Abstract
The cancer stem cell hypothesis posits that tumors are derived from a single cancer-initiating cell with stem cell properties. The task of identifying and characterizing a single cancer-initiating cell with stem cell properties has proven technically difficult because of the scarcity of the cancer stem cells in the tissue of origin and the lack of specific markers for cancer stem cells. Here we show that a single LA7 cell derived from rat mammary adenocarcinoma has the following properties: the differentiation potential to generate all of the cell lineages of the mammary gland; the ability to generate branched duct-like structures that recapitulate morphologically and functionally the ductal–alveolar-like architecture of the mammary tree; and the capacity to initiate heterogeneous tumors in nonobese diabetic-SCID mice. In addition, we show that cultured cells derived from tumors generated by a single LA7 cell-injection have properties similar to LA7 cells, can generate all of the cell lineages of the mammary gland, and recapitulate the ductal–alveolar-like architecture of the mammary tree. The properties of self-renewal, extensive capacity for proliferation, multilineage differentiation potential, and single-cell tumor-initiation potential suggest that LA7 cells are cancer stem cells and can be used as a model system to study the dynamics of tumor formation at the single-cell level.
Keywords: p21/WAF1, mammary gland differentiation, single cell injection
Increasingly, experimental evidence and clinical data suggest that somatic stem cells may be the targets of transformation during carcinogenesis and that virtually all cancers are clonal and represent the progeny of a single cell (1). This suggests that tumorigenic cancer cells may undergo the processes of self-renewal and differentiation as normal stem cells.
We show here that mammary gland LA7 cells isolated by R.D. (2) from a mammary adenocarcinoma induced in rat using DMBA (3) have the stem cell properties of self-renewal, have the capacity to differentiate into all of the cell lineages of the mammary gland, and form heterogeneous tumors in nonobese diabetic (NOD)-SCID mice at a single-cell level. Demonstrating that a single LA7 cell has the capacity to initiate cancer gives strong support to the hypothesis that recurrence of cancer after remedial therapy can occur if even a single malignant cell survives. Characterization of cells with the property of single-cell tumor-formation potential should contribute to the advancement of appropriate therapies that can effectively target specific individual cancer stem cells that have the capacity to initiate tumors at the single-cell level. Previously, LA7 cells have been used to study mammary gland differentiation (4–10). Exposure of LA7 cells to lactogenic hormones, lipids, or differentiating agents results in the formation of hemispherical polarized dome-shaped structures representing cellular changes that occur in vivo in the mammary gland at pregnancy when alveoli are formed (4, 9).
We show here that a single LA7 cell can generate mammospheres for >63 passages; we also show that outgrowths from a single mammosphere (generated by a single LA7 cell) contain cells of the ductal epithelial, myoepithelial, and alveolar lineages, as indicated by the coexpression of specific-lineage markers. We show that a single LA7 cell, grown in three-dimensional cultures, generates complex branched duct-like structures and cysts, reminiscent of the entire tubuloalveolar architecture of the mammary tree. Moreover, we demonstrate that a single LA7 cell can generate heterogeneous tumors in NOD-SCID mice and that the tumor heterogeneity is due to the clonal expansion of a single LA7 cell into all cell lineage types of the tissue of origin. We show, in addition, that a subset of cells derived from tumors generated from a single LA7 cell retains the multipotential differentiation capacity of the parental LA7 cells and recapitulates the ductal–alveolar-like structure of the mammary tree in vitro. These results suggest that LA7 cells are cancer stem cells and are a model system for the study of the dynamics of solid tumor formation at the single-cell level.
Results
A Single LA7 Cell Has Trilineage Differentiation Potential.
To assess the multilineage differentiation potential of LA7 cells, recently subcloned cells (LA73F12ms) were grown as mammosphere as described in ref. 11. The first generation of LA7-cell-derived mammospheres, obtained at 5–7 days, was mechanically and enzymatically dissociated into a single-cell suspension and used to generate serially passaged mammospheres (11). Mammospheres were continuously generated at 7–10 day intervals for at least 63 passages; Fig. 1A shows mammospheres at passages 8 (p8) and 47 (p47). To test the frequency of the mammosphere-forming unit, single mammospheres generated from single LA7 cells were disrupted, and cells were plated again in 96-well plates at a density of one cell per well to derive new mammospheres. Single-cell-derived mammospheres were serially regenerated with an 80–85% frequency of mammosphere-forming units for each passage tested.
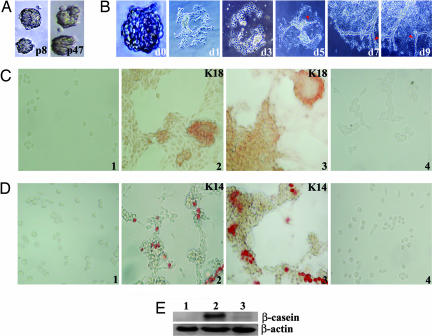
Fig. 1.
Analysis of mammospheres generated by LA7 cells. (A) Self-renewing mammospheres generated by LA7 cells at passages 8 (p8) and 47 (p47). (Magnification: p8, ×20; p47, ×32.) (B) Mammosphere from a single LA7 cell at passage 13 (d0), plated onto collagen over a 10-day time period, generates morphologically differentiated cells (d1–d9) and forms tubular-like structures at days 5–9 (d5–d9, red arrowheads). (Magnification: d0, ×40; d1 and d3, ×20; d5, d7, and d9, ×10.) (C) Mammosphere-generated outgrowths on collagen express K18. (C1–C3) Images show K18 staining on cells derived from the disaggregated mammosphere before outgrowth generation (C1), a mammosphere outgrowth generated from a single LA7 cell at passage 13 (C2), and a mammosphere outgrowth generated from a single LA7 cell at passage 36 (C3). (C4) A negative control stained using the secondary antibody only. (Magnification: ×40.) Specifically, a single outgrowth from a single-cell-generated mammosphere was sectioned into portions, and individual portions were used for marker expression analysis for C2–C4 and D2–D4 and for E (lane 2). (D) Mammosphere-generated outgrowths on collagen express K14. Shown is K14 staining on cells derived from the disaggregated mammosphere before outgrowth generation (D1), a section of the mammosphere outgrowth generated from a single LA7 cell at passage 13 (D2), and a section of the mammosphere outgrowth generated from a single LA7 cell at passage 36 (D3). (D4) A negative control stained with the secondary antibody only. (Magnification: D2, ×32; D1, D3, and D4, ×40.) (E) Mammosphere-generated outgrowths express β-casein. After incubation using lactogenic hormones, the remaining portion of the 10-day outgrowth at early passage, used in Fig. 2 C and D, was used to test for the expression of β-casein with Western blot analysis. Lanes: 1, Cells derived from mammospheres; 2, outgrowth from a mammosphere at passage 13 after hormone induction; 3, outgrowth from a mammosphere at passage 13 without induction.
For differentiation studies, single-cell-generated mammospheres were plated on collagen-coated wells. Fig. 1B shows that an outgrowth from asingle-cell-generated mammosphere at passage 13 produces morphologically differentiated cells with tubular-shape structures (Fig. 1B, red arrowheads); similar morphology was observed from mammosphere outgrowth generated at higher passages (data not shown). Multilineage differentiation potential of LA7 cells was assessed by plating single-cell-generated mammospheres (at low and high passages) onto collagen over a 10-day period and by testing the outgrowths for expression of the mammary gland cell-lineage-specific markers K18, K14, and β-casein, which are markers for the ductal epithelial, myoepithelial, and alveolar lineages, respectively (Fig. 1 C–E) (11, 12). Disaggregated mammospheres generated from single LA7 cells, when plated onto collagen before outgrowth formation, show no staining for K18 and K14 (Fig. 1 C1 and D1). For outgrowth differentiation analysis, single-cell-generated mammosphere outgrowths at early and late passages were divided into portions: one was stained with K18 (Fig. 1C2, early passage, and C3, late passage); a second portion was stained with K14 (Fig. 1D2, early passage, and D3, late passage); and a third portion of cells, collected from the same single-cell-generated mammosphere outgrowths after incubation with lactogenic hormones, was assayed for the expression of β-casein (Fig. 1E, lane 2, shows β-casein expression from mammosphere outgrowth at early passage; late passage not shown).
These results show that a single mammosphere generated by a single LA7 cell has the potential for self-renewal and that a mammosphere outgrowth can generate rudimentary tubular-like structures on two-dimensional cultures and can differentiate into luminal, myoepithelial, and alveolar cells.
A Single LA7 Cell Has the Potential to Generate in Vitro Complex Functional Structures Reminiscent of the Mammary Tree.
Three-dimensional cultivation systems provide the physiological signals necessary for mammary morphogenesis in vitro and enable mammary cells to recapitulate the spatial orientation and architecture of the mammary tree in vivo (13). The differentiation potential of LA7 cells to form the mammary architecture was assessed by determining the ability of a single LA7 cell to develop functional ductal–alveolar-like structures using rat tail collagen or Matrigel (Becton Dickinson, Franklin Lakes, NJ). Complex tubular, branched structures reminiscent of the mammary ductal tree were generated (Fig. 2 A and B). Fig. 2A shows the progression of tubuli formation from a single cell; Fig. 2 A2 and A4 (red arrowheads) show emerging tubuli and terminal end buds; and Fig. 2A3 (red arrowhead) shows intermediate and advanced tubuli containing multiple branches. Nuclear staining and confocal microscopy show elongated hollow tubular structures (Fig. 2 B2–B4). Fig. 2C shows LA7 cells that, when grown on Matrigel, retain the ability to generate cyst-like structures that recapitulate the formation of mammary gland alveoli at pregnancy that secrete β-casein (data not shown) (11). Cells obtained from the tubular structures generated from a single LA7 cell were collected from collagen at day 14 and tested for the expression of K18 and K14 proteins. Fig. 2D shows that the tubular structures expressed both luminal and myoepithelial markers (lane 2).
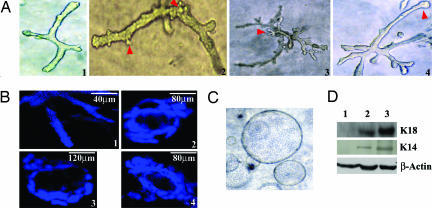
Fig. 2.
LA7 cells are able to clonally reproduce in vitro complex functional tubular structures expressing K18 and K14. (A) Tubuli formation from a single LA7 cell in collagen, including emerging ductal trees (day 5–8) (A1 and A2) and intermediate ductal trees (day 10–14) (A3 and A4) containing multiple branches of tubular elongated structures. Red arrowheads show brunching and putative terminal end buds. (Magnification: A3, ×20; A1, A2, and A4, ×32.) (B) Confocal microscopy of nuclear-stained cells showing hollow tubular structures. (B1) An elongated tubular structure. (B2–B4) Cross-sections of hollow tubuli. (C) Cyst-like structures generated by LA7 cells at day 10 in Matrigel. (Magnification: ×20.) (D) Cells collected from tubular structures generated from a single LA7 cell at day 14 show expression of K18 and K14. Lanes: 1, LA7 undifferentiated cells; 2, cells collected from tubular structures generated at day 10; 3, DMSO-induced LA7 cells.
These results indicate that a single LA7 cell has the potential to morphologically differentiate along all three mammary gland cell lineages and structurally and functionally recapitulate the architecture of the mammary gland.
A Single LA7 Cell Has the Potential to Generate Heterogeneous Tumors.
To assess the ability of LA7 cells to form tumors, 100,000, 1,000, and 100 LA7 cells were injected into fat pads of NOD-SCID mice. Tumor formation was detected in six of six (100%) fat pads for mice with 100,000 cells injected, in five of six (83%) for those with 1,000 cells, and in four of six (67%) for those with 100 cells injected (Fig. 3A). It was observed that tumors generated from injection of 100,000 LA7 cells were first detected at ≈ day 7 and those generated from injections of 100 LA7 cells were detected at ≈ day 24. In addition, it was observed that at day 52, for mice with 100,000 and 100 cells injected (1,000-fold difference in cell number), the difference in tumor mass was 5.7 times (104 mm3/18 mm3). These results suggested that transplantation of even one LA7 cell might result in tumor formation. Mice were therefore injected with one LA7 cell. Tumor formation occurred in six of nine fat pads (67%), as shown in Fig. 3A.
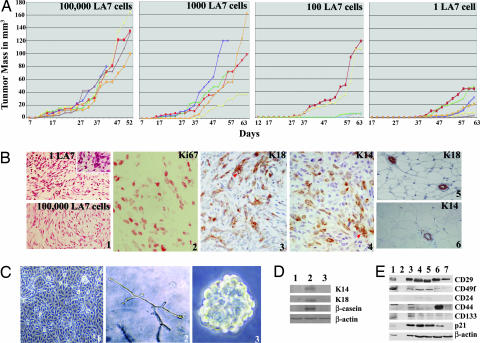
Fig. 3.
LA7 cell tumor initiation potential and tumor cell analysis. (A) LA7 cell tumor formation potential. Shown are the results when amounts of 100,000, 1,000, 100, or 1 LA7 cell generate tumors after injection into the fat pads of NOD-SCID mice. The vertical axis portrays tumor mass in cubed millimeters, and the horizontal axis portrays time at tumor mass measurement. (B) Histology and marker expression of the tumor generated from one LA7 cell. (B1) Histological sections of tumors formed from injections of one (Upper) and 100,000 (Lower) LA7 cells. (B2) Ki67 staining shows cell proliferation. (B3) K18 staining shows the presence of differentiated luminal cells. (B4) K14 staining shows the presence of differentiated myoepithelial cells. (B5) K18 staining of mouse normal mammary gland tubuli. (B6) K14 staining of mouse normal mammary gland tubuli. Red arrowheads in B3 and B4 show cross-sections of rudimentary and partially formed tubuli. (C) Cells established from tumors generated by injection of a single LA7 cell have properties similar to the parental LA7 cells. (C1) Established cultures obtained from a tumor generated by a single LA7 cell are morphologically similar to LA7 cells. (C2) When plated at clonogenic density, 3–5% of the cells derived from cultures shown in C1 are able to generate in vitro complex branched tubular structures similar to those generated by the parental LA7 cells. (C3) Cells derived from cultures shown in C1 were used to generated mammospheres for at least eight passages. (D) Cells established from tumors generated by a single LA7 cell have the same trilineage differentiation potential as the parental LA7 cells. K14, K18, and β-casein expression analysis on established cultures isolated from a tumor generated from a single LA7 cell. Lanes: 1, established cells isolated from a tumor generated by a single LA7 cell; 2, the same cells after induction with DMSO; 3, uninduced LA7 cells. (E) Stem and cancer stem cell marker-expression. Expression analysis for CD29, CD49f, CD24, CD44, CD133, and p21/WAF1: mouse embryonal carcinoma cell line F9, used as a control (lane 1), an empty lane (lane 2), LA7 cells (lane 3), single-LA7-cell-derived mammospheres at early passage (lane 4), single-LA7-cell-derived mammospheres at late passage (lane 5), established cells as shown in Fig. 3C1 (lane 6), and DMSO-induced LA7 cells (lane 7).
Histological sections of the tumors generated in mice that had one, 100, 1,000, and 100,000 LA7 cells injected were very similar and showed large areas of cellular necrosis and highly proliferating cells with several mitotic structures. For all tumor masses, two cell types were distinguishable: rounded epithelium-like cells and elongated mesenchymal-like cells. Fig. 3B1 shows the tumor histology from mice with one cell (upper image) and 100,000 cells injected (lower image). The tumor mass generated from one LA7 cell shows cells expressing Ki67 (Fig. 3B2), indicating cell proliferation (14) and K18- and K14-expressing cells (Fig. 3 B3 and B4, respectively), suggesting the presence of differentiated luminal epithelial and myoepithelial cells (11, 12). The cells staining for K18 and K14 are not randomly dispersed but show a partial higher order organization and identify structures that are suggestive of the presence of interspersed rudimentary tubuli (Fig. 3 B3 and B4, red arrowheads). Staining for K18 and K14 proteins of normal mammary gland tubular structures is indicated in panels 5 and 6, respectively, of Fig. 3B. The tumors from mice with 100, 1,000, and 100,000 LA7 cells injected similarly show expression of K18, K14, and the partial ultrastructural organization indicative of the presence of rudimentary tubuli (data not shown).
These results show that tumors generated from a single LA7 cell contain highly proliferating cells, expressing Ki67, and terminally differentiated cells, expressing luminal (K18) and myoepithelial (K14) markers, and show partial higher order structural organization, reminiscent of the architecture of the tissue of origin.
Differentiation of LA7 Cells Results in a Cellular Hierarchy in Which Most Cells Do Not Contribute to Tumor Initiation and Growth.
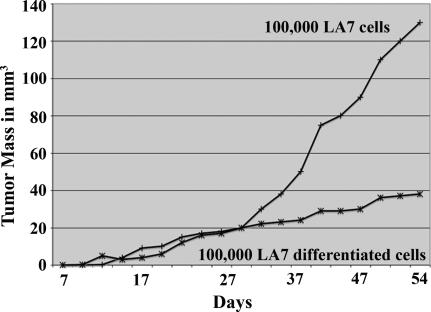
To test whether differentiated LA7 cells also form tumors, we cultivated LA7 cells by using DMSO for 60 h (2, 5). After verifying the expression of the differentiation markers K18 and K14 (Fig. 2D, lane 3), we injected 100,000 DMSO-induced cells into NOD-SCID mice and compared the resulting average tumor-mass growth curve with that obtained from the injection of 100,000 undifferentiated LA7 cells. The growth curve for 100,000 undifferentiated LA7 cells (Fig. 4) is similar to that shown in Fig. 3A for 100,000 LA7 cells. In contrast, the growth curve obtained from 100,000 DMSO-induced LA7 cells is more similar to the curves obtained from the injection of one and 100 LA7 cells than to the curve obtained from the injection of 100,000 LA7 cells (Fig. 3A), suggesting that DMSO-induced differentiation results in a cell population hierarchy in which most cells have lost tumor initiation potential.
Fig. 4.
Differentiation of LA7 cells using DMSO results in a cell population hierarchy in which most cells are not tumor initiating. Undifferentiated LA7 cells (100,000) were injected into NOD-SCID mice in parallel with 100,000 LA7 cells differentiated using DMSO for 60 h. Both conditions were performed in triplicate, and the average tumor mass growth curves are shown.
Cells Derived from Tumors Generated from Injection of a Single LA7 Cell Have Stem Cell Properties Similar to LA7 Cells.
Cells derived from the tumors generated from a single LA7 cell were cultured and established under selective conditions so that mouse cells, rat tumor differentiating, and rat tumor differentiated cells were removed (Fig. 3C1) (15). Primers that amplify different size fragments for mouse and rat genomic DNA were used for PCR to prove that mouse cells were not present in established tumor cell cultures (data not shown). The established cell cultures were found to contain cells that were morphologically similar to LA7 cells (Fig. 3C1) and that, like LA7 cells, can be induced to differentiate into all three lineage types of the mammary gland (Fig. 3D, lane 2). Cells derived from the established cell cultures shown in Fig. 3C1, when plated at clonogenic density, show that ≈3–5% of the total number of cells are able to generate complex branched tubular structures (Fig. 3C2), similar to the structures obtained from LA7 cells (Fig. 2). Mammospheres were also generated from the established cell culture shown in Fig. 3C1. Early and late passage cells shown in Fig. 3C1, reformed mammospheres for up to eight generations at the time of publication (Fig. 3C3); data not shown for early passage. The ability of the cultured cells derived from the tumor mass to produce serially passaged mammospheres and tubuli is indicative of the stem cell potential of the cultured cells.
Recently, use of specific cell-surface markers has allowed for cell enrichment of normal stem cells (12, 16) and tumor-initiating cells from tissues (17). Normal mouse mammary stem/progenitor cell enrichment was based on selecting cell fractions that were CD29+/CD49f+ (12, 16). Human mammary tumor-initiating cells enrichment was based on selecting cell fractions that were CD24−/low/CD44+ (17). We tested undifferentiated LA7 cells, mammospheres generated from LA7 cells at early and late passage, cells from the tumor established culture (shown in Fig. 3C1), and DMSO-differentiated LA7 cells for these markers. The markers CD29 and CD49f were found expressed in LA7 cells, in early and late passage mammospheres, in cells established from tumors, and in DMSO-differentiated cells (Fig. 3E). The presence of marker CD24 was low or not detected in LA7 cells, early and late mammospheres, and the established tumor cells. The marker CD44 appears to be predominantly expressed in the established tumor cell line (Fig. 3E, lane 6).
In addition, CD133 was recently implicated in the maintenance of the stem/progenitor cell state (18) and associated with human colon cancer-initiating cells (19, 20). We therefore tested the expression of CD133 by using the same culture assays as above. Like the expression of CD29 and CD49f, CD133 is found expressed in LA7 cells, early and late passage mammospheres, and cells established from LA7 tumors and is moderately down-regulated in DMSO-induced cells. Another factor implicated in tumor development and in the maintenance of stem cell potential is the protein p21/WAF1 (21). The protein p21/WAF1 is found expressed in LA7 cells, early and late passage mammospheres, and cells established from tumors and is completely down-regulated in DMSO-differentiated LA7 cells (Fig. 3E, lane 7). It was proposed that loss of p21/WAF1 is accompanied by accelerated depletion of the stem cell compartment and that p21 activity might contribute to cancer development by preventing the depletion of tumor-initiating cells (21).
Conclusion
Xenogenic cancer models generated by injecting cell lines or implanting pieces of primary tumor into immunodeficient mice have commonly been used for studies in human tumor biology. In the emerging field of cancer stem cell biology, quantitative assays for single-cell isolation and analysis are essential to fully understand the tumorigenic properties and potential of cancer stem cells in solid tumors. Typically, cell lines have been of limited benefit in quantitative assays because cell lines usually do not recapitulate all aspects of the primary tumor.
The comprehensive characterization of the tumorigenic properties of cells with single-cell tumor formation potential depends on identifying and isolating single cancer-initiating cells. The task of identifying and characterizing a single cancer-initiating cell with stem cell properties has proven to be technically difficult because of the lack of universal and specific markers for cancer stem cells.
Here we demonstrate that a single undifferentitated LA7 cell has the potential to produce a tumor. Cultures of LA7 cells differentiated by using DMSO, conversely, retain only a subpopulation of tumor-initiating cells. The resulting average tumor mass growth curve obtained with the injection of 100,000 LA7 cells differentiated by using DMSO (Fig. 4) was most similar to that obtained with the injection of 1–100 undifferentiated LA7 cells (Fig. 3A), suggesting that ≈1–100 cells of 100,000 total in vitro differentiated LA7 cells (Fig. 4) are tumor-initiating cells and that the differentiation of multipotent LA7 cells results in a cellular hierarchy in which most cells do not contribute to tumor initiation and growth. Although this evidence is consistent with the conclusion that the differentiated cells contain a reduced number of tumor-initiating cells compared with undifferentiated cells, an alternative explanation is that the differentiated cells proliferate more slowly but still retain tumor-initiating capacity. We found that the first conclusion is more likely true because the DMSO-differentiated cells do not retain the capacity to form tumor spheres, and the sphere-forming ability is associated with stem cells.
Analyses of individual cells and cell lines show that the tumorgenic/cancer stem cell behavior of LA7 cells was not due to mutations acquired while establishing the cell lines or from continuous culturing of these cells and lines (2, 5–10). Therefore, LA7 cells are a model system that can be used to study the cancer stem cell properties associated with tumor formation at the single-cell level. This model system can significantly contribute to the understanding of cancer-specific pathways that operate within the bulk tumor cell population and to the development of appropriate therapies to target individual cancer stem cells.
In addition to their ability to form tumors at the single-cell level, LA7 cells have retained significant stem cell potential, which is typical of normal mammary gland stem cells, as shown by their expression of the mammary gland stem cell markers CD29 and CD49f (12, 16) and by their ability to form the entire ductal–alveolar-like architecture of the mammary tree in vitro and to express and secrete milk proteins (9).
Materials and Methods
Cells, Media, and Differentiation Inducers.
LA7 cells (LA7/3F12ms) that were recently subcloned from the LA7AA10 cell line were cultured in DMEM (2). For differentiation experiments in adherent cultures, LA7 cells were exposed to 1.8% DMSO (2, 5). Three-dimensional cultures were performed by using rat tail collagen (22) or Matrigel (Becton Dickinson, San Diego, CA) according to the manufacturer's specification.
Mammosphere Cultures.
For generation of mammospheres, LA7 cells were grown in low-attachment plates (Bibby Sterilin, Staffordshire, United Kingdom) at clonogenic density (11) or at the dilution of one cell per well into 96-well plates (Nunc, Rochester, NY) by using DMEM media supplemented with B27 (Invitrogen, Carlsbad, CA), 20 ng/ml EGF (Sigma, St. Louis, MO), 20 ng/ml basic fibroblast growth factor (Sigma), and 4 μg/ml heparin (Sigma). For testing mammosphere-regeneration capability, each single-cell-generated mammosphere was mechanically and enzymatically dissociated with 0.05% trypsin and 0.53 mM EDTA (Invitrogen), and the cells were plated again in 96-well plates at a density of one cell per well. Efficiency of single cell dissociation was monitored microscopically.
Differentiation Culture Conditions, Cell Fixation, Staining, and Confocal Microscopy Analysis.
Single mammospheres generated by a single cell were plated onto collagen-coated plates and cultured in DMEM for 2 weeks. Cells plated onto collagen or cells embedded in rat tail collagen were washed with 1× PBS, fixed for 10 min with 4% paraformaldehyde-PBS, and then stained for K18, K14, or Hoechst nuclear dye 33342 (Sigma). Confocal microscopy analysis was performed by using a Leica confocal microscope (Bannockburn, IL). For the alveolar differentiation assays, cells were induced for an additional 10 days with lactogenic hormones (9) and harvested for Western blot analysis for β-casein.
Immunohistochemistry.
For histological examination, the tumors were surgically removed and fixed in 10% paraformaldehyde-PBS, dehydrated, and embedded in paraffin. Serial sections were stained with H&E or processed for immunohistochemistry. After hydration, the heat-induced antigen was retrieved in citrate buffer at pH 6.0. Endogenous peroxidases were blocked with 3% H2O2 in water. Nonspecific binding was inhibited by incubating the samples in 10% normal goat serum (NGS; Sigma–Aldrich, St. Louis, MO). The following primary antibodies were used: rabbit monoclonal anti-Ki67 (NeoMarkers, Fremont, CA), rabbit anti-K14 (NeoMarkers), and mouse monoclonal anti-K18 (Sigma). Negative controls were run in parallel with preimmune rabbit serum. Slides were washed and incubated with either biotin-conjugated goat anti-rabbit or anti-mouse (BioSpa, Milan, Italy). After washing, HRP-conjugated streptavidin (BioSpa) was added and incubated for 10 min. Slides were then stained with the 3-amino-9-ethylcarbazole or diaminobenzidine chromogen and then analyzed using the Ultravision Detection System (Lab Vision, Fremont, CA). Counterstaining was performed with hematoxylin.
Western Blot Analysis.
Cells were lyzed on ice as described (5). Blots were probed by using the manufacturer's recommended dilutions with a mouse monoclonal anti-K14 (Sigma), mouse monoclonal anti-K18 (Becton Dickinson), mouse monoclonal anti-p21 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal anti-α6 integrin (Santa Cruz Biotechnology), rabbit polyclonal anti-β1 integrin (Santa Cruz Biotechnology), anti-β casein (a gift from Giorgio Merlo, Dulbecco Telethon Institute/CNR-ITB, Milan, Italy), and goat polyclonal anti-β actin (Santa Cruz Biotechnology). Secondary antibodies were all from Santa Cruz Biotechnology (rabbit anti-goat, goat anti-mouse, and goat anti-rabbit).
Cell Transplantation in NOD-SCID Mice.
Either 100,0000, 1,000, 100, or 1 LA7 cell was injected into the fat pads of NOD-SCID mice. Subconfluent cells were trypsinized, counted, and diluted in 1× PBS at the concentration necessary to inject the desired cell numbers in a volume not exceeding 30 μl. For the injection of single LA7 cells, cells at low concentration were placed in a bacterial Petri dish in 1× PBS. A single cell was drawn in a volume of 30 μl into a syringe verified by using a stereomicroscope and injected into the mouse fat pad. Six-week-old females were anesthetized with ketamine/xilazine. Cells were then injected into the fourth mammary gland of both sides. Three females (six fat pads) were used for the injection of higher cell numbers (100 cells or more), and five females (10 fat pads) were used for single-cell injections. Tumor growth was monitored three times per week, and visible nodules were measured with a caliper. The animals were killed after 63 days of observation (or earlier), and a biopsy was performed to score for possible metastasis. The tumors removed were used to establish cell lines or fixed in formalin and embedded in paraffin for further analysis.
Mammary Tumor Dissociation.
Tumors generated by LA7 cell injection were mechanically and enzymatically dissected as described in ref. 23.
Acknowledgments
We thank Prof. Hans R. Schöler (Max Planck Institute, Muenster, Germany) for guidance; Dr. Benjamin Neel (Harvard University, Cambridge, MA), Dr. Roberto Montesano (University of Geneva, Geneva, Switzerland) for advice and suggestions; and Drs. Giovanni Bertalot and U. Fascio for help with confocal microscopy. This work was supported by Italy–USA Project Grant N.527 B-B7 (to I.Z.), Fondazione Cariplo Grant N.2003/1656 (to I.Z.), Fondo per gli Investimenti della Ricerca di Base Internazionali Grant RBIN04CBSM_000 (to I.Z.), and Ministero dell'Università e della Ricerca–Fondo per gli Investimenti della Ricerca di Base Internazionali Grant RBIP0695BB_003 (to I.Z.). R.A.R. was supported by Bundesministerium für Bildung und Forschung Grant 01GN0539 (to Dr. H. Schöler). This manuscript is no. 1 of the Network Operativo per la Biomedicina di Eccellenza in Lombardia Grant funded by Fondazione Cariplo (to I.Z.).
Abbreviations
- K18
cytokeratin 18
- K14
cytokeratin 14
- NOD
nonobese diabetic.
Footnotes
The authors declare no conflict of interest.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Dulbecco R, Bologna M, Unger M. Proc Natl Acad Sci USA. 1979;76:1256–1260. doi: 10.1073/pnas.76.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett DC, Peachey LA, Durbin H, Rudland PS. Cell. 1978;15:283–298. doi: 10.1016/0092-8674(78)90104-6. [DOI] [PubMed] [Google Scholar]
- 4.Dulbecco R, Bologna M, Unger M. Proc Natl Acad Sci USA. 1980;77:1551–1555. doi: 10.1073/pnas.77.3.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zucchi I, Montagna C, Susani L, Vezzoni P, Dulbecco R. Proc Natl Acad Sci USA. 1998;95:1079–1084. doi: 10.1073/pnas.95.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zucchi I, Montagna C, Susani L, Montesano R, Affer M, Zanotti S, Redolfi E, Vezzoni P, Dulbecco R. Proc Natl Acad Sci USA. 1999;96:13766–13770. doi: 10.1073/pnas.96.24.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zucchi I, Bini L, Valaperta R, Ginestra A, Albani D, Susani L, Sanchez JC, Liberatori S, Magi B, Raggiaschi R, et al. Proc Natl Acad Sci USA. 2001;98:5608–5613. doi: 10.1073/pnas.091101898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zucchi I, Dulbecco R. J Mammary Gland Biol Neoplasia. 2002;7:373–384. doi: 10.1023/a:1024081914634. [DOI] [PubMed] [Google Scholar]
- 9.Zucchi I, Bini L, Albani D, Valaperta R, Liberatori S, Raggiaschi R, Montagna C, Susani L, Barbieri O, Pallini V, et al. Proc Natl Acad Sci USA. 2002;99:8660–8665. doi: 10.1073/pnas.132259399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zucchi I, Prinetti A, Scotti M, Valsecchi V, Valaperta R, Mento E, Reinbold R, Vezzoni P, Sonnino S, Albertini A, Dulbecco R. Proc Natl Acad Sci USA. 2004;101:1880–1885. doi: 10.1073/pnas.0307292101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 13.Simian M, Hirai Y, Navre M, Werb Z, Lochter A, Bissell MJ. Development (Cambridge, UK) 2001;128:3117–3131. doi: 10.1242/dev.128.16.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerdes J, Li L, Schlueter C, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, Flad HD. Am J Pathol. 1991;138:867–873. [PMC free article] [PubMed] [Google Scholar]
- 15.Sacco MG, Gribaldo L, Barbieri O, Turchi G, Zucchi I, Collotta A, Bagnasco L, Barone D, Montagna C, Villa A, et al. Breast Cancer Res Treat. 1998;47:171–180. doi: 10.1023/a:1005988715285. [DOI] [PubMed] [Google Scholar]
- 16.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li H, Eaves J. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 17.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee BC, Cheng T, Adams GB, Attar EC, Miura N, Lee SB, Saito Y, Olszak I, Dombkowski D, Olson DP, et al. Genes Dev. 2003;17:1592–1604. doi: 10.1101/gad.1071503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Brien CA, Pollett A, Gallinger S, Dick JE. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 20.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 21.Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 22.Soriano JV, Pepper MS, Nakamura T, Orci L, Montesano R. J Cell Sci. 1995;108:413–430. doi: 10.1242/jcs.108.2.413. [DOI] [PubMed] [Google Scholar]
- 23.Stingl J, Eaves CJ, Kuusk U, Emerman JT. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]