Abstract
IQGAP1 modulates several cellular functions, including cell–cell adhesion, transcription, cytoskeletal architecture, and selected signaling pathways. We previously documented that IQGAP1 binds ERK and MAPK kinase (MEK) and regulates EGF-stimulated MEK and ERK activity. Here we characterize the interaction between IQGAP1 and B-Raf, the molecule immediately upstream of MEK in the Ras/MAPK signaling cascade. B-Raf binds directly to IQGAP1 in vitro and coimmunoprecipitates with IQGAP1 from cell lysates. Importantly, IQGAP1 modulates B-Raf function. EGF is unable to stimulate B-Raf activity in IQGAP1-null cells and in cells transfected with an IQGAP1 mutant construct that is unable to bind B-Raf. Interestingly, binding to IQGAP1 significantly enhances B-Raf activity in vitro. Our data identify a previously unrecognized interaction between IQGAP1 and B-Raf and suggest that IQGAP1 is a scaffold necessary for activation of B-Raf by EGF.
Keywords: EGF, MAP kinase, signalling
The Ras/Raf/MAPK kinase (MEK)/ERK module is a ubiquitously expressed signaling pathway that conveys mitogenic and differentiation signals from the cell membrane to the interior of the cell (1, 2). This cascade, which is the best studied of the five MAPK pathways, regulates cell growth, proliferation, and differentiation. In response to a stimulus, such as growth factors, cytokines, or hormones, the guanine nucleotide exchange factor Sos induces the exchange of GDP for GTP on Ras, thereby activating Ras. In turn, Ras recruits Raf from the cytosol to the membrane for activation (3, 4). Raf catalyzes the phosphorylation of MEK, which phosphorylates and activates ERK. Activated ERK modulates the function of multiple substrates in all cellular compartments including the nucleus, cytoplasm, and cytoskeleton (2).
C-Raf was originally identified as the protein product of the retroviral oncogene v-Raf (5). The Raf family of protein kinases comprises three isoforms, A-Raf, B-Raf, and C-Raf (also known as Raf-1) (4, 6). The Raf proteins share a common architecture, and all function as serine/threonine kinases. Evidence derived by several approaches, including genetic studies in mice, indicates that the proteins have distinct functions. The specific mechanism by which Raf proteins are activated is not known, but oligomerization, binding to other proteins, and multiple phosphorylation events are important (4, 6). Although A-Raf, B-Raf, and C-Raf are all regulated by phosphorylation, the presence of different phosphorylation sites indicates that the proteins can be independently regulated (4). C-Raf is the best characterized and most intensively studied of the Raf isoforms (4). More recently, the identification that B-Raf is an important oncogene (7) has resulted in considerable attention being directed toward B-Raf. Notwithstanding these investigations, much remains to be learned about B-Raf regulation.
IQGAP1 is a multidomain molecule that contains several protein-interacting motifs (for reviews, see refs. 8–11). IQGAP1 binds to diverse targets, thereby participating in numerous fundamental cellular activities (9). These binding partners include active Cdc42 and Rac1 (but not RhoA or H-Ras) (12–14), actin (15–17), calmodulin (14, 16), E-cadherin (18, 19), β-catenin (19, 20), CLIP-170 (21), and adenomatous polyposis coli (APC) (22).
IQGAP1 had been postulated to be a scaffold protein that integrates signaling pathways and coordinates several fundamental cellular activities (9, 16). It has become widely recognized over the last few years that scaffolds are important in MAPK signaling (23, 24). In this regard, several scaffold proteins, such as kinase suppression of Ras (KSR1), SUR8, β-arrestin, and MAGUIN, that modulate MEK/ERK signaling have been identified (23, 24). Recent work from our laboratory demonstrates that IQGAP1 functions as a scaffold in the MEK/ERK signal transduction pathway (25, 26). We documented direct binding between IQGAP1 and both MEK and ERK, which modifies the ability of EGF to activate MEK and ERK. Because B-Raf is the predominant activator of MEK (24), we explored a possible interaction between B-Raf and IQGAP1. We observed a direct interaction between IQGAP1 and B-Raf, which modulates activation of B-Raf by EGF.
Results
IQGAP1 Binds to B-Raf in Vitro.
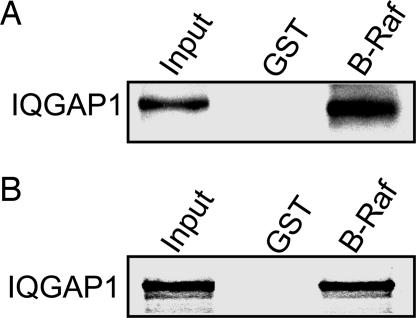
In vitro analysis with pure proteins revealed that IQGAP1 binds to GST-B-Raf (Fig. 1A). Binding is specific because no IQGAP1 is present in the samples incubated with GST alone. This observation was confirmed with the transcription and translation (TNT) system; IQGAP1 labeled with [35S]methionine binds specifically to B-Raf (Fig. 1B). These data reveal a direct interaction between IQGAP1 and B-Raf.
Fig. 1.
IQGAP1 binds to B-Raf in vitro. (A) GST alone or GST-B-Raf (B-Raf) bound to glutathione-Sepharose was incubated with equal amounts of purified IQGAP1. Complexes were isolated, processed by Western blotting, and probed with anti-IQGAP1 monoclonal antibody. Input is 20 ng of purified IQGAP1. (B) GST or GST-B-Raf was incubated with equal amounts of [35S] methionine-labeled IQGAP1. Complexes were isolated with glutathione-Sepharose and processed by SDS/PAGE and autoradiography. Input is ≈10% of the amount subjected to pull-down. Data are representative of three to five independent experiments.
B-Raf Coimmunoprecipitates with IQGAP1.
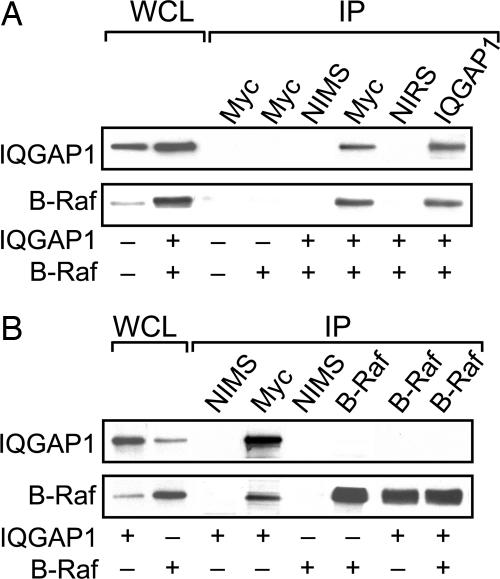
To investigate whether IQGAP1 and B-Raf interact in a normal cell milieu, HEK-293H cells were cotransfected with myc-IQGAP1 and HA-B-Raf. B-Raf specifically coimmunoprecipitates with IQGAP1 (Fig. 2A). Neither protein is detected in samples precipitated with nonimmune serum. In addition, endogenous B-Raf coimmunoprecipitates with transfected myc-IQGAP1 (Fig. 2B), confirming that IQGAP1 and B-Raf interact in cells. However, IQGAP1 does not coimmunoprecipitate with B-Raf (Fig. 2B). The reason for this result is not known, but it may be due to masking of the (B-Raf) antibody recognition epitope by IQGAP1 or conformation of the protein when bound. Alternatively, a relatively small fraction of B-Raf may be bound to IQGAP1 in quiescent cells. This situation would not be unexpected for a scaffold because <5% of total C-Raf coprecipitated with the scaffold KSR (27).
Fig. 2.
B-Raf coimmunoprecipitates with IQGAP1. 293H cells were transiently transfected with pcDNA3 vector, myc-IQGAP1, and/or HA-B-Raf. Equal amounts of protein lysate were resolved by Western blotting (WCL) or immunoprecipitated (IP) with nonimmune mouse (NIMS) or rabbit (NIRS) serum or anti-myc monoclonal (all IQGAP1 constructs are myc-tagged), anti-IQGAP1 polyclonal, or anti-B-Raf monoclonal antibodies. Complexes were isolated and analyzed by Western blotting and probed with anti-myc (upper blots) or anti-B-Raf (lower blots) antibodies. Data are representative of two to eight independent experiments.
IQGAP1 Modulates Activation of B-Raf by EGF.
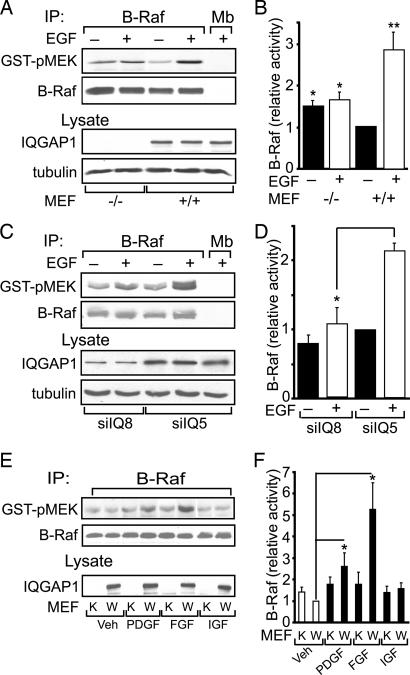
We previously documented that IQGAP1 regulates the activity of MEK and ERK (25, 26). To determine whether the interaction between IQGAP1 and B-Raf has functional sequelae, we measured B-Raf kinase activity. Two complementary strategies were adopted. In the first, we compared EGF-stimulated B-Raf kinase activity in IQGAP1−/− and IQGAP1+/+ mouse embryonic fibroblasts (MEFs). We immortalized MEFs from IQGAP1-null mice (28). No IQGAP1 is detected in IQGAP1−/− MEFs by Western blotting, whereas MEFs from littermate controls express IQGAP1 (Fig. 3A, lysate). Quantification of B-Raf kinase activity reveals that EGF enhances B-Raf activity in IQGAP1+/+ MEFs by 2.8-fold (Fig. 3B). In contrast, EGF is unable to promote B-Raf kinase activity in MEFs lacking IQGAP1. Knockdown of endogenous IQGAP1 in 293H cells yields similar findings. EGF is unable to significantly increase B-Raf kinase activity when IQGAP1 levels are specifically reduced by siRNA (Fig. 3 C and D). Interestingly, basal B-Raf kinase activity is slightly higher in cells lacking IQGAP1 (Fig. 3 A and B and Fig. 4 A and B). The reason for this observation is not known.
Fig. 3.
IQGAP1 is necessary for EGF to stimulate B-Raf activity. (A) Serum-starved IQGAP1−/− (−/−) and IQGAP1+/+ (+/+) MEFs were incubated without (−) or with (+) 100 ng/ml EGF for 10 min. After lysis, equal amounts of protein were immunoprecipitated (IP) with anti-B-Raf antibody. Anti-myoglobin (Mb) antibody was used as control. The B-Raf kinase activity in immunoprecipitates was determined using inactive GST-MEK as substrate. Samples were resolved by SDS/PAGE, probed with anti-phospho-MEK antibody (GST-pMEK), and then reprobed with anti-B-Raf antibody. Equal amounts of unfractionated lysate were probed with anti-IQGAP1 and anti-β-tubulin (as loading control) antibodies. (B) B-Raf kinase activity (as phospho-MEK) was quantified by densitometry and corrected for the amount of B-Raf protein in the corresponding sample. Data are expressed relative to the B-Raf activity in untreated IQGAP1+/+ MEFs and are expressed as the means ± SE (n = 6). ∗, P < 0.01; ∗∗, P < 0.001. (C) 293H cells, transfected with siRNA directed against IQGAP1 (siIQ8) or a nonsilencing siRNA (siIQ5), were incubated with or without EGF. B-Raf kinase activity was assayed as described for A. (D) B-Raf kinase activity (quantified as in B) is expressed relative to that in untreated nonsilenced cells and represents means ± SE (n = 2). ∗, P < 0.05. (E) Serum-starved wild-type (W) and IQGAP1−/− (K) MEFs were incubated for 10 min without (Veh) or with 10 ng/ml PDGF, 50 nmol/liter insulin-like growth factor (IGF), or 40 ng/ml FGF (5 min). B-Raf kinase activity was assayed as described for A. (F) B-Raf kinase activity is expressed relative to that in vehicle-treated wild-type cells and represents means ± SE (n = 4–5). ∗, P < 0.05.
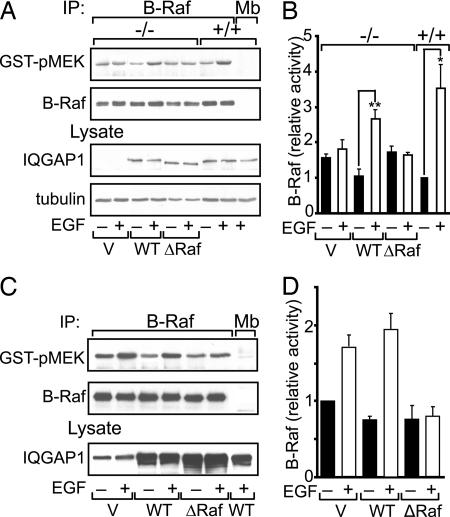
Fig. 4.
Effect of wild-type and mutant IQGAP1 on B-Raf kinase activity. (A) IQGAP1−/− (−/−) MEFs were transiently transfected with 15 μg of empty vector (V), wild type (WT) IQGAP1, or IQGAP1ΔB-Raf (ΔRaf). Untransfected IQGAP1+/+ (+/+) MEFs were processed in parallel. Serum-starved cells were incubated without (−) or with (+) EGF. Equal amounts of protein lysate were immunoprecipitated (IP) with anti-B-Raf or anti-myoglobin (Mb) (control) monoclonal antibodies. The B-Raf kinase activity in immunoprecipitates was determined using inactive GST-MEK as substrate. Western blots were probed with anti-phospho-MEK antibody (GST-pMEK) and then reprobed with anti-B-Raf antibody. Unfractionated lysates (Lysate) were probed for IQGAP1 and β-tubulin. (B) The amount of phospho-MEK was quantified by densitometry and corrected for the amount of B-Raf protein in the corresponding sample. Data are expressed relative to B-Raf activity in untreated IQGAP1+/+ cells and are expressed as means ± SE (n = 3). ∗, P < 0.05; ∗∗, P < 0.005. (C) HEK-293H cells were transiently transfected with vector, wild-type IQGAP1, or IQGAP1ΔB-Raf. Cells were incubated with EGF and processed as described for A. (D) B-Raf kinase activity was quantified as described for B. Data are means ± SE (n = 4).
Several stimuli activate MAPK signaling (1). To determine whether the participation of IQGAP1 in B-Raf activation is confined to EGF, we examined other growth factors. PDGF and basic FGF activate B-Raf in control MEFs, whereas insulin-like growth factor (Fig. 3 E and F), vascular endothelial growth factor, nerve growth factor, and transforming growth factor β (data not shown) have no effect. Neither PDGF, FGF, nor any of the other growth factors tested significantly activate B-Raf in IQGAP1−/− cells (Fig. 3 E and F and data not shown). Thus, IQGAP1 appears to be necessary for several stimuli to activate B-Raf.
To verify the requirement for IQGAP1 in activation of B-Raf by EGF, we examined its ability to “rescue” activation. Transient transfection of wild-type IQGAP1 into IQGAP1−/− MEFs enabled EGF to significantly increase B-Raf kinase activity (Fig. 4 A and B). These data strongly suggest that IQGAP1 couples EGF receptor signaling to B-Raf.
Identification of the B-Raf Binding Domain of IQGAP1.
We used the TNT system to identify the B-Raf binding site on IQGAP1. B-Raf binds to the N- but not the C-terminal half of IQGAP1 [see supporting information (SI) Fig. 6]. Similarly, B-Raf coimmunoprecipitates with full-length IQGAP1 and the N-terminal fragment but not the C terminus (see SI Fig. 6C). The binding site was narrowed to a peptide comprising amino acids 763–964 of IQGAP1 (see SI Fig. 7). Specificity of binding was confirmed with constitutively active GST-Cdc42V12, which binds the C-terminal half of IQGAP1 but not amino acids 763–964 of IQGAP1 (see SI Fig. 7). [Active Cdc42 is known to bind exclusively to IQGAP1-C (12).] Collectively, these data reveal that the B-Raf-binding region on IQGAP1 is located between amino acids 763 and 863. To ascertain whether these residues are necessary for B-Raf binding, we deleted residues 746–860 from IQGAP1. B-Raf is unable to bind to this mutant IQGAP1 construct (see SI Fig. 8), which we term IQGAP1ΔB-Raf.
Effect of IQGAP1ΔB-Raf on EGF-Stimulated B-Raf Activity.
Our data suggest that IQGAP1 is necessary for EGF to activate B-Raf. One would therefore anticipate that an IQGAP1 construct that is unable to bind B-Raf would fail to couple the EGF receptor to B-Raf. Consistent with this hypothesis, EGF does not stimulate B-Raf activity in IQGAP1−/− MEFs transfected with IQGAP1ΔB-Raf (Fig. 4 A and B). We also examined transfection in HEK-293H cells, which contain endogenous IQGAP1. Overexpression of wild-type IQGAP1 slightly reduces basal B-Raf kinase activity, consistent with the increased basal activity seen in IQGAP1-null MEFs, and appears to slightly augment the ability of EGF to promote B-Raf kinase activity (Fig. 4 C and D). In contrast, transfection of IQGAP1ΔB-Raf, which is unable to bind B-Raf, abrogates EGF-induced activation of B-Raf. These data imply that IQGAP1ΔB-Raf functions as a dominant negative construct for activation of B-Raf by EGF.
The Effect of IQGAP1 on B-Raf Kinase Activity In Vitro.
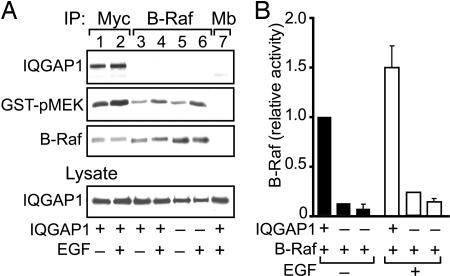
B-Raf is a serine/threonine kinase (6). We wanted to ascertain whether binding to IQGAP1 alters the kinase activity of B-Raf. Ideally, an in vitro kinase assay with pure proteins would be the easiest and most direct way to evaluate this concept. However, B-Raf is maintained in an autoinhibited state (29), precluding this approach. We resolved this problem using the knowledge that IQGAP1 does not coimmunoprecipitate with B-Raf under our assay conditions (Figs. 2B and 5A, lanes 3–6) to our advantage. By separately immunoprecipitating IQGAP1 and B-Raf, we obtained complexes that contain both IQGAP1 and B-Raf (anti-IQGAP1 immunoprecipitates, Fig. 5A, lanes 1 and 2) or B-Raf alone (anti-B-Raf immunoprecipitates, Fig. 5A, lanes 3–6). Therefore, we were able to determine the effect of IQGAP1 on B-Raf kinase activity in vitro. IQGAP1 produces a dramatic effect; B-Raf activity is significantly higher in the presence of IQGAP1 than that in assays lacking IQGAP1 (Fig. 5). IQGAP1 increases basal B-Raf kinase activity by ≈15-fold. Similarly, the in vitro kinase activity of B-Raf obtained from cells stimulated with EGF is markedly augmented by IQGAP1 (Fig. 5). These data strongly suggest that binding to IQGAP1 enhances B-Raf kinase activity in vitro.
Fig. 5.
IQGAP1 modulates the activity of B-Raf kinase in vitro. (A) HEK-293H cells were transfected with HA-B-Raf and either myc-IQGAP1 (+) or empty vector (−). Serum-starved cells were treated without (−) or with (+) 100 ng/ml EGF for 10 min. Lysates were immunoprecipitated (IP) with anti-myc, anti-B-Raf, or anti-myoglobin (Mb, control) antibodies. The B-Raf kinase activity in immunoprecipitates was analyzed using inactive GST-MEK (GST-pMEK). Blots of equal amounts of unfractionated lysate were probed with anti-IQGAP1 antibody (Lysate). (B) B-Raf kinase activity in assays with (+) or without (−) IQGAP1 was quantified by densitometry and corrected for the amount of immunoprecipitated B-Raf protein in the corresponding sample. Data are expressed relative to the B-Raf activity in anti-myc immune complexes without EGF stimulation (lane 1) and represent means ± SE. Assays were performed at least three times, except for lanes 3 and 4, which were only done once.
Discussion
MAPK modules are evolutionarily conserved biological circuits that connect cell-surface receptors to critical regulatory targets within cells (2). B-Raf and C-Raf are differentially regulated and differ in their ability to activate MAPK in different cell systems (30). One of the mechanisms by which specificity of Raf/MEK/ERK signaling is achieved is via scaffold proteins (23, 24). Although a number of scaffolds for C-Raf, including KSR1, β-arrestin, and MAGUIN, have been identified (23, 24), to the best of our knowledge, no scaffolds for B-Raf have previously been described (4). The fundamental role of IQGAP1 in diverse signaling pathways (9), coupled with our prior observations that IQGAP1 functions as a scaffold in the MEK/ERK pathway (25, 26), led us to evaluate a possible interaction between IQGAP1 and B-Raf. We document here direct binding of IQGAP1 to B-Raf. An interaction was observed both in vitro and by coimmunoprecipitation from cell lysates. In conjunction with the prior observations that IQGAP1 binds directly to MEK1/2 and ERK1/2 (25, 26), these data further support the hypothesis that IQGAP1 is a scaffold in the Ras/Raf/MEK/ERK pathway.
The association between B-Raf and IQGAP1 has significant functional sequelae. Analogous to its effects on other targets (9, 31), IQGAP1 alters B-Raf function. Binding to IQGAP1 substantially increases B-Raf kinase activity in vitro. The molecular mechanism underlying this observation is not known. The N-terminal region of B-Raf acts as an autoinhibitory domain (29). Therefore, it is possible that binding to IQGAP1 alters the conformation of B-Raf, releasing it from the autoinhibited state and facilitating kinase activity. Another conformational change in B-Raf that may be induced by IQGAP1 is disruption of the intramolecular interaction between the glycine-rich loop and the activation segment (32). Solving the structure of B-Raf bound to IQGAP1 is necessary to test this hypothesis. A second possibility is that IQGAP1 promotes the coupling of B-Raf to its substrate MEK, thereby enhancing B-Raf kinase activity. The latter hypothesis is consistent with the model of IQGAP1 serving as a scaffold to link B-Raf to MEK. These are not mutually exclusive possibilities. Regardless of the mechanism, our data may provide an explanation for the demonstration by Richard Hynes et al. (33) that IQGAP1 is one of only 32 genes that are overexpressed in metastatic melanoma. The increased IQGAP1 concentrations in the melanoma cells could augment B-Raf kinase activity, contributing to the aggressive phenotype.
Our data also suggest another role for IQGAP1 in B-Raf function. EGF is unable to activate B-Raf in MEF cells lacking IQGAP1. Similar results are obtained when endogenous IQGAP1 is specifically reduced by siRNA. Moreover, reconstitution of IQGAP1 in IQGAP1−/− MEFs enables EGF to increase B-Raf activity, whereas IQGAP1ΔB-Raf has no effect. These observations imply that IQGAP1 also participates upstream of B-Raf in the Ras/Raf/MEK pathway linking B-Raf to EGF receptor signaling. The mechanism has not been identified.
Previous studies reported that H-Ras does not associate with IQGAP1 (12, 34), implying that IQGAP1 functions further upstream. Consistent with this hypothesis, EGF has been shown to induce the recruitment of IQGAP1 to a Grb2-EGF receptor complex (35). It is noteworthy that although EGF is unable to activate B-Raf in cells lacking IQGAP1, basal B-Raf activity is slightly higher in these cells than in the control cells. The reason for this is not known, but conceivably the level and/or activity of (an)other protein(s), e.g., kinase or phosphatase, that contributes to basal B-Raf activity is altered in the IQGAP1−/− cells. In this context, IQGAP1 has been shown to bind to several protein kinases (including protein kinase A and protein kinase C) and protein phosphatases (such as protein phosphatase 2A and protein tyrosine phosphatase μ) (9).
Interestingly, IQGAP1ΔB-Raf abrogates activation of B-Raf by EGF in 293H cells. This last observation is consistent with a role for IQGAP1 as a scaffold that links the EGF receptor to B-Raf. One would anticipate that a mutant IQGAP1 construct that is unable to bind B-Raf, but which binds normally to upstream components of the EGF/Ras/B-Raf cascade, will sequester upstream signaling molecules away from endogenous IQGAP1. By this process, the IQGAP1 mutant should uncouple signaling from the EGF receptor to B-Raf and reduce the ability of EGF to stimulate B-Raf activation. This is exactly what we observed.
In the current study, we document a previously unrecognized interaction between IQGAP1 and B-Raf, which has substantial impact on B-Raf activation. These findings further support the concept that IQGAP1 is a scaffold in the Ras/B-Raf/MEK/ERK pathway. Finally, the identification of an additional regulatory component of B-Raf adds another level of complexity to the intricate mechanisms that modulate B-Raf function.
Materials and Methods
Plasmid Construction and Preparation of Fusion Proteins.
Myc-tagged constructs of human IQGAP1, IQGAP1-N, IQGAP1-C, IQGAP1-N1, IQGAP1-N2, IQGAP1-(763–964) (the numbers indicate the amino acids residues of IQGAP1 that are included), and IQGAP1ΔB-Raf (amino acids 746–860 deleted) and the siRNAs were described (16, 25, 36, 37). HA-B-Raf (38) and GST-V12-Cdc42 were provided by Kun-Lian Guan (University of Michigan Medical School, Ann Arbor, MI) and Anne Ridley (University College London, London, U.K.), respectively.
To construct GST-B-Raf, PCR was performed on full-length pEBG HA-B-Raf using primers 5′-GCGCCCGGGTGCGGCGCTGAGCGGTGGC-3′ (forward, XmaI site included) and 5′-CGCGCGGCCGCTCAGTGGACAGGAAACGC-3′ (reverse, NotI site and a stop codon were included). The product was cut with XmaI and NotI and subcloned into the XmaI-NotI site of pGEX4T-1. GST fusion proteins were expressed in Escherichia coli and isolated with glutathione-Sepharose essentially as described (16). The GST tag was cleaved from GST-IQGAP1 using tobacco etch virus protease as described (36).
Isolation of MEF Cells.
MEFs were isolated from embryonic day 14 embryos of IQGAP1−/− mice (28) and normal littermate controls and grown in primary culture (39). IQGAP1 knockout mice were provided by Wadie Bahou (State University of New York, Stony Brook, NY) and Andre Bernards (Harvard Medical School). For immortalization, primary MEFs were transfected with simian virus 40 large T antigen (40) [a gift from Judith Tevethia (Pennsylvania State University, Hershey, PA)] using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and cultured until single colonies formed.
Cell Culture and Transfection.
HEK-293H and MEF cells were maintained as described (41). Cells were transfected using Lipofectamine 2000 essentially as described (37, 41). Where necessary, pcDNA3 was added to ensure that total DNA transfected was constant.
TNT Product Production.
[35S]methionine-labeled TNT products were produced with the TNT Quick Coupled Transcription/Translation system (Promega, Madison, WI) as described (36, 42). Briefly, 2 μg of IQGAP1 plasmid was incubated with 40 μl of TNT Quick Master Mix (Promega) and 2 μCi (1 Ci = 37 GBq) of [35S]methionine (New England Nuclear, Boston, MA) at 30°C for 90 min. Products were identified by SDS/PAGE and autoradiography.
In Vitro Binding Assays.
Equal amounts of [35S]methionine-labeled IQGAP1 constructs were incubated for 3 h at 4°C with 5 μg GST-B-Raf in 1 ml of buffer A (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mM EGTA/1% Triton X-100) containing 1 mM PMSF and protease inhibitor mixture. Complexes were isolated with glutathione-Sepharose, resolved by SDS/PAGE, and processed by autoradiography.
Immunoprecipitation.
Subconfluent 293H cells were transfected for 24 h with myc-IQGAP1 and/or HA-B-Raf using Lipofectamine 2000 as described (36). pcDNA3 vector was added to ensure that the total amount of plasmid was the same in each sample. EGF treatment (100 ng/ml for 10 min) and immunoprecipitation were performed as described (26). Cells were lysed in 20 mM Tris (pH 8.0), 137 mM NaCl, 10% glycerol, and 0.2% Triton X-100. Anti-myc (9E10.2) and anti-B-Raf (Santa Cruz, Santa Cruz, CA) monoclonal antibodies or anti-IQGAP1 polyclonal antibodies (16) were incubated with protein G- or protein A-Sepharose beads (Amersham, Piscataway, NJ) for 2 h at 4°C, washed, and incubated for 3 h at 4°C with equal amounts of precleared protein lysate. Nonimmune mouse or rabbit serum (Sigma, St. Louis, MO) or anti-myoglobin monoclonal antibody (a gift of Jack Ladenson, Washington University, St. Louis, MO) were used as controls. Samples were resolved by SDS/PAGE and Western blotting and probed with anti-IQGAP1 monoclonal antibody (26) and anti-B-Raf antibodies. Antigen–antibody complexes were visualized with the appropriate (rabbit or mouse) horseradish peroxidase-conjugated secondary antibodies (GE Healthcare, Piscataway, NJ) and visualized by enhanced chemiluminescence (Millipore, Bedford, MA).
In Vitro B-Raf Kinase Assays.
B-Raf kinase activity was quantified with an in vitro coupled kinase assay (Upstate Cell Signaling Solutions, Charlottesville, VA). After serum-starving for 16 h, cells were incubated with or without 100 ng/ml EGF for 10 min at 22°C and lysed in RIPA buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl/1% Nonidet P-40/1 mM EDTA/0.25% deoxycholic acid) containing protease and phosphatase inhibitor cocktails. Lysates were immunoprecipitated with anti-B-Raf or anti-myc antibodies, washed, and incubated with 500 μM ATP and 1 μg inactive GST-MEK in a kinase assay buffer at 30°C for 30 min. GST-MEK phosphorylation was quantified by blotting with anti-phospho-MEK1/2 antibody and corrected for the amount of B-Raf immunoprecipitated.
Miscellaneous.
Determination of protein concentrations, densitometry, and statistical analysis with Student's t test was performed as described (26, 41).
Supplementary Material
Acknowledgments
We thank Monideepa Roy for performing initial in vitro binding experiments and Wadie Bahou, Andre Bernards, Judith Tevethia, Kun-Lian Guan, Anne Ridley, Michael White (University of Texas Southwestern Medical Center, Dallas, TX), and Jack Ladenson for reagents. We appreciate Rob Krikorian's expert help in the preparation of the manuscript and are grateful to members of our laboratory for insightful comments and suggestions. This study was supported in part by grants from the National Institutes of Health (to D.B.S.).
Abbreviations
- MEK
MAPK kinase
- MEF
mouse embryonic fibroblast
- TNT
transcription and translation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611308104/DC1.
References
- 1.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 2.Roux PP, Blenis J. Microbiol Mol Biol Rev. 2004;68:320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morrison DK, Cutler RE. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 4.Wellbrock C, Karasarides M, Marais R. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 5.Rapp UR, Goldsborough MD, Mark GE, Bonner TI, Groffen J, Reynolds FH, Jr, Stephenson JR. Proc Natl Acad Sci USA. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong H, Vikis HG, Guan KL. Cell Signal. 2003;15:463–469. doi: 10.1016/s0898-6568(02)00139-0. [DOI] [PubMed] [Google Scholar]
- 7.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 8.Briggs MW, Sacks DB. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown MD, Sacks DB. Trends Cell Biol. 2006;16:242–249. doi: 10.1016/j.tcb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Mateer SC, Wang N, Bloom GS. Cell Motil Cytoskeleton. 2003;55:147–155. doi: 10.1002/cm.10118. [DOI] [PubMed] [Google Scholar]
- 11.Noritake J, Watanabe T, Sato K, Wang S, Kaibuchi K. J Cell Sci. 2005;118:2085–2092. doi: 10.1242/jcs.02379. [DOI] [PubMed] [Google Scholar]
- 12.Hart MJ, Callow MG, Souza B, Polakis P. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- 13.Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. J Biol Chem. 1997;272:29579–29583. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- 14.Joyal JL, Annan RS, Ho YD, Huddleston ME, Carr SA, Hart MJ, Sacks DB. J Biol Chem. 1997;272:15419–15425. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- 15.Erickson JW, Cerione RA, Hart MJ. J Biol Chem. 1997;272:24443–24447. doi: 10.1074/jbc.272.39.24443. [DOI] [PubMed] [Google Scholar]
- 16.Ho YD, Joyal JL, Li Z, Sacks DB. J Biol Chem. 1999;274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 17.Mateer SC, McDaniel AE, Nicolas V, Habermacher GM, Lin MJ, Cromer DA, King ME, Bloom GS. J Biol Chem. 2002;277:12324–12333. doi: 10.1074/jbc.M109535200. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Kim SH, Higgins JM, Brenner MB, Sacks DB. J Biol Chem. 1999;274:37885–37892. doi: 10.1074/jbc.274.53.37885. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al. Science. 1998;281:832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- 20.Briggs MW, Li Z, Sacks DB. J Biol Chem. 2002;277:7453–7465. doi: 10.1074/jbc.M104315200. [DOI] [PubMed] [Google Scholar]
- 21.Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 23.Morrison DK, Davis RJ. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 24.Kolch W. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 25.Roy M, Li Z, Sacks DB. J Biol Chem. 2004;279:17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- 26.Roy M, Li Z, Sacks DB. Mol Cell Biol. 2005;25:9740–9752. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kortum RL, Lewis RE. Mol Cell Biol. 2004;24:4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Wang Q, Chakladar A, Bronson RT, Bernards A. Mol Cell Biol. 2000;20:697–701. doi: 10.1128/mcb.20.2.697-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran NH, Wu X, Frost JA. J Biol Chem. 2005;280:16244–16253. doi: 10.1074/jbc.M501185200. [DOI] [PubMed] [Google Scholar]
- 30.Papin C, Denouel-Galy A, Laugier D, Calothy G, Eychene A. J Biol Chem. 1998;273:24939–24947. doi: 10.1074/jbc.273.38.24939. [DOI] [PubMed] [Google Scholar]
- 31.Briggs MW, Sacks DB. FEBS Lett. 2003;542:7–11. doi: 10.1016/s0014-5793(03)00333-8. [DOI] [PubMed] [Google Scholar]
- 32.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 33.Clark EA, Golub TR, Lander ES, Hynes RO. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- 34.McCallum SJ, Wu WJ, Cerione RA. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- 35.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 36.Ren JG, Li Z, Crimmins DL, Sacks DB. J Biol Chem. 2005;280:34548–34557. doi: 10.1074/jbc.M507321200. [DOI] [PubMed] [Google Scholar]
- 37.Mataraza JM, Briggs MW, Li Z, Entwistle A, Ridley AJ, Sacks DB. J Biol Chem. 2003;278:41237–41245. doi: 10.1074/jbc.M304838200. [DOI] [PubMed] [Google Scholar]
- 38.Zhang BH, Guan KL. EMBO J. 2000;19:5429–5439. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodnar MS, Meneses JJ, Rodriguez RT, Firpo MT. Stem Cells Dev. 2004;13:243–253. doi: 10.1089/154732804323099172. [DOI] [PubMed] [Google Scholar]
- 40.Karjalainen HE, Tevethia MJ, Tevethia SS. J Virol. 1985;56:373–377. doi: 10.1128/jvi.56.2.373-377.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swart-Mataraza JM, Li Z, Sacks DB. J Biol Chem. 2002;277:24753–24763. doi: 10.1074/jbc.M111165200. [DOI] [PubMed] [Google Scholar]
- 42.Li L, Li Z, Sacks DB. J Biol Chem. 2005;280:13097–13104. doi: 10.1074/jbc.M410642200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.