Abstract
During pancreas development, both the exocrine and endocrine lineages differentiate from a common pool of progenitor cells with similarities to mature pancreatic duct cells. A small set of transcription factors, including Tcf2, Onecut1, and Foxa2, has been identified in these pancreatic progenitor cells. The Sry/HMG box transcription factor Sox9 is also expressed in the early pancreatic epithelium and is required for normal pancreatic exocrine and endocrine development in humans. In this study, we found Sox9 in mice specifically expressed with the other progenitor transcription factors in both pancreatic progenitor cells and duct cells in the adult pancreas. Sox9 directly bound to all three genes in vitro and in intact cells, and regulated their expression. In turn, both Foxa2 and Tcf2 regulated Sox9 expression, demonstrating feedback circuits between these genes. Furthermore, Sox9 activated the expression of the proendocrine factor Neurogenin3, which also depends on the other members of the progenitor transcription network. These studies indicate that Sox9 plays a dual role in pancreatic progenitor cells: both maintaining a stable transcriptional network and supporting the programs by which these cells differentiate into distinct lineages.
The distinct cell types that populate the mature pancreas derive from a common pool of undifferentiated progenitor cells that form the early pancreatic bud. Although recent work has begun to define the gene-expression cascades that drive the differentiation of the mature pancreatic cell lineages (1), the gene-expression programs that maintain the progenitor cells and also permit their proper differentiation are not well understood.
The pancreas first appears as a cluster of cells budding from the dorsal aspect of the gut endoderm at embryonic day (e)9.5 in the mouse embryo, and a ventral bud appears approximately a day later. The endoderm cells that make up the buds rapidly divide and form a branching epithelium surrounded by mesenchyme. As this epithelial progenitor cell population expands during the first 3 days of pancreatic growth, a small number of cells delaminate from this epithelium and stop dividing. Most of these postmitotic cells express the glucagon gene, and the remainder express insulin, but they lack many key characteristics of mature pancreatic endocrine cells (2–5).
Starting at e13 in the mouse, the embryonic pancreas undergoes a dramatic transformation in a synchronized wave of differentiation termed the secondary transition. The branching epithelium organizes into ducts, with the ends of the ducts differentiating into exocrine cells, whereas the cells lining the ducts retain progenitor cell characteristics and the ability to generate endocrine cells. In scattered progenitor cells along the ducts, notch signaling is inactivated, allowing the transient expression of the proendocrine bHLH transcription factor Neurogenin3 that initiates endocrine differentiation (6, 7).
The epithelial progenitor cells that populate the pancreatic buds before the secondary transition differ from the progenitor cells along the ducts from which the major wave of differentiated cells originates after e13. Both cell types express the transcription factors Tcf2, Onecut1, and Foxa2 (8–11), but the early population of progenitor cells also express a number of transcription factors that later become restricted to the endocrine lineage, including Hb9, Pdx1, Nkx2.2, Nkx6.1, and Sox4 (4, 12–17) and at least one factor that is later restricted to the exocrine cells, Ptf1a (18). Interestingly, Tcf2, Onecut1, and Foxa2 are simultaneously expressed in several regions in the early endoderm, including the developing gut and liver, and persist in the duct cells of the adult pancreas.
Recently, several members of the SRY/HMG box (Sox) family of transcription factors (19–21) have been detected during pancreatic development (14, 17, 22, 23). Among these, Sox9 is of particular interest because of its high level of expression in the early pancreatic epithelium (14), In addition, mice carrying lacZ inserted into the Sox9 gene have demonstrated that Sox9 is expressed in the adult pancreatic ductal epithelium, and lineage tracing in the same study revealed that pancreatic cells from all lineages derive from Sox9-expressing precursors (24). Haploinsufficiency of SOX9 causes campomelic dysplasia (CD) in humans, which is characterized by a severe skeletal dysplasia, male (XY) sex reversal (25), and defects in the development of both the exocrine and endocrine pancreas (26).
To better understand the mechanisms by which Sox9 regulates pancreatic development, we explored its role in the transcriptional network that maintains gene expression in the pancreatic progenitors and its role in the transcriptional cascade that drives the progenitor cells to differentiate into endocrine cells. Here, we demonstrate that Sox9 regulates a network of transcription factors that both controls progenitor cell identity and supports endocrine cell differentiation within the developing organ.
Results
Pancreatic Expression of Sox9.
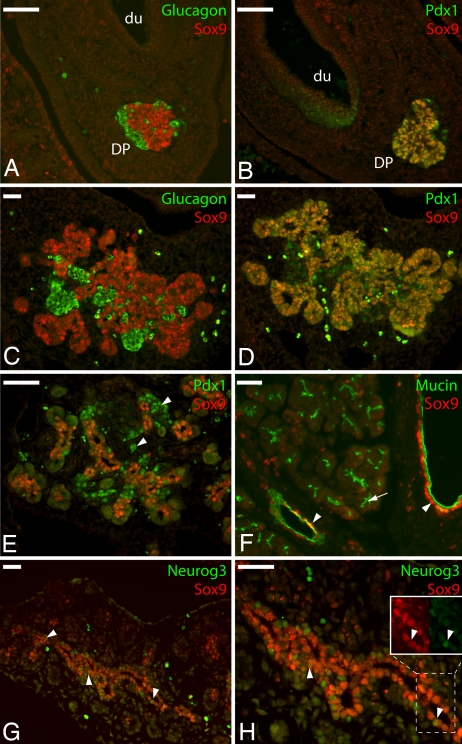
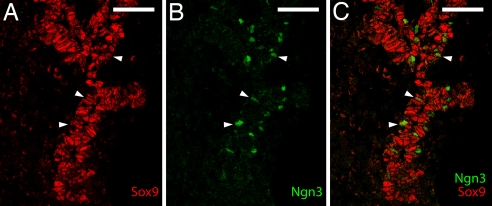
Immunohistochemical staining of sections from developing mouse pancreas detected Sox9 immunoreactivity at e10.5 and e12.5 in the dorsal (Fig. 1 A–D) and ventral (data not shown) pancreas along with Pdx1 in primary pancreatic progenitor cells but not in differentiated glucagon-expressing cells or in Pdx1-expressing cells in the adjacent duodenum. At e14.5, after the secondary transition, Sox9 persisted along with lowered levels of Pdx1 in the secondary pancreatic progenitor cells along the ducts but not in the intensely Pdx1-staining differentiated cells (Fig. 1E). At e15.5, the proendocrine transcription factor Neurogenin3 was detected in a subset of Sox9-positive cells scattered along the ducts, as well as Sox9-negative cells adjacent to the ducts (Fig. 1 G and H). Confocal microscopy verified the coexpression of Sox9 with Neurogenin3 in a subset of pancreatic duct cells at e14.5 (Fig. 2) but not with more mature islet markers Nkx2.2 and Isl1 (data not shown).
Fig. 1.
Sox9 expression in the mouse pancreas by immunofluorescence. Staining for Sox9 (red) is shown with Pdx1 (green; B, D, and E), glucagon (green; A and C), mucin (green; F), and Neurogenin3 (green, G and H) at e10.5 (A and B), e12.5 (C and D), e15.5 (G and H), and adult (F). In E, arrowheads indicate examples of cells with high expression of Pdx1. In F, arrowheads indicate examples of duct cells coexpressing Sox9 and mucin; arrow indicates an example of intraacinar cells expressing mucin but not Sox9. In G and H, arrowheads indicate examples of cells coexpressing Sox9 and Neurogenin3. (H Inset) Red and green channels are shown separately. Du, duodenum; DP, dorsal pancreas. (Scale bars, 50 μM.)
Fig. 2.
Sox9 coexpression with Neurogenin3 in the mouse embryonic pancreas. Immunofluorescence staining for Sox9 (red) is shown with Neurogenin3 (green) at e14.5 imaged by confocal microscopy. Arrowheads indicate examples of cells coexpressing Sox9 and Neurogenin3. (Scale bars, 50 μM.)
In the adult pancreas, Sox9 expression persisted in the cells along the major ducts but not in the small distal intraacinar ducts (Fig. 1F). Interestingly, we also observed patchy Sox9 immunoreactivity in the cytoplasm of β-cells in the adult islets of Langerhans (data not shown).
Sox9 Regulates the Proendocrine neurog3 Gene.
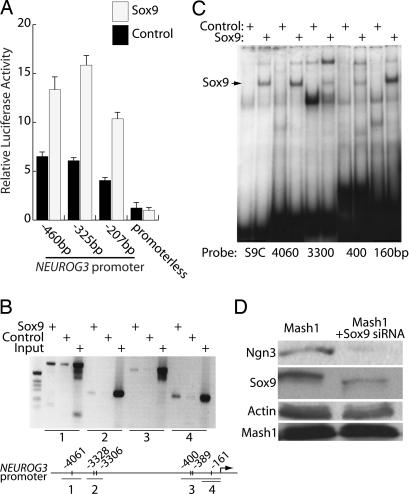
The expression of Neurogenin3 in Sox9-positive duct cells suggests that Sox9 could play a role in initiating Neurog3 gene expression and, thereby, islet cell differentiation. To test this hypothesis, the activities of human NEUROG3 promoter fragments cloned upstream of a luciferase reporter gene were assessed after cotransfection with either a plasmid vector expressing Sox9 or an empty vector (Fig. 3A). Sox9 significantly increased expression from all of the NEUROG3 reporter constructs, with the −325 promoter construct showing the greatest effect.
Fig. 3.
Sox9 regulates Neurog3 expression. (A) Fragments of the human NEUROG3 promoter with the indicated 5′ endpoints driving luciferase expression were cotransfected into αTC1.6 cells together with plasmids expressing either the Sox9 cDNA or no cDNA (Control). Luciferase activities were determined 48 h after transfection and expressed relative to the activity in cells transfected with the promoterless reporter construct and the empty expression plasmid. Results are expressed as the mean ± SEM of data from experiments performed in triplicate on at least three separate occasions. (B) Chromatin IP studies were performed by immunoprecipitating cross-linked chromatin with antiserum against Sox9 or with control IgG. Four fragments of the mouse Neurog3 promoter outlined in the line drawing were amplified by PCR from the precipitates or the input DNA. (C) DNA binding of in vitro-translated Sox9 or luciferase (Control) protein was tested by EMSA. Double-stranded, radiolabeled oligonucleotide probes contained the binding sites shown in the line figure in B and a consensus Sox9-binding site (S9C). (D) Stable mPAC L20 cell lines expressing an shRNA directed against Sox9 or a control shRNA were infected with a Mash1 adenovirus to induce Neurogenin3 expression. Neurogenin3 protein levels were assessed by Western blot 48 h after infection.
The human and mouse Neurog3 gene promoters contain several conserved sequences that conform to the Sox9 consensus binding site (27). We tested for binding by Sox9 to these sites by ChIP in the pancreatic duct cell line mPAC L20. After chemical cross-linking of mPAC L20 pancreatic ductal cells and immunoprecipitation of the cell extracts with antiserum directed against Sox9, selective immunoprecipitation of regions of the Neurog3 promoter containing putative Sox9-binding sites was demonstrated by PCR (Fig. 3B). The Sox9 antiserum did not immunoprecipitate the insulin promoter, the upstream stimulatory factor (USF) promoter, or the phosphoenolpyruvate carboxykinase promoter (data not shown).
EMSA mapped these binding sites in more detail. In vitro-translated mouse Sox9 bound to sequences at four distinct sites within the NEUROG3 promoter with affinities similar to a consensus Sox9-binding site (28) (Fig. 3C). It should be noted that some lanes also contain a slower mobility band. This larger complex may result from dimerization of Sox9 on the DNA, which has been shown to affect the function of Sox9 (29).
Finally, we used RNA interference technology to test the importance of Sox9 in normal Neurogenin3 expression. By stably expressing an shRNA targeting Sox9, we established mPAC L20 mouse pancreatic cells with reduced Sox9 expression. Because the mPAC L20 cell line does not normally express Neurogenin3, an adenovirus expressing the bHLH protein Mash1 was used to induce Neurogenin3 expression (30). In mPAC L20 cells containing normal levels of Sox9, Mash1 activated Neurogenin3 expression as described (30); however, Mash1 could not induce Neurogenin3 expression in the Sox9-deficient cells (Fig. 3D).
Sox9 Regulates a Network of Factors Upstream of Neurogenin3.
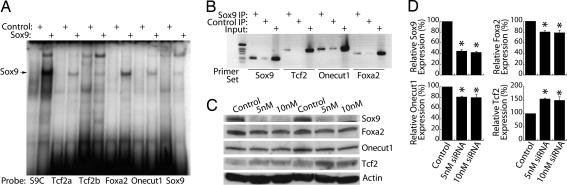
Because Sox9 expression parallels that of other pancreatic progenitor and ductal cell transcription factors Tcf2, Onecut1, and Foxa2, all of which have been implicated in the expression of Neurogenin3, we tested the hypothesis that Sox9 coordinates the expression of these factors and establishes an environment that is permissive for Neurogenin3 induction in pancreatic progenitor cells. Scanning the upstream sequences of these genes identified conserved consensus Sox9-binding sequences in each. Binding to each of these sites was confirmed by EMSA (Fig. 4A) and ChIP from mPAC L20 cells (Fig. 4B) and embryonic pancreas [Onecut1 only, supporting information (SI) Fig. 7]. Interestingly, a conserved consensus binding site in the Sox9 gene itself was also verified by EMSA and ChIP (Fig. 4 A and B), suggesting that Sox9 may also autoregulate its own expression in the pancreas.
Fig. 4.
Sox9 regulates the other progenitor cell transcription factors. (A) DNA binding of in vitro-translated Sox9 or luciferase (Control) protein was tested by EMSA. Double-stranded, radiolabeled oligonucleotide probes contained binding sites from the indicated promoters and a consensus Sox9-binding site (S9C). (B) chromatin IP studies were performed by immunoprecipitating cross-linked chromatin with antiserum against Sox9 or with control IgG. 4 fragments of the indicated promoters were amplified by PCR from the precipitates or the input DNA. (C) mPAC L20 cells were transfected with synthetic double-stranded siRNA directed against Sox9 or with a control siRNA. Protein levels were assessed by Western blot 48 h after transfection. (D) Quantification is shown for Western blots. Data represent the mean ± SEM of three independent experiments; statistical analyses were carried out by using one-way ANOVA, followed by the Newman–Keuls post hoc test. Asterisks indicate significant difference (P < 0.001) from control conditions.
To test whether Sox9 regulates these genes in intact cells, Sox9 expression was reduced by RNAi with synthetic double-stranded RNA oligonucleotides transfected into mPAC L20 cells. Transient knockdown of Sox9 levels by ≈50% resulted in a 20–30% reduction in the expression of Foxa2 and Oncut1 and a 50% increase in the expression of Tcf2 (Fig. 4 C and D).
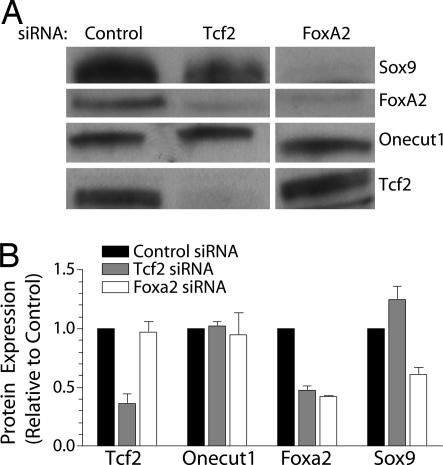
Finally, we used the same siRNA approach to test the role of Tcf2 and Foxa2 in regulating Sox9 and the other members of the network (Fig. 5). Targeted reduction of Tcf2 expression by siRNA using transfected double-stranded RNA oligonucleotides in mPAC L20 cells caused modest reductions in the expression of Foxa2 but no significant change in Onecut1 expression. Foxa2 knockdown produced a more dramatic decrease in Sox9 expression but did not affect Onecut1 or Tcf2 expression. These experiments demonstrate that Foxa2 is necessary for the maintenance of Sox9 expression in these cells and that Sox9 participates in a network of factors including Foxa2, Tcf2, and Onecut1 that are required for normal pancreatic organogenesis.
Fig. 5.
Foxa2 and Tcf2 regulate Sox9 expression. mPAC L20 cells were transfected with a control siRNA or siRNAs against Foxa2 and Tcf2 at a concentration of 10 nM. Protein levels were assessed by Western blot 48 h after transfection. (B) Quantification for Western blots shown in A. Data represent the mean ± SEM of four independent experiments.
Discussion
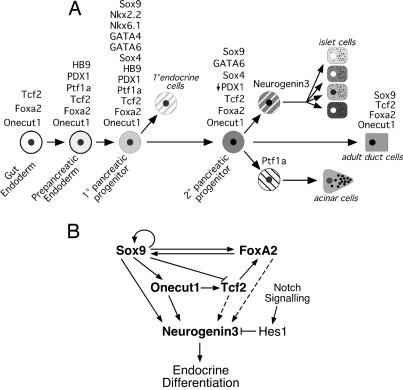
During embryonic development, the differentiated cells of the pancreas derive from a transient pool of stable, replicating progenitor cells. The data presented here demonstrate that Sox9 plays a central role in both maintaining a gene network that functions in these progenitor cells and activating the cascade of gene-expression events that leads to their differentiation into endocrine cells. Sox9 is coexpressed with a network of transcription factors, including Foxa2, Tcf2, and Onecut1, in the primary progenitor cells before e13, in the secondary progenitor cells from which the mature exocrine and endocrine cells differentiate, and in the duct cells in the adult pancreas (Fig. 6A). Here, we demonstrate that Sox9 binds to and regulates the genes encoding each of these factors, and in turn, these factors regulate Sox9 and each other in a complex interlocking network. Sox9, along with other members of the network, also binds to and activates the gene encoding the proendocrine factor Neurogenin3, which initiates the differentiation of the endocrine cells (Fig. 6B).
Fig. 6.
Model for the role of Sox9 in pancreatic progenitor cells. The model in A shows the transcription factors expressed at different steps of differentiation from gut endoderm to mature pancreatic duct cells (modified from refs. 6 and 31). The model in B shows the regulatory relationships among Sox9 and the other transcription factors in pancreatic progenitor cells. Dashed lines indicate relationships supported only by in vitro DNA-binding studies.
The observation of a gene network functioning in pancreatic progenitor cells conforms to the previously proposed roles of the members of the network in the pancreas. Onecut1 has been shown to be necessary for expression of Pdx1 and Tcf1 as well as Neurogenin3 expression and endocrine cell differentiation but not for expression of Foxa2 or Shh (9, 10, 31). Foxa2 expression is necessary for normal endoderm formation, and development of α-cells and maintenance of Pdx1 expression in the mature β-cell and has been implicated in the expression of Neurogenin3 (6, 11, 32, 33). Tcf2 expression initiates early in endoderm specification and closely parallels the expression of Sox9 in the fetal and adult pancreas (8), and it has also been implicated in expression of Neurogenin3 (6, 31). Interestingly, mutations in Tcf2 cause dramatic reduction of Pdx1 expression within the developing pancreatic endoderm in zebrafish (34) and lead to diabetes (MODY-5) in humans (35).
Onecut1, Tcf2, and Foxa2 also function in a self-maintaining gene network along with other transcription factors in the developing and adult liver (36–39). Foxa2 also participates in a transcriptional network in adult islets that includes Tcf1, HNF4α, and Pdx1 (37, 40). Transcriptional networks can become self-maintaining by building a complex web of direct and indirect feedback interactions, but can function in a potent feed-forward fashion as well through cooperative interactions of multiple network factors on the target genes. In this regard, Sox9 and its network partners function in two distinct roles: First, they maintain the network and stabilize progenitor cell gene expression through multiple feedback loops; but, in addition, they cooperate in binding and activating the gene encoding Neurogenin3, thereby leading to the disruption of the network and the differentiation to postmitotic islet cells. It is interesting to speculate that the network partners may function similarly in supporting Ptf1a expression and acinar cell differentiation. In support of this idea, we found that Sox9 can bind to the Ptf1a gene promoter (see SI Fig. 8).
In conclusion, Sox9 plays an essential role in coordinating gene expression during pancreatic development and activating the endocrine cell-differentiation program. Understanding how the early Sox9-expressing pancreatic cells differentiate from the surrounding gut endoderm and then how they mature into the secondary progenitor cells from which the mature endocrine cells differentiate will help guide efforts to develop methods for generating β-cells from progenitor cells for patients with diabetes.
Methods
Animals.
Mice were housed on a 12-hr light–dark cycle in a controlled climate according to the University of California, San Francisco, regulations. Timed matings were carried out, with e0.5 being set as midday of the day of discovery of a vaginal plug. CD-1 control mice and were obtained from Charles River Laboratories (Wilmington, MA).
Immunfluorescence Analysis.
Harvesting and processing of embryonic and adult mouse tissues were performed as described (4, 12–17). Immunostaining was performed overnight at 4°C in PBS containing 1% goat serum with the following primary antisera: 1:2,000 rabbit anti-pdx1, 1:2,000 guinea pig anti-pdx1, 1:2,000 guinea pig anti-neurogenin3 (41), 1:4,000 guinea pig anti-glucagon (Linco, St Charles, MO), 1:200 rabbit anti-sox9 (Santa Cruz Biotechnology, Santa Cruz, CA), 1:800 rabbit anti-sox9 (a gift from M. Wegner (Universität Erlangen–Nürnberg, Nürnberg, Germany), and 1:200 hamster anti-mucin (NeoMarkers, Fremont, CA). All secondary antibodies were obtained from Jackson ImmunoResearch (West Grove, PA) and were used at 1:200 (FITC) or 1:800 (Cy3) dilutions, and coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, CA). Slides were imaged on a Axioskop microscope (Zeiss, Thornwood, NY) with an attached camera (Hamamatsu, Hammamatsu City, Japan) using Openlab software (Fig. 1) or on a TCS SL confocal microscope (Leica, Bannockburn, IL) (Fig. 2).
Cell Culture and Transfection.
Culture of mouse mPAC L20 and αTC1.6 cells, transfection of αTC1.6 cells and luciferase assays were performed as described (6). For stable lines, 10 cm2 dishes of mPAC L20 cells were transfected with 5 μg of plasmid DNA and Lipofectamine 2000 (Invitrogen, Carlsbad, CA) per the manufacturer's recommendations. Cells recovered overnight before the addition of puromycin (2 μg/ml). After 1 week of selection, puromycin-resistant cells were trypsinized, and pooled clones were used for further experiments.
For adenovirus experiments, mPAC cells were infected at a MOI of 50:1 with adenovirus encoding rat Mash1 or control adenovirus encoding β-galactosidase as described (30). Cells were allowed to recover for 2 days before assay for Neurogenin3.
RNA Interference.
For knockdown of Sox9 in pooled stable mPAC L20 lines, a short hairpin (GATCCCCGGAACAGACTCACATCTCTTTCAAGAGAAGAGATGTGAGTCTGTTCC-TTTTTGGAAA) against mouse Sox9 was cloned into the pSilencerTM-4.1-CMV vector (Ambion, Austin, TX). Transient knockdown was performed by transfecting small interfering RNA duplexes targeting Sox9 (AAAGAGATGTGAGUCUGUUCCGGGGAUC), Foxa2 (GGUCUCGGGUCU G A UUUAATT), Tcf2 (GAGAGUAUGGAAAACCGGCUU), or Control siRNA #1 (Ambion) into mPAC L20 cells by using HiPerfect (Qiagen, Valencia, CA). Low concentrations (5–10 nM) of siRNA duplexes were used to minimize potential off-target effects.
EMSAs.
Synthetic DNA oligonucleotides were labeled, and EMSAs were performed in the presence of 500 ng of poly(deoxyinosine-deoxycytidine) per 10 μl of binding mix as described (6). Protein was generated in vitro with the TNT-coupled Reticulocyte Lysate System (Promega) in 50 μl of total volume from 1 μg of DNA; 1 μl (≈5 ng of protein) of the reaction mix was then used per binding reaction.
The oligonucleotides sequences are in SI Table 1.
ChIP Assays.
Mouse mPAC L20 cells grown to 70% confluence were fixed at room temperature with 1% formaldehyde for 10 min. Cross-linking was quenched by the addition of glycine (125 mM). Cells were then washed in ice-cold PBS, scraped from the growth surface, and pelleted. The pellets were washed and lysed as described (37). Chromatin was then sheared to ≈1-kb fragments by using sonication and cleared by centrifugation. Antibody binding was carried out overnight at 4°C by using 200 μg of chromatin and normal rabbit IgG (sc-2027; Santa Cruz Biotechnology) or rabbit anti-Sox9 antibody (sc-20095; Santa Cruz Biotechonolgy). Antibody-bound complexes were coupled for 1 h to previously blocked (1 mg/ml BSA, 0.1 mg/ml salmon sperm DNA) protein A-Sepharose beads (GE Healthcare/Amersham, Piscataway, NJ). Coupled beads were washed in 1.5 ml each of: TSE1 [0.1% SDS/1% Triton X-100/2 mM EDTA/20 mM Tris·HCl (pH 8.1)/150 mM NaCl], TSE2 (same as TSE1 but with 500 mM NaCl), ChIP Buffer 3 [0.25M LiCl/1% Nonidet P-40/1% sodium deoxycholate/1 mM EDTA/10 mM Tris·HCl (pH 8.1)] and TE, all containing protease inhibitors (Roche, Indianapolis, IN) and PMSF. Bound material was then eluted from the beads in 50 mM NaHCO3 containing 1% SDS; cross-linking was reversed by incubation at 67°C for 5 h; and DNA was precipitated in ethanol. DNA pellets were then resuspended in TE and treated successively with 200 μg/ml RNase A and with 75 μg/ml proteinase K for 2 h each at 37°C and 55°C, respectively. DNA was then phenol/chloroform/isoamyl alcohol-extracted and precipitated with 5 μg of glycogen and then resuspended in 20 μl of water.
Five to 15 ng of immunoprecipitated DNA per reaction was assayed by PCR with the primers in SI Table 1 to test for the precipitation of specific promoter fragments.
Western Blotting.
Whole-cell lysates were harvested with 100°C SDS/PAGE loading buffer [62.5 mM Tris (pH 6.8)/1 mM sodium vanadate/1 mM sodium fluoride/2% SDS/10% glycerol], incubated for 10 min at 100°C, and sonicated by using an ultrasonic needle tip processor (Tekmar, Cincinnati, OH) for 1 min at 70% maximal power (4–6 W). Cellular debris was removed from the lysate by centrifugation at 10,000 × g for 10 min at 20°C. Supernatants were collected, and proteins were separated by electrophoresis through 10% Tris·HCl polyacrylamide gels. Proteins were transferred to PVDF membranes (Hybond-P; Amersham Biosciences, Piscataway, NJ) and incubated with rabbit anti-neurogenin3 [1:1,000; (41)], rabbit anti-sox9 (1:500; Santa Cruz Biotechnology), rabbit anti-onecut1 (1:500; Santa Cruz Biotechnology), and rabbit anti-tcf2 (1:1,000, Chemicon, Temecula, CA) for 3 h at room temperature. After washing, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Finally, membranes were imaged by using the enhanced chemiluminescence kit (ECL; Amersham).
Supplementary Material
Acknowledgments
We thank members of the M.S.G. and Hebrok laboratories, John Rubenstein, and Didier Stainier for helpful advice and criticism. This work was supported by National Institutes of Health (NIH) Grants DK21344 and DK61245, cores from NIH Grant P30 DK063720, Larry L. Hillblom Foundation Grant 2002/1(E), the Nora Eccles Treadwell Foundation, and Juvenile Diabetes Research Foundation Fellowship Awards 3-98-341 (to M.E.W.) and 3-2004-276 (to F.C.L.).
Abbreviation
- e(n)
embryonic day (n).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704054104/DC1.
References
- 1.Wilson ME, Scheel D, German MS. Mech Dev. 2003;120:65–80. doi: 10.1016/s0925-4773(02)00333-7. [DOI] [PubMed] [Google Scholar]
- 2.Wilson ME, Kalamaras JA, German MS. Mech Dev. 2002;115:171–176. doi: 10.1016/s0925-4773(02)00118-1. [DOI] [PubMed] [Google Scholar]
- 3.Teitelman G, Alpert S, Polak JM, Martinez A, Hanahan D. Development (Cambridge, UK) 1993;118:1031–1039. doi: 10.1242/dev.118.4.1031. [DOI] [PubMed] [Google Scholar]
- 4.Sander M, Sussel L, Conners J, Scheel D, Kalamaras J, Dela Cruz F, Schwitzgebel V, Hayes-Jordan A, German M. Development (Cambridge, UK) 2000;127:5533–5540. doi: 10.1242/dev.127.24.5533. [DOI] [PubMed] [Google Scholar]
- 5.Oster A, Jensen J, Serup P, Galante P, Madsen OD, Larsson LI. J Histochem Cytochem. 1998;46:707–715. doi: 10.1177/002215549804600602. [DOI] [PubMed] [Google Scholar]
- 6.Lee JC, Smith SB, Watada H, Lin J, Scheel D, Wang J, Mirmira RG, German MS. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- 7.Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U, Edlund H. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- 8.Coffinier C, Barra J, Babinet C, Yaniv M. Mech Dev. 1999;89:211–213. doi: 10.1016/s0925-4773(99)00221-x. [DOI] [PubMed] [Google Scholar]
- 9.Jacquemin P, Lemaigre FP, Rousseau GG. Dev Biol. 2003;258:105–116. doi: 10.1016/s0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 10.Jacquemin P, Durviaux SM, Jensen J, Godfraind C, Gradwohl G, Guillemot F, Madsen OD, Carmeliet P, Dewerchin M, Collen D, et al. Mol Cell Biol. 2000;20:4445–4454. doi: 10.1128/mcb.20.12.4445-4454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee CS, Sund NJ, Behr R, Herrera PL, Kaestner KH. Dev Biol. 2005;278:484–495. doi: 10.1016/j.ydbio.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Sussel L, Kalamaras J, Hartigan-O'Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS. Development (Cambridge, UK) 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 13.Ahlgren U, Jonsson J, Edlund H. Development (Cambridge, UK) 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 14.Lioubinski O, Muller M, Wegner M, Sander M. Dev Dyn. 2003;227:402–408. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Arber S, Jessell TM, Edlund H. Nat Genet. 1999;23:67–70. doi: 10.1038/12669. [DOI] [PubMed] [Google Scholar]
- 16.Harrison KA, Thaler J, Pfaff SL, Gu H, Kehrl JH. Nat Genet. 1999;23:71–75. doi: 10.1038/12674. [DOI] [PubMed] [Google Scholar]
- 17.Wilson ME, Yang KY, Kalousova A, Lau J, Kosaka Y, Lynn FC, Wang J, Mrejen C, Episkopou V, Clevers HC, German MS. Diabetes. 2005;54:3402–3409. doi: 10.2337/diabetes.54.12.3402. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 19.Wegner M, Stolt CC. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Wegner M. Pigment Cell Res. 2005;18:74–85. doi: 10.1111/j.1600-0749.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 21.Wegner M. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mavropoulos A, Devos N, Biemar F, Zecchin E, Argenton F, Edlund H, Motte P, Martial JA, Peers B. Dev Biol. 2005;285:211–223. doi: 10.1016/j.ydbio.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 23.Lee YH, Saint-Jeannet JP. Int J Dev Biol. 2003;47:459–462. [PubMed] [Google Scholar]
- 24.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Proc Natl Acad Sci USA. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E, et al. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 26.Piper K, Ball SG, Keeling JW, Mansoor S, Wilson DI, Hanley NA. Mech Dev. 2002;116:223–226. doi: 10.1016/s0925-4773(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 27.Mertin S, McDowall SG, Harley VR. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harley VR, Lovell-Badge R, Goodfellow PN. Nucleic Acids Res. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernard P, Tang P, Liu S, Dewing P, Harley VR, Vilain E. Hum Mol Genet. 2003;12:1755–1765. doi: 10.1093/hmg/ddg182. [DOI] [PubMed] [Google Scholar]
- 30.Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. Proc Natl Acad Sci USA. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maestro MA, Boj SF, Luco RF, Pierreux CE, Cabedo J, Servitja JM, German MS, Rousseau GG, Lemaigre FP, Ferrer J. Hum Mol Genet. 2003;12:3307–3314. doi: 10.1093/hmg/ddg355. [DOI] [PubMed] [Google Scholar]
- 32.Lee CS, Sund NJ, Vatamaniuk MZ, Matschinsky FM, Stoffers DA, Kaestner KH. Diabetes. 2002;51:2546–2551. doi: 10.2337/diabetes.51.8.2546. [DOI] [PubMed] [Google Scholar]
- 33.Lantz KA, Kaestner KH. Clin Sci (London) 2005;108:195–204. doi: 10.1042/CS20040309. [DOI] [PubMed] [Google Scholar]
- 34.Sun Z, Hopkins N. Genes Dev. 2001;15:3217–3229. doi: 10.1101/gad946701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn BN, Lindner T, Yamagata K, Ogata M, Tomonaga O, et al. Nat Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 36.Zaret KS. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 37.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Genes Dev. 2006;20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costa RH, Kalinichenko VV, Holterman AX, Wang X. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 40.Friedman JR, Kaestner KH. Cell Mol Life Sci. 2006;63:2317–2328. doi: 10.1007/s00018-006-6095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwitzgebel VM, Scheel DW, Conners JR, Kalamaras J, Lee JE, Anderson DJ, Sussel L, Johnson JD, German MS. Development (Cambridge, UK) 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.