Abstract
Previously, a small molecule, reversine, was identified that reverses lineage-committed murine myoblasts to a more primitive multipotent state. Here, we show that reversine can increase the plasticity of C2C12 myoblasts at the single-cell level and that reversine-treated cells gain the ability to differentiate into osteoblasts and adipocytes under lineage-specific inducing conditions. Moreover, reversine is active in multiple cell types, including 3T3E1 osteoblasts and human primary skeletal myoblasts. Biochemical and cellular experiments suggest that reversine functions as a dual inhibitor of nonmuscle myosin II heavy chain and MEK1, and that both activities are required for reversine's effect. Inhibition of MEK1 and nonmuscle myosin II heavy chain results in altered cell cycle and changes in histone acetylation status, but other factors also may contribute to the activity of reversine, including activation of the PI3K signaling pathway.
Keywords: myoblasts, multipotent, small molecule, osteoblast, adipocyte
A growing body of evidence suggests that lineage-restricted somatic cells are capable of gaining increased plasticity. This can occur through somatic cell nuclear transfer, cell fusion, ectopic gene expression, and/or treatment with exogenous factors. For example, somatic cells can be reprogrammed to a state of developmental totipotency or pluripotency, respectively, by somatic cell nuclear transfer (1, 2) or fusion with ES cells (3, 4). More recently, it has been shown that the combined expression of four transcription factors, i.e., Oct3/4, Sox2, c-Myc, and Klf4, is sufficient to reprogram primary mouse embryonic and adult fibroblast culture to pluripotent cells, which can express pluripotency markers and contribute to teratomas (5). In addition, enforced expression of C/EBPα and C/EBPβ in differentiated B cells leads to their rapid and efficient reprogramming into macrophages (6); and ectopic expression of Msx1 (7) or treatment with newt regeneration extract (8) has been reported to induce dedifferentiation of terminally differentiated myotubes.
The identification of small molecules that increase the cellular plasticity of mammalian cells would provide further insights into the mechanisms that control this complex process and may ultimately provide chemical reagents that make it possible to use healthy, abundant, and easily accessible adult cells to generate different types of stem/progenitor cells for therapeutic applications. Previously, myoseverin, a small molecule, was shown to convert myotubes to proliferative myoblast-like cells by disrupting microtubule assembly (9, 10). More recently, a purine derivative, reversine (Fig. 1A), was discovered in a cell-based screen, which can increase the plasticity of lineage-committed murine C2C12 myoblasts. The reversine-treated cells gained the ability to redifferentiate into osteoblasts and adipocytes, respectively, under lineage-specific inducing conditions (LSICs) (11). In addition, it was reported that reversine treatment reprograms primary murine and human dermal fibroblasts into myogenic-competent cells, which can further differentiate into skeletal muscle both in vitro and in vivo (12). Here, we further characterize biological activities of this small molecule in multiple cell types and the mechanism of action of reversine by affinity chromatography and additional biochemical and genetic experiments.
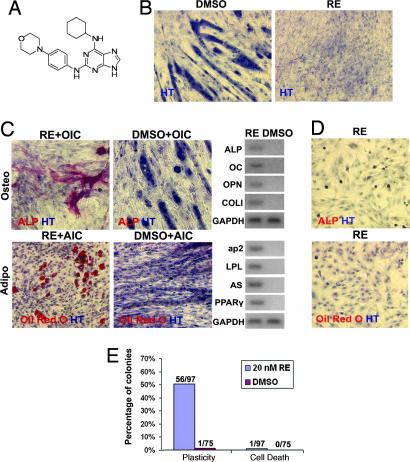
Fig. 1.
Reversine (RE) increases the cellular plasticity of C2C12 myoblasts. (A) Chemical structure of reversine. (B) Reversine blocks the terminal differentiation of C2C12 myoblasts. C2C12 myoblasts (plated at 6,000 cells per cm2) were cultured in GM in the presence or absence of 20 nM reversine for 4 days and then stained with hematoxylin (HT). (C) C2C12 myoblasts gain multipotency after 48-h treatment with 20 nM reversine. C2C12 myoblasts (plated at 6,000 cells per cm2) were cultured in GM supplemented with 20 nM reversine for 48 h. After removal of compound, cells were cultured in OI or AI conditions (OIC or AIC) for an additional 6 days and analyzed by histocytochemistry and RT-PCR. (D) Reversine does not induce C2C12 myoblasts to directly transdifferentiate into osteoblasts or adipocytes without LSICs. After 48-h treatment with 20 nM reversine, cells were fixed and analyzed by histocytochemistry. (E) Clonal analysis. Reversine increases the cellular plasticity of C2C12 myoblasts at the single-cell level. C2C12 myoblasts (plated at 1 cell per well in 96-well plate with GM) were grown in the presence of 20 nM reversine for 2 weeks. The resulting colonies were split into two and cultured with OI or AI conditions for an additional 6 days. Plasticity, cells with increased plasticity that can differentiate into adipocytes and osteoblasts; Cell Death, the colonies that underwent apoptosis during the expanding process. DMSO was used as a negative control in all experiments. Red, ALP, Oil red O; blue, HT.
Results and Discussion
Characterization of the Cellular Activity of Reversine.
Treatment of C2C12 myoblasts (plated at 6,000 cells per cm2) with 20 nM reversine in growth media (GM, DMEM supplemented with 10% FBS) for 4 days leads to striking differences between the reversine-treated and the control cells (treated with DMSO). Myotube formation is completely inhibited in the presence of 20 nM reversine, in contrast to DMSO treated cells, in which >60% myoblasts fuse into multinucleated myotubes (Fig. 1B). To characterize the multipotency of the reversine-treated cells, C2C12 myoblasts (plated at 6,000 cells per cm2) were treated with 20 nM reversine for 48 h and then cultured under osteoblast-inducing (OI) conditions (DMEM supplemented with 10% FBS, 50 μg/ml ascorbic acid-2-phosphate, 0.1 μM dexamethasone, and 10 mM α-glycerophosphate) or adipocyte-inducing (AI) conditions (DMEM supplemented with 10% FBS, 0.5 mM isobutyl-methylxanthine, 1 μM dexamethasone, and 10 μg/ml insulin) for an additional 6 days in the absence of reversine. Reversine-treated cells differentiate into osteoblasts (reversine, 20.1 ± 3.0%; DMSO, 0%) and adipocytes (reversine, 21.2 ± 4.3%; DMSO, 0%), under the respective conditions. The osteoblasts positively stain for alkaline phosphatase (ALP) and express multiple osteogenic markers, including osteocalcin (OC), osteoponin (OPN), and collagen type-I (COLI) (Fig. 1C); the adipocytes positively stain with Oil red O (which detects the lipid content of adipocytes) and express multiple adipogenic markers, including adipocyte fatty acid-binding protein 2 (ap2), lipoprotein lipase (LPL), adiposin (AS), and peroxisome proliferator-activated receptor γ (PPARγ) (Fig. 1C) [see supporting information (SI) Materials and Methods]. In contrast, the C2C12 myoblasts (plated at 6,000 cells per cm2) treated with DMSO fuse into multinucleated myotubes even in the presence of OI or AI conditions (Fig. 1C). In addition, when C2C12 myoblasts are treated with 20 nM reversine for up to 8 days without additional culture under OI or AI conditions, the cells do not directly differentiate into osteoblasts or adipocytes (Fig. 1D), suggesting that C2C12 myoblasts gain increased plasticity, but do not transdifferentiate into osteoblasts or adipocytes directly.
Because C2C12 myoblasts cultured in vitro for long term may contain a less differentiated stem-cell-like side population (13, 14), clonal analysis was carried out to verify that reversine does not function through a selection process. C2C12 myoblasts were grown from single cells (single cell per well in 96-well plate) in the presence of 20 nM reversine in GM for 2 weeks. The resulting colonies were split into two (plated at 6,000 cells per cm2) and cultured under OI or AI conditions in the absence of reversine to test their ability to undergo osteogenesis or adipogenesis, respectively. Six days later, cells were stained for ALP and with Oil red O; colonies that can differentiate into both lineages are defined as having increased plasticity. In the case of the control (DMSO-treated) cells, only 1 of 75 colonies shows the ability to differentiate into both osteoblasts and adipocytes under LSICs at very low efficiency (osteoblasts, 3.4%; adipocytes, 3.1%); whereas 56 of 96 colonies expanded under reversine treatment gain multipotency (the efficiency of osteogenesis varies from 20% to 50%; the efficiency of adipogenesis varies from 15% to 42%, Fig. 1E). These results suggest that reversine indeed increases the plasticity of C2C12 myoblasts with reasonable efficiency.
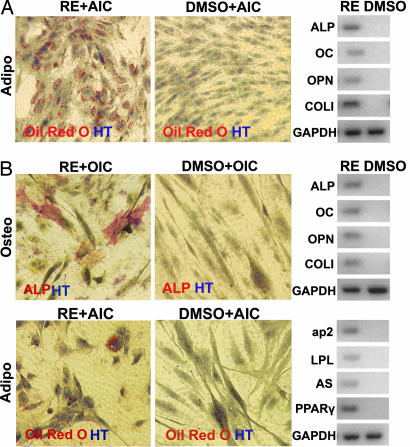
The activity of reversine on additional cell types, including 3T3E1 osteoblasts and human primary skeletal myoblasts (hSMs), was then tested. After 4-day incubation with 300 nM reversine in GM, 3T3E1 osteoblasts (plated at 6,000 cells per cm2) gain the ability to differentiate into adipocytes under AI conditions (reversine, 35.6 ± 2.3%); whereas the control 3T3E1 cells (treated with DMSO) do not differentiate into adipocytes under AI conditions (DMSO, 0%; Fig. 2A). In addition, after 4-day culture with 300 nM reversine in GM, hSMs (plated at 3,000 cells per cm2) can redifferentiate into osteoblasts (reversine, 13.6 ± 1.2%; DMSO, 0%) or adipocytes (reversine, 8.2 ± 1.7%; DMSO, 0%), respectively, under LSICs; whereas the control hSMs (treated with DMSO) fuse into myotubes even in the presence of OI or AI conditions (Fig. 2B). Finally, it has been reported that reversine also reprograms primary murine and human dermal fibroblasts into a myogenic-competent state that can further differentiate into skeletal muscle at high frequency both in vitro and in vivo (12). Thus, reversine has activity with multiple cell types, although the full generality of its effects remains to be determined.
Fig. 2.
Reversine increases cellular plasticity of 3T3E1 osteoblasts and hSMs. (A) The 3T3E1 osteoblasts (plated at 6,000 cells per cm2 in GM) were cultured in the presence of 300 nM reversine (RE) for 4 days. The compound was then removed, and cells were grown in AI conditions for an additional 6 days. (B) hSMs cells (plated at 3,000 cells per cm2 in GM) were cultured in the presence of 300 nM reversine for 4 days. The compound was then removed, and cells were grown in OI or AI conditions (OIC or AIC) for an additional 9 days. The cells were analyzed by histocytochemistry and RT-PCR. DMSO was used as a negative control in all experiments. Red, ALP, Oil red O; blue, HT.
To determine whether any component in the GM used in these experiments is involved in reversine's activity, C2C12 myoblasts (plated at 6,000 cells per cm2) were treated with 20 nM reversine in serum-free ITS (insulin–transferrin–selenium) media (versus GM) followed by differentiation under OI or AI conditions. Interestingly, more osteoblasts and adipocytes were generated in ITS media (osteoblasts, 29.5 ± 2.5%; adipocytes, 28.8 ± 1.9%) than serum-containing GM (osteoblasts, 20.1 ± 3.0%; adipocytes, 21.2 ± 4.3%). Next, insulin was removed from culture media, and a caspase inhibitor (50 μM z-VAD.fmk) was added to the insulin-free media to inhibit cellular apoptosis. It was found that, in the absence of insulin, viable C2C12 myoblasts do not increase cellular plasticity after reversine treatment; whereas the addition of 10 μg/ml insulin can restore reversine's activity. PI3K is a major kinase activated by insulin signaling and involved in diverse cellular processes, including proliferation, cell division, and apoptosis (15). The PI3K inhibitor, LY294002 (50 μM), blocks the effect of 20 nM reversine: no osteoblasts or adipocytes are found after C2C12 myoblasts (plated at 6,000 cells per cm2) were treated with 20 nM reversine and 50 μM LY294002 for 48 h, followed by 6-day culture in LSICs (SI Fig. 5). Collectively, these results suggest that the activation of the PI3K signaling pathway is essential for the reversine-induced increase in cellular plasticity.
Target Identification.
To identify the cellular target(s) of reversine, a derivative was linked through the purine C6 position to an agarose-affinity matrix via a polyethyleneglycol linker (AM·reversine+; Fig. 3A). A previous structure–activity relationship study had shown that the C6 position tolerates a wide range of substitution without loss of activity. To distinguish specific binding to reversine from nonspecific interactions with the affinity matrix, an affinity resin was also synthesized from an inactive analog (N9 methylated version of reversine) as a negative control (AM·reversine−; Fig. 3A). Whole-cell lysates isolated from C2C12 myoblasts were pretreated with AM·reversine− and incubated with AM·reversine+. SDS/PAGE analysis revealed that three bands of ≈60, 120, and 200 kDa are bound specifically by AM·reversine+; addition of 50 μM reversine to the lysates blocks binding of these proteins to the affinity matrix (Fig. 3B). These three bands were identified as mitogen activated extra-cellular signal regulated kinase (MEK1, 60 kDa), focal adhesion kinase (FAK, 120 kDa), and nonmuscle myosin II heavy chain (NMMII, 200 kDa), respectively, by liquid chromatography/mass spectrometry (LC/MS); these results were confirmed independently by Western blotting analysis (see SI Materials and Methods). Subsequent experiments showed that overexpression of FAK had no effect on reversine's activity; therefore, we focused our attention on MEK1 and NMMII.
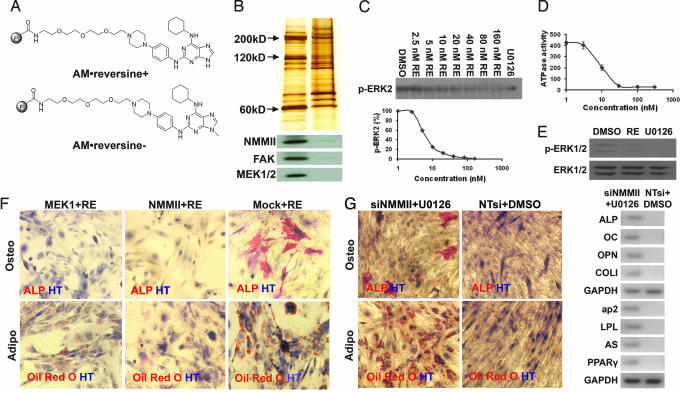
Fig. 3.
Affinity chromatography and target identification. (A) Structures of positive (AM·reversine+) and negative (AM·reversine−) affinity matrices. (B) Silver staining and Western blot analysis of proteins retained by affinity supports. (C and D) Reversine inhibits the enzymatic activity of active MEK1 in a dose-dependent fashion as indicated by ERK2-phosphorylation (C) and reversine inhibits the ATPase activity of myosin II (heavy chain) in vitro (D). (E) Reversine (20 nM) blocks MEK-dependent ERK1/2-phosphorylation in C2C12 cells. C2C12 cells were treated with 20 nM reversine for 1 h, and cells were lysed and analyzed by Western blot. DMSO was used as a negative control. U0126 (10 μM) was used as a positive control. (F) Overexpression of MEK1 or NMMII can partially block reversine's activity. C2C12 myoblasts (plated at 6,000 cells per cm2 in GM) transiently transfected with MEK1 or NMMII (with FuGENE6) were cultured in the presence of 20 nM reversine for 48 h. The compound was then removed, and cells were cultured in OI or AI conditions for an additional 6 days and analyzed by histocytochemistry. Empty vector was used as a negative control (Mock). (G) C2C12 myoblasts transfected with siNMMIIs (with X-treme) and cultured in the presence of 10 μM U0126 in GM for 48 h gain multipotency. C2C12 myoblasts (plated at 6,000 cells per cm2 in GM) transiently transfected with siNMMIIs were cultured in the presence of 10 μM U0126 for 48 h. The compound was then removed, and the cells were cultured in OI or AI conditions for an additional 6 days and analyzed by histocytochemistry. Cells transfected with NTsi and treated with DMSO were used as a negative control. Red, ALP, Oil red O; blue, HT.
Biochemical experiments were carried out to confirm the interaction between reversine and MEK1 or NMMII. Reversine inhibits the enzymatic activity of purified active MEK1 in vitro as determined by an ERK2 phosphorylation assay (IC50 = 8 nM; Fig. 3C). Reversine also blocks the ATPase activity of Myosin II (heavy chain) in vitro as determined by release of inorganic phosphate (IC50 = 10 nM; Fig. 3D; and see SI Materials and Methods). And finally, 20 nM reversine blocks MEK-dependent signaling as determined by the inhibition of ERK1/2 phosphorylation in C2C12 myoblasts (Fig. 3E). Because in some cases MEK signaling and PI3K signaling have been shown to play opposite roles in stem cell fate determination (16), the inhibition of MEK-dependent signaling by reversine and the activation of the PI3K signaling by insulin may function interactively to contribute to the effects of reversine. Indeed, the PI3K inhibitor, LY294002 (50 μM), partially reverses the inhibition of ERK1/2 phosphorylation induced by 20 nM reversine (SI Fig. 6).
To confirm that the effects of reversine are mediated by its interaction with MEK1 and NMMII, additional genetic and pharmacological studies were performed. MEK1 and NMMII (in pCMV·SPORT6 mammalian expression vector under the control of CMV promoter) were transiently overexpressed in C2C12 myoblasts (FuGENE6, transfection efficiency is ≈70%). After overnight transfection, 20 nM reversine was added; 48 h later, reversine and media were removed, and cells were assayed for their ability to undergo osteogenesis or adipogenesis under LSICs. Compared with the control cells transfected with the empty vector (Mock), there are significantly less osteoblasts (MEK1, 6.7 ± 1.8%; NMMII, 4.8 ± 1.9%; Mock, 18.8 ± 1.6%), and adipocytes (MEK1, 9.6 ± 1.4%; NMMII, 8.8 ± 1.8%; Mock, 19.1 ± 4.3%) found in the wells of cells overexpressing MEK1 or NMMII (Fig. 3F). Thus, ectopic expression of either MEK1 or NMMII can partially block the activity of reversine. To further demonstrate that the activity of reversine is mediated through its interaction with MEK1 and NMMII, C2C12 myoblasts (plated at 6,000 cells per cm2) were transiently transfected with an siRNA against NMMII (NMMII expression was reduced >80% by Western blot analysis; SI Fig. 7) and treated with 10 μM MEK inhibitor U0126 for 48 h. The treated cells can redifferentiate into osteoblasts (9.8 ± 1.2%) under OI conditions and adipocytes (26.5 ± 2.1%) under AI conditions (Fig. 3G). In contrast, cells transfected with nontargeting siRNAs (NTsi) and treated with DMSO did not gain multipotency and fused into myotubes even in the presence of LSICs. Additionally, C2C12 myoblasts treated with either siNMMIIs or U0126 (10 μM) alone did not gain the ability to undergo osteogenesis or adipogenesis under the same LSICs (SI Fig. 8), indicating that inhibition of both MEK1 and NMMII is required for reversine's activity.
Mechanism of Action Studies.
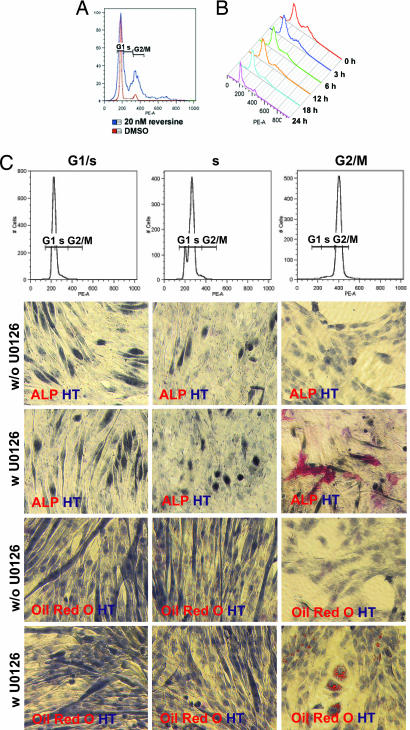
NMMII is a cytoskeleton protein that plays a major role in cytokinesis, especially cleavage furrow formation and ingression (17–19). Two small-molecule inhibitors of NMMII, BTS (20) and blebbistatin (21), have been shown to block cytokinesis and induce G2/M phase arrest. Although reversine (20 nM) does not lead to cell cycle arrest (SI Fig. 9), it does induce G2/M phase accumulation in C2C12 myoblasts in GM (G2/M, reversine, 23.4%; DMSO, 10.6%; plated at 6,000 cells per cm2; Fig. 4A). After removal of reversine, the proportion of cells in G2/M phase begins to decrease (Fig. 4B), indicating that the cell cycle effects of reversine are reversible. To determine whether G2/M phase accumulation is involved in reversine's activity, C2C12 myoblasts (plated at 6,000 cells per cm2) were synchronized at G1/S phase by 1 mM hydroxyurea (HU) treatment, at S phase by 1 mM HU treatment plus an additional 2-h culture in GM after removal of HU, or at G2/M phase with nocodazole (100 ng/ml) (Fig. 4C). After release from synchronization, cells were treated with GM in the presence or absence of 10 μM of the MEK inhibitor U0126 for 4 h and assayed for their ability to undergo osteogenesis or adipogenesis after an additional 6-day culture under OI or AI conditions. Only the cells arrested at G2/M phase and treated with U0126 gain the ability to differentiate into both osteoblasts (without U0126: G1/S, 0%; S, 0%; G2/M, 0%; with U0126: G1/S, 0%; S, 0%; G2/M; 8.7 ± 2.0%, Fig. 4C) or adipocytes (without U0126: G1/S, 0%; S, 0%; G2/M, 0%; with U0126: G1/S, 0%; S, 0.9 ± 0.4%; G2/M, 19.7 ± 3.3%; Fig. 4C) under LSICs. Moreover, C2C12 myoblasts treated with 10 μM U0126 plus 50 μM monastrol (22), an Eg5 inhibitor that induces G2/M phase accumulation, gain multipotency (osteoblasts, 19.9 ± 2.5%; adipocytes, 13.5 ± 3.1%; SI Fig. 10). These results suggest that the effects of reversine on cell cycle may contribute to the increase in cellular plasticity.
Fig. 4.
Cell cycle effects. (A) Treatment with reversine induces G2/M phase accumulation in C2C12 myoblasts. C2C12 myoblasts treated with 20 nM reversine in GM for 48 h were analyzed by FACS based on DNA content (by using PI staining). (B) C2C12 myoblasts were cultured in GM in the presence of 20 nM reversine for 48 h. After removal of reversine (0 h), cells cultured in GM were analyzed by FACS. (C) C2C12 myoblasts synchronized at G2/M phase and treated with 10 μM U0126 gain multipotency. C2C12 myoblasts synchronized at G1/S, S, or G2/M phase were analyzed by FACS. After release from synchronization by the addition of fresh GM in the presence or absence of 10 μM U0126, cells were incubated for 4 h. The compound was then removed, and the cells were cultured in OI or AI conditions for an additional 6 days and analyzed by histocytochemistry. Red, ALP, Oil red O; blue, HT.
Although reversine leads to accumulation of cells in G2/M phase, it is not clear whether the increase in cellular plasticity is a direct or indirect consequence of this activity. In Drosophila, imaginal disk transdetermination (TD), a transient cell cycle shift, is observed at the initial stage (23), suggesting that this cell cycle shift may be involved in the cellular reprogramming of TD process. However, NMMII also plays a key role in cytoskeleton regulation by tensioning cortical actin structure (24), which in turn is linked to focal adhesions that provide the pathway of force transmission from inside of the cell to the elastic matrix (25, 26). Emerging evidence indicates that cell fate is not only controlled by soluble factors, but also modified by both extracellular matrix (ECM) and intracellular cytoskeletal dynamics (27). Indeed, several studies have observed that changes in cytoskeleton and cell shape themselves can alter the differentiation of precommitted mesenchymal lineages (28–32). More recent studies have indicated that physical forces such as matrix (33), cell shape, and cytoskeletal tension (34) also play a role in cell fate determination, and notably, NMMII has been implicated in these processes (33).
Reversine also blocks MEK1 activity, which is part of the evolutionarily conserved MEK/ERK pathway that is involved in the control of many cellular processes including cell proliferation, survival, apoptosis, motility, metabolism, and stem cell fate determination (35). For example, sustained ERK1/2 activation leads mES cells to differentiate into a neural lineage (36), and inhibition of ERK1/2 contributes to maintenance of mES cell self-renewal (37, 38). In addition, ERK activation blocks adipogenic differentiation of preadipocyte 3T3L1 cells (39), and inhibition of ERK phosphorylation by PD98059, a specific inhibitor of the MEK/ERK pathway, blocks osteogenic differentiation in human mesenchymal stem cells (resulting in adipogenic differentiation) (40). Moreover, the MEK/ERK pathway has been shown to be involved in reprogramming of Schwann cells (41). Although MEK/ERK signaling controls the transcription of specific genes by regulating transcription factor activity, it also regulates histone acetylation directly by phosphorylating histone acetyltransferase (HAT) (42, 43), and indirectly by modifying signaling pathways affecting HAT activity (44, 45). Consistent with these reports, treatment with either reversine or U0126 results in decreased acetylation of histone H3 at lysine 9 (SI Fig. 11A). We therefore determined the effects of Trichostatin A (TSA, a histone deacetylase inhibitor that induces hyperacetylation of histone H3) on reversine's activity in C2C12 myoblasts. TSA (400 nM) blocks the effect of 20 nM reversine: no osteoblasts or adipocytes are found after C2C12 myoblasts (plated at 6,000 cells per cm2) were treated with 20 nM reversine for 48 h, followed by 6-day culture in LSICs (SI Fig. 11B). One possible mechanism whereby changes in histone acetylation may be involved in the reversine's activity is through the suppression of cell-fate-determining genes. Consistent with this notion, inhibition of HAT activity by an antibody or by mutation inhibits myogenesis by blocking MyoD or MEF2 activity (46, 47), and overexpression of HDAC-4 and -5 efficiently blocks MyoD-dependent fate conversion of fibroblasts into muscles (48).
Conclusion
In summary, we have characterized the effects of a previously identified small molecule, reversine. Reversine-treated cells gain the ability to undergo osteogenesis and adipogenesis under LSICs. In addition, reversine increases the plasticity of multiple cell types, including 3T3E1 osteoblasts and hSMs. Reversine appears to act as a dual inhibitor on NMMII and MEK1. By blocking NMMII, reversine induces G2/M phase accumulation of C2C12 myoblasts and, at the same time, modulates acetylation of histone H3 by inhibiting MEK-dependent signaling. Moreover, PI3K signaling is shown to be essential in reversine's activity. However, the detailed mechanism whereby reversine affects cellular plasticity has yet to be established, and whether other mechanisms related to the inhibition of NMMII and MEK1, such as cytoskeletal reorganization, play a role in reversine's activity remains to be determined.
Materials and Methods
Cell Culture.
C2C12 myoblasts and 3T3E1 cells are from American Type Culture Collection (Manassas, VA) and maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Invitrogen) at 37°C in 5% CO2. hSMs and culture media are from Cambrex (East Rutherford, NJ).
Culture media.
Growth media (GM) used were DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen).
ITS media.
DMEM/F12 (Invitrogen) containing 3 mM sodium pyruvate (Sigma, St. Louis, MO), 0.2% BSA fraction V (Invitrogen), 0.1 mM ascorbic acid (Sigma), 1% insulin-transferrin-selenium (Invitrogen), and 2 mM l-glutamine (Invitrogen).
OI conditions.
DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 50 μg/ml ascorbic acid-2-phosphate (Sigma), 0.1 μM dexamethasone (Sigma), and 10 mM α-glycerophosphate (Sigma).
AAI conditions.
DMEM (Invitrogen) supplemented with 10% FBS (Invitrogen), 0.5 mM isobutyl-methylxanthine (Sigma), 1 μM dexamethasone (Sigma), and 10 μg/ml insulin (Sigma).
Affinity Chromatography.
C2C12 myoblasts were lysed with homogenization buffer [60 mM β-glycerophosphate (Sigma), 15 mM p-nitrophenyl phosphate (Sigma) 25 mM Mops (pH 7.2; Sigma), 15 mM EGTA (Sigma), 15 mM MgCl2 (Sigma), 1 mM DTT (Invitrogen), protease inhibitors (Sigma), and 0.5% Nonidet P-40 (Sigma)]. Cell lysates were centrifuged at 16,000 × g for 20 min at 4°C, and the supernatant was collected. The total protein concentration in the supernatant was determined by using a BCA protein assay kit (Pierce, Rockford, IL). The lysates (1 mg) were then added to the packed affinity matrix (30 μl) and the bead buffer [50 mM Tris·HCl (pH 7.4)/5 mM NaF/250 mM NaCl/5 mM EDTA/5 mM EGTA/protease inhibitors/0.1% Nonidet P-40 (all from Sigma)] was added up to a final volume of 1 ml (for the competition experiment, reversine was added to a final concentration of 50 μM). After rotating at 4°C for 1 h, the mixture was centrifuged at 16,000 × g for 1 min at 4°C, and the supernatant was removed. The affinity matrix was then washed (six times) with cold bead buffer and eluted by boiling with Laemmli sample buffer (Invitrogen) at 95°C for 3 min. Samples were loaded and separated on a 4–20% Tris-glycine gel (Invitrogen), and a standard silver staining protocol was used to visualize proteins.
For Western blot analysis, samples were electroblotted on a nitrocellulose membrane, the membrane was blocked for 1 h at room temperature with 5% nonfat milk in DPBS, and immunoblotted overnight at 4°C with rabbit anti-NMMII (1:1,000; Covance, Princeton, NJ) and rabbit anti-MEK1/2 (1:1,000; Cell Signaling Technology, Beverly, MA). The membrane was then washed, incubated with anti-rabbit peroxidase-conjugated affinity-purified secondary antibody (1:1,000; Pierce) at room temperature for 1 h and developed by SuperSignal chemiluminescence (Pierce).
Transfection.
siNMMII (target sequence: CTCCTCTCGATTCGGTAAA) and nontargeting siRNA were purchased from Dharmacon (Lafayette, CO). Transfections of cDNAs or siRNAs were performed by using FuGENE6 (Roche, Indianapolis, IN) or X-treme (Roche), respectively, as directed by the manufacturer. For each well of 96-well plate, 180 ng of cDNA and 0.72 μl of FuGENE6 (Roche) or 50 ng of siRNA and 0.5 μl of X-treme (Roche) were used. After overnight incubation, 10 μM U0126 or 20 nM reversine was added. After 48-h treatment, compound was removed, and media was changed to OI or AI conditions. Six days later, cells were fixed and analyzed by histocytochemistry.
Cell Cycle Analysis.
C2C12 myoblasts were treated with 20 nM reversine in GM for 48 h and analyzed by FACS based on DNA content (by using propidium iodide staining). After removal of reversine, C2C12 myoblasts were cultured in GM and analyzed with FACS every 2 h.
Synchronization.
G1/S and S phase arrest.
C2C12 myoblasts (plated at 6,000 cells per cm2 in GM) were rinsed twice with PBS at 24 h after plating, and then media was changed to DMEM supplemented with 1% FBS. At 36 h later, media was changed to DMEM supplemented with 1% FBS and 1 mM HU and maintained for an additional 14 h. Cells were then extensively washed with PBS and cultured in GM. Maximal S phase was obtained at 2 h after release of HU.
G2/M phase arrest.
C2C12 myoblasts (plated at 6,000 cells per cm2 in GM) were rinsed twice with PBS at 24 h after plating and then media was changed to GM supplemented with 2 mM thymidine. After 18 h, the media was changed to GM and maintained for four hours; the media was then changed to GM supplemented with 100 ng/ml nocodazole and maintained for 12 h. Cell cycle was analyzed based on DNA content by using PI staining.
Cell cycle and reversine.
C2C12 myoblasts were synchronized at G1/S, S, or G2/M phase. After released from synchronization by the addition of fresh GM in the presence or absence of 10 μM U0126, cells were incubated for 4 h. The compound was then removed, and the cells were cultured in OI or AI conditions for an additional 6 days and analyzed by histocytochemistry. C2C12 myoblasts (plated at 6,000 cells per cm2) synchronized at G1/S phase were treated with 20 nM reversine (DMSO was used as a negative control) in the presence or absence of 1 mM HU for 48 h. Cells were analyzed by FACS to detect DNA content by using propidium iodide staining. The compound was then removed, and the cells were cultured in OI or AI conditions for an additional 6 days and analyzed by histocytochemistry.
Supplementary Material
Acknowledgments
This work was supported by the Novartis Research Foundation (P.G.S.) and National Institutes of Health/National Institutes of Child Health and Human Development Grant R21HD47452 (to S.D.). This is manuscript 18653 of the Scripps Research Institute.
Abbreviations
- ap2
adipocyte fatty acid-binding protein
- AI
adipocyte-inducing
- AS
adiposin
- COLI
collagen type-I
- GM
growth media
- HT
hematoxylin
- HU
hydroxyurea
- hSMs
human primary skeletal myoblasts
- ITS
insulin–transferrin–selenium
- LPL
lipoprotein lipase
- LSICs
lineage-specific inducing conditions
- MEK
mitogen-activated extracellular signal-regulated kinase
- Mon
monastrol
- NMMII
nonmuscle myosin II heavy chain
- NTsi
nontargeting siRNA
- OC
osteocalcin
- OI
osteoblast-inducing
- OPN
osteoponin
- PPARγ
peroxisome proliferator-activated receptor γ
- TD
transdetermination.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704360104/DC1.
References
- 1.Hochedlinger K, Jaenisch R. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- 2.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 3.Cowan CA, Atienza J, Melton DA, Eggan K. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 4.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Xie H, Ye M, Feng R, Graf T. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 7.Odelberg SJ, Kollhoff A, Keating MT. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 8.McGann CJ, Odelberg SJ, Keating MT. Proc Natl Acad Sci USA. 2001;98:13699–13704. doi: 10.1073/pnas.221297398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosania GR, Chang YT, Perez O, Sutherlin D, Dong H, Lockhart DJ, Schultz PG. Nat Biotechnol. 2000;18:304–308. doi: 10.1038/73753. [DOI] [PubMed] [Google Scholar]
- 10.Perez OD, Chang YT, Rosania G, Sutherlin D, Schultz PG. Chem Biol. 2002;9:475–483. doi: 10.1016/s1074-5521(02)00131-x. [DOI] [PubMed] [Google Scholar]
- 11.Chen S, Zhang Q, Wu X, Schultz PG, Ding S. J Am Chem Soc. 2004;126:410–411. doi: 10.1021/ja037390k. [DOI] [PubMed] [Google Scholar]
- 12.Anastasia L, Sampaolesi M, Papini N, Oleari D, Lamorte G, Tringali C, Monti E, Galli D, Tettamanti G, Cossu G, Venerando B. Cell Death Differ. 2006;3:2042–2051. doi: 10.1038/sj.cdd.4401958. [DOI] [PubMed] [Google Scholar]
- 13.Benchaouir R, Rameau P, Decraene C, Dreyfus P, Israeli D, Pietu G, Danos O, Garcia L. Exp Cell Res. 2004;294:254–268. doi: 10.1016/j.yexcr.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Decraene C, Benchaouir R, Dillies MA, Israeli D, Bortoli S, Rochon C, Rameau P, Pitaval A, Tronik-Le Roux D, Danos O, et al. Physiol Genomics. 2005;23:132–149. doi: 10.1152/physiolgenomics.00141.2004. [DOI] [PubMed] [Google Scholar]
- 15.Engelman JA, Luo J, Cantley LC. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 16.Hong CC, Peterson QP, Hong JY, Peterson RT. Curr Biol. 2006;16:1366–1372. doi: 10.1016/j.cub.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young PE, Richman AM, Ketchum AS, Kiehart DP. Genes Dev. 1993;7:29–41. doi: 10.1101/gad.7.1.29. [DOI] [PubMed] [Google Scholar]
- 18.Matsumura F, Ono S, Yamakita Y, Totsukawa G, Yamashiro S. J Cell Biol. 1998;140:119–129. doi: 10.1083/jcb.140.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straight AF, Field CM, Mitchison TJ. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheung A, Dantzig JA, Hollingworth S, Baylor SM, Goldman YE, Mitchison TJ, Straight AF. Nat Cell Biol. 2002;4:83–88. doi: 10.1038/ncb734. [DOI] [PubMed] [Google Scholar]
- 21.Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ. Science. 2003;299:1743–1747. doi: 10.1126/science.1081412. [DOI] [PubMed] [Google Scholar]
- 22.Mayer TU, Kapoor TM, Haggarty SJ, King RW, Schreiber SL, Mitchison TJ. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 23.Sustar A, Schubiger G. Cell. 2005;120:383–393. doi: 10.1016/j.cell.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 24.Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. J Biol Chem. 2006;281:35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- 25.Beningo KA, Dembo M, Kaverina I, Small JV, Wang YL. J Cell Biol. 2001;153:881–888. doi: 10.1083/jcb.153.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamada M, Sheetz MP, Sawada Y. Dev Cell. 2004;7:709–718. doi: 10.1016/j.devcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Geiger B, Bershadsky A, Pankov R, Yamada KM. Nat Rev Mol Cell Biol. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- 28.Spiegelman BM, Ginty CA. Cell. 1983;35:657–666. doi: 10.1016/0092-8674(83)90098-3. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez Fernandez JL, Ben-Ze'ev A. Differentiation. 1989;42:65–74. doi: 10.1111/j.1432-0436.1989.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 30.Carvalho RS, Schaffer JL, Gerstenfeld LC. J Cell Biochem. 1998;70:376–390. doi: 10.1002/(sici)1097-4644(19980901)70:3<376::aid-jcb11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 31.Thomas CH, Collier JH, Sfeir CS, Healy KE. Proc Natl Acad Sci USA. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavalko FM, Chen NX, Turner CH, Burr DB, Atkinson S, Hsieh YF, Qiu J, Duncan RL. Am J Physiol. 1998;275:C1591–C1601. [PubMed] [Google Scholar]
- 33.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 34.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 35.Kolch W. Nat Rev Mol Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Li L. Dev Biol. 2005;284:1–11. doi: 10.1016/j.ydbio.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Do JT, Zhang Q, Yao S, Yan F, Peters EC, Scholer HR, Schultz PG, Ding S. Proc Natl Acad Sci USA. 2006;103:17266–17271. doi: 10.1073/pnas.0608156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Dev Biol. 1999;210:30–43. doi: 10.1006/dbio.1999.9265. [DOI] [PubMed] [Google Scholar]
- 39.Hu E, Kim JB, Sarraf P, Spiegelman BM. Science. 1996;274:2100–2103. doi: 10.1126/science.274.5295.2100. [DOI] [PubMed] [Google Scholar]
- 40.Jaiswal RK, Jaiswal N, Bruder SP, Mbalaviele G, Marshak DR, Pittenger MF. J Biol Chem. 2000;275:9645–9652. doi: 10.1074/jbc.275.13.9645. [DOI] [PubMed] [Google Scholar]
- 41.Harrisingh MC, Perez-Nadales E, Parkinson DB, Malcolm DS, Mudge AW, Lloyd AC. EMBO J. 2004;23:3061–3071. doi: 10.1038/sj.emboj.7600309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ait-Si-Ali S, Carlisi D, Ramirez S, Upegui-Gonzalez LC, Duquet A, Robin P, Rudkin B, Harel-Bellan A, Trouche D. Biochem Biophys Res Commun. 1999;262:157–162. doi: 10.1006/bbrc.1999.1132. [DOI] [PubMed] [Google Scholar]
- 43.Kawasaki H, Schiltz L, Chiu R, Itakura K, Taira K, Nakatani Y, Yokoyama KK. Nature. 2000;405:195–200. doi: 10.1038/35012097. [DOI] [PubMed] [Google Scholar]
- 44.Park GY, Joo M, Pedchenko T, Blackwell TS, Christman JW. Am J Physiol. 2004;286:L956–L962. doi: 10.1152/ajplung.00338.2003. [DOI] [PubMed] [Google Scholar]
- 45.Koch A, Giembycz M, Ito K, Lim S, Jazrawi E, Barnes PJ, Adcock I, Erdmann E, Chung KF. Am J Respir Cell Mol Biol. 2004;30:342–349. doi: 10.1165/rcmb.2003-0122OC. [DOI] [PubMed] [Google Scholar]
- 46.Puri PL, Avantaggiati ML, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sartorelli V, Huang J, Hamamori Y, Kedes L. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu J, McKinsey TA, Zhang CL, Olson EN. Mol Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.