experiments, except the noninduced control, which was an average from two experiments.
The response of cultured prostate cancer cells to androgen has been extensively studied (23, 25). We find that DHT induced the expression of a set of 199 transcripts by at least 4-fold (P ≤ 0.01). Of this set, 70 were also inhibited by polyamide 1 by at least 2-fold (P ≤ 0.01). For comparison, polyamide 2 inhibited 20, and bicalutamide inhibited 186, of the 199 DHT-induced transcripts with the same thresholds (Fig. 4C). DHT repressed the expression of a set of 88 transcripts by at least 4-fold (P ≤ 0.01). Of this set, eight were also derepressed, as compared with DHT-treated controls, by polyamide 1 by at least 2-fold (P ≤ 0.01). For comparison, polyamide 2 derepressed 3, and bicalutamide derepressed 87, of the 88 transcripts repressed by DHT with the same thresholds (Fig. 4C). A complete list of the DHT-induced transcripts and those affected by 1 is provided in SI Tables 3 and 4. It is not known what proportions of these genes are direct targets of AR. Table 2 displays the effects of each treatment on the expression of a few selected genes that were observed to be induced by DHT and are known to be targets of AR (26, 27). Effects on the expression of KLK2 and TMPRSS2 were verified by quantitative real-time RT-PCR (SI Fig. 5).
Table 2.
Fold-changes of selected AR-target genes relative to DHT-induced controls
| Treatment | DHT | KLK2 | KLK3 (PSA) | TMPRSS2 | FKBP5 |
|---|---|---|---|---|---|
| — | — | −23.0 | −6.1 | −6.2 | −42.9 |
| B | + | −14.7 | −3.2 | −4.1 | −36.4 |
| 1 | + | −2.4 | −3.3 | −2.3 | −3.1 |
| 2 | + | −1.1 | −1.4 | −1.4 | 1.5 |
—, No treatment.
Discussion
Because numerous signaling pathways converge on a smaller number of transcription factors to exert their effects on gene expression, it has been proposed that transcription factors could be among the most appropriate drug targets in oncology (28, 29). This possibility has underscored the challenge to design small molecules capable of selectively disrupting protein–protein interactions between coactivators as well as protein–DNA interactions between transcription factors and their target sites in gene regulatory sequences.
Prostate cancer cells depend on stimulation by circulating androgens that exert their effects through the AR signaling axis. Hormone therapies that block AR activity by starving it of androgens or inhibiting ligand binding are initially successful but ultimately fail to control disease (12). This failure can occur through up-regulation of AR, mutations in the ligand-binding pocket, and ligand-independent activation from upstream signaling proteins (13, 30, 31). It is thought, however, that intact activity of AR signaling is necessary for disease progression (9). Inhibition of the AR–DNA interaction by a sequence-specific DNA-binding molecule could be expected to interfere with AR signaling under both hormone-sensitive and hormone-refractory conditions.
Polyamide 1 binds to a half-site of the ARE of the PSA promoter with a subnanomolar Kd and inhibits expression of ≈35% of transcripts that are observed to be induced at least 4-fold by DHT in LNCaP cells. Down-regulation of PSA by this polyamide is comparable to that produced by the synthetic antiandrogen bicalutamide at the same concentration. Control polyamide 2, which targets a different DNA sequence, 5′-WGWCGW-3′, had significantly less effect on androgen-induced gene expression. Expression of PSA (KLK3), KLK2, TMPRSS2, and FKBP5, which are direct AR targets, were all affected by 1. TMPRSS2 encodes a transmembrane protease and can undergo a chromosomal deletion in which a member of the ETS transcription factor family is placed under control of the strongly androgen-responsive TMPRSS2 5′ regulatory region (27, 32).
At the same concentration, polyamide 1 and bicalutamide affected a comparable number of transcripts, whereas polyamide 2 affected significantly fewer. When using bicalutamide as a point of reference, the overall effects on genomic transcription by 1 and 2 are relatively modest. Although it is difficult to compare across experimental conditions, the observation that a limited number of genes are affected by each polyamide in this study is consistent with previous reports (21). A comparison of the expression data for cells treated with polyamide 1 or 2 reveal that some transcripts are similarly affected, but many are differentially affected by the two polyamides (Fig. 4A), which is consistent with previous comparisons of gene expression profiles of cells treated with polyamides of different target sequence (21, 33).
The AR, glucocorticoid receptor, and estrogen receptor share a highly conserved DNA-binding domain (34–36). This domain, related to the classical Cys-2-His-2 zinc finger motifs (37), contains two modules of zinc coordinated by four cysteines. Previously, a polyamide targeted to the estrogen receptor response element inhibited binding of estrogen receptors α and β in gel-shift assays (38). In separate in vitro experiments, minor groove-binding polyamides have been shown to inhibit the major groove binding of Zif268 and other zinc finger proteins to their target sites on DNA by an allosteric mechanism (39). In light of this observation, it is not unexpected that a polyamide targeted to the ARE would inhibit AR binding.
The ARE is sufficiently degenerate such that a single polyamide is not likely to affect all AR-regulated genes simultaneously. The identities of the particular AR target genes involved in prostate cancer progression are not fully known. In the absence of this knowledge, it was our goal to target the ARE broadly to maximize the number of AR target genes affected by using a single polyamide. However, the programmability of polyamides might allow selective inhibition of a predetermined subset of AR target genes by one or a small mixture of tailored polyamide molecules. The utility of disrupting the AR–ARE interface with DNA-binding small molecules will depend on continued experimentation in small animal models of hormone refractory prostate cancer and AR-regulated gene expression (40–42).
Materials and Methods
Synthesis of Polyamides.
Polyamides 1 and 2 were synthesized by solid-phase methods on Kaiser oxime resin (Nova Biochem, Darmstadt, Germany) according to established protocols (43). Polyamides were cleaved from resin with 3,3′-diamino-N-methyl-dipropylamine and purified by reverse-phase HPLC. Isophthalic acid was activated with PyBOP (Nova Biochem) and conjugated to the polyamides as described (22). Purities and identities of the polyamides were assessed by HPLC, UV-visible spectroscopy, and MALDI-TOF MS.
Determination of DNA-Binding Affinity and Sequence Specificity.
Plasmid pAR-PSA was constructed by inserting a 70-bp sequence from the PSA promoter containing the ARE into pUC19 plasmid. Quantitative DNase I footprint titration experiments were used to measure the binding affinities of 1 and 2 on a 5′-32P-labeled fragment of pAR-PSA that contains the PSA promoter ARE. Detailed experimental protocols are reported elsewhere (44).
Electrophoretic Mobility Shift Assay.
The oligonucleotide 5′-GCATTGCAGAACAGCAAGTGCTAGCTCTCCC-3′ containing the PSA promoter ARE (underlined) was end-labeled with 32P and annealed to its complement. Polyamides 1 and 2 were incubated with the duplex for 3 h in previously optimized buffer conditions (45). Nuclear extract from DHT-treated LNCaP cells (Genetex, San Antonio, TX) was then added for an additional 45 min. Complexes were run on a 5% polyacrylamide gel and visualized on a phosphorimager.
Measurement of Androgen-Induced PSA mRNA and Protein.
LNCaP cells (ATCC) were plated in 24-well plates at a density of 40–50 × 103 cells per well (80–100 × 103 cells per ml) in RPMI medium 1640 (ATCC) supplemented with 10% FBS (Irvine Scientific, Santa Ana, CA). After 72 h, the medium was replaced with RPMI medium 1640 containing 10% charcoal stripped FBS with or without polyamides at the designated concentrations. Cells were grown for an additional 48 h and then treated with 1 nM DHT for 16 h. When appropriate, bicalutamide was added 2 h before DHT stimulation. Isolation of RNA and cDNA synthesis was performed as described (21). Quantitative real-time RT-PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA) on an ABI 7300 instrument. PSA mRNA was measured relative to β-glucuronidase as an endogenous control. Primer sequences are available upon request. Cell-culture supernatants were collected for an ELISA (R & D Systems, Minneapolis, MN) to measure PSA protein according to the manufacturer's protocol.
Chromatin Immunoprecipitation.
LNCaP cells were plated in 15-cm diameter plates at a density of 2 × 106 cells per plate. Media, polyamide treatment, time course, and DHT stimulation were the same as described above. After the 16-h DHT treatment, cells were treated with 1% formaldehyde for 10 min. Chromatin was isolated and sheared. Antibodies to AR (AR-20, Santa Cruz Biotechnology, Santa Cruz, CA) were used to immunoprecipitate AR-bound DNA fragments. Crosslinks were reversed, and PCRs using primers targeted to the regions of interest were used to assess enrichment of bound fragments as compared with mock-precipitated (no antibody) controls. PCRs were monitored with SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7300 instrument. Primer sequences and a more detailed experimental protocol are available upon request.
Analysis of Gene Expression with Oligonucleotide Microarrays.
LNCaP cells were plated in 12-well plates at a density of 80–100 × 103 cells per well. Media, polyamide treatments, and time courses were the same as described above. Bicalutamide was added 2 h before DHT stimulation. RNA was isolated as described in ref. 21. From this point, experiments were carried out at the Millard and Muriel Jacobs Gene Expression Facility at the California Institute of Technology. Labeled mRNA was hybridized to Affymetrix high-density Human Genome U133 Plus 2.0 arrays according to established protocols. Gene expression was analyzed by using Resolver (Rosetta Biosoftware, Seattle, WA). Data were uploaded to the Gene Expression Omnibus repository (accession no. GSE7708).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM57148. Mass spectrometry analyses were performed in the Mass Spectrometry Laboratory of the Division of Chemistry and Chemical Engineering at the California Institute of Technology, supported in part by the National Science Foundation Materials Research Science and Engineering program. Oligonucleotide microarray experiments were performed in the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology.
Abbreviations
- AR
androgen receptor
- ARE
androgen response element
- PSA
prostate-specific antigen
- DHT
dihydrotestosterone.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE7708).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704217104/DC1.
References
- 1.Tsai MJ, Omalley BW. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- 2.Tyagi RK, Lavrovsky Y, Ahn SC, Song CS, Chatterjee B, Roy AK. Mol Endocrinol. 2000;14:1162–1174. doi: 10.1210/mend.14.8.0497. [DOI] [PubMed] [Google Scholar]
- 3.Roche PJ, Hoare SA, Parker MG. Mol Endocrinol. 1992;6:2229–2235. doi: 10.1210/mend.6.12.1491700. [DOI] [PubMed] [Google Scholar]
- 4.Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J. Mol Endocrinol. 1997;11:148–161. doi: 10.1210/mend.11.2.9883. [DOI] [PubMed] [Google Scholar]
- 5.Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- 6.Huang WB, Shostak Y, Tarr P, Sawyers C, Carey M. J Biol Chem. 1999;274:25756–25768. doi: 10.1074/jbc.274.36.25756. [DOI] [PubMed] [Google Scholar]
- 7.Shang YF, Myers M, Brown M. Mol Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 8.Wang QB, Carroll JS, Brown M. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Scher HI, Sawyers CL. J Clin Oncol. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 10.Huggins C, Hodges CV. Cancer Res. 1941;1:293–297. [Google Scholar]
- 11.Huggins C, Stevens RE, Hodges CV. Arch Surg (Chicago) 1941;43:209–223. [Google Scholar]
- 12.Oefelein MG, Agarwal PK, Resnick MI. J Urol. 2004;171:1525–1528. doi: 10.1097/01.ju.0000118294.88852.cd. [DOI] [PubMed] [Google Scholar]
- 13.Xin L, Teitell MA, Lawson DA, Kwon A, Mellinghoff IK, Witte ON. Proc Natl Acad Sci USA. 2006;103:7789–7794. doi: 10.1073/pnas.0602567103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, Rosenfeld MG, Sawyers CL. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 15.Han GZ, Buchanan G, Ittmann M, Harris JM, Yu XQ, DeMayo FJ, Tilley W, Greenberg NM. Proc Natl Acad Sci USA. 2005;102:1151–1156. doi: 10.1073/pnas.0408925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohl CE, Gao WQ, Miller DD, Bell CE, Dalton JT. Proc Natl Acad Sci USA. 2005;102:6201–6206. doi: 10.1073/pnas.0500381102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CCC, Dervan PB. Tetrahedron. 2007 doi: 10.1016/j.tet. 2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dervan PB, Edelson BS. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 19.Kielkopf CL, Baird EE, Dervan PD, Rees DC. Nat Struct Biol. 1998;5:104–109. doi: 10.1038/nsb0298-104. [DOI] [PubMed] [Google Scholar]
- 20.White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Nature. 1998;391:468–471. doi: 10.1038/35106. [DOI] [PubMed] [Google Scholar]
- 21.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Dervan PB. Proc Natl Acad Sci USA. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePrimo SE, Diehn M, Nelson JB, Reiter RE, Matese J, Fero M, Tibshirani R, Brown PO, Brooks JD. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0032. research0032.1–research0032.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magee JA, Chang LW, Stormo GD, Milbrandt J. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 25.Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin BY. Proc Natl Acad Sci USA. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell SH, Murtha PE, Zhang SB, Zhu W, Young CYF. Mol Cell Endocrinol. 2000;168:89–99. doi: 10.1016/s0303-7207(00)00319-1. [DOI] [PubMed] [Google Scholar]
- 27.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao XH, Tchinda J, Kuefer R, et al. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 28.Darnell JE. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 29.Pandolfi PP. Oncogene. 2001;20:3116–3127. doi: 10.1038/sj.onc.1204299. [DOI] [PubMed] [Google Scholar]
- 30.Mellinghoff IK, Vivanco I, Kwon A, Tran C, Wongvipat J, Sawyers CL. Cancer Cell. 2004;6:517–527. doi: 10.1016/j.ccr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 31.Chen TS, Wang LH, Farrar WL. Cancer Res. 2000;60:2132–2135. [PubMed] [Google Scholar]
- 32.Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgesson BE, Cao XH, Wei JT, Rubin MA, Shah RB, et al. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 33.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, Dervan PB, Gottesfeld JM. Proc Natl Acad Sci USA. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. Proc Natl Acad Sci USA. 2004;101:4758–4763. doi: 10.1073/pnas.0401123101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 36.Schwabe JWR, Chapman L, Finch JT, Rhodes D. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- 37.Pavletich NP, Pabo CO. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 38.Gearhart MD, Dickinson L, Ehley J, Melander C, Dervan PB, Wright PE, Gottesfeld JM. Biochemistry. 2005;44:4196–4203. doi: 10.1021/bi047872o. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen-Hackley DH, Ramm E, Taylor CM, Joung JK, Dervan PB, Pabo CO. Biochemistry. 2004;43:3880–3890. doi: 10.1021/bi030223c. [DOI] [PubMed] [Google Scholar]
- 40.Klein KA, Reiter RE, Redula J, Morad H, Zhu XL, Brothman AR, Lamb DJ, Marcelli M, Belldegrun A, Witte ON, et al. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 41.Ellwood-Yen K, Wongvipat J, Sawyers C. Cancer Res. 2006;66:10513–10516. doi: 10.1158/0008-5472.CAN-06-1397. [DOI] [PubMed] [Google Scholar]
- 42.Iyer M, Salazar FB, Lewis X, Zhang L, Wu L, Carey M, Gambhir SS. Transgenic Res. 2005;14:47–55. doi: 10.1007/s11248-004-2836-1. [DOI] [PubMed] [Google Scholar]
- 43.Belitsky JM, Nguyen DH, Wurtz NR, Dervan PB. Bioorg Med Chem. 2002;10:2767–2774. doi: 10.1016/s0968-0896(02)00133-5. [DOI] [PubMed] [Google Scholar]
- 44.Trauger JW, Dervan PB. Methods Enzymol. 2001;340:450–466. doi: 10.1016/s0076-6879(01)40436-8. [DOI] [PubMed] [Google Scholar]
- 45.Zhang JY, Zhang SB, Murtha PE, Zhu W, Hou SSM, Young CYF. Nucleic Acids Res. 1997;25:3143–3150. doi: 10.1093/nar/25.15.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.