Abstract
In plants and in the nematode Caenorhabditis elegans, an RNAi signal can trigger gene silencing in cells distant from the site where silencing is initiated. In plants, this signal is known to be a form of dsRNA, and the signal is most likely a form of dsRNA in C. elegans as well. Furthermore, in C. elegans, dsRNA present in the environment or expressed in ingested bacteria is sufficient to trigger RNAi (environmental RNAi). Ingestion and soaking delivery of dsRNA has also been described for other invertebrates. Here we report the identification and characterization of SID-2, an intestinal luminal transmembrane protein required for environmental RNAi in C. elegans. SID-2, when expressed in the environmental RNAi defective species Caenorhabditis briggsae, confers environmental RNAi.
Keywords: dsRNA, transmembrane, intestine lumen
Translocation of nucleic acids across cellular membranes is associated with viral infection, bacterial conjugation, and transport of nuclear encoded tRNAs into mitochondria (1–3). In these cases, specific machinery acts to translocate a specific RNA or DNA across one or more membranous barriers. However, observations in plants suggest the routine intercellular transport of cellular mRNAs as well as processed, likely short, dsRNAs associated with RNAi-related phenomena that mediate systemic virus resistance (4, 5). In the nematode Caenorhabditis elegans, RNA-induced gene silencing is also systemic, spreading from the site of injection or expression to silence the targeted gene throughout the animal and in its progeny (6, 7). Furthermore, RNAi can be initiated by soaking animals in solutions of dsRNA or feeding worms bacteria expressing dsRNA (8, 9). Thus dsRNA can enter the animal from the environment. RNAi triggered by environmental exposure to dsRNA has also been documented in planaria, moth, tick, hydra, and numerous parasitic nematodes (10–15), suggesting that many invertebrates possess mechanisms to transport sequence nonspecific dsRNA into and between cells.
To identify cellular components required for systemic RNAi in C. elegans, we isolated systemic RNAi defective (Sid) mutants defective for spreading of RNAi (7). The first characterized gene, sid-1, encodes a transmembrane protein expressed in all cells sensitive to systemic RNAi and is required for uptake of dsRNA (7, 16). Furthermore, SID-1 expressed in Drosophila S2 cells is sufficient to mediate passive uptake of dsRNA from the growth media, indicating that SID-1 most likely acts as a channel for diffusion of dsRNA into cells (16). These observations provide strong support for the notion that dsRNA is systemically transported in C. elegans.
A SID-1::GFP fusion reporter construct that rescues sid-1 mutant worms was expressed at highest levels in cells directly exposed to the environment (7). This observation suggested that environmental dsRNA might enter the animal via these sid-1 expressing cells. Here we describe sid-2, a gene specifically required for uptake of silencing information (hereafter assumed to be dsRNA) from the environment. We find that SID-2 is a transmembrane protein expressed in the intestine and localized to the apical (luminal) membrane, indicating that the gut is the only significant route of entry for dsRNA into the animal. Remarkably, SID-2 function is not conserved in the related nematode Caenorhabditis briggsae, but expression of C. elegans SID-2 is sufficient to confer environmental RNAi on C. briggsae. Based on our analysis of the roles of sid-1 and -2, we propose that environmental RNAi in C. elegans is composed of at least two distinct steps: sid-2-dependent uptake of dsRNA from the environment and sid-1-dependent spreading of dsRNA throughout the animal.
Results and Discussion
sid-2 Is Required for Environmental RNAi but Not for Spreading Among Cells.
sid-2 mutants were isolated as animals resistant to bacteria-mediated systemic RNAi of a GFP reporter but sensitive to transgene-mediated systemic RNAi of the same reporter (7). One sid-2 mutant (qt13) was selected for initial analysis, mapping, and cloning. We found that sid-2(qt13) worms were completely resistant to bacteria-mediated RNAi targeting endogenous somatic and germ-line-expressed genes (Tables 1 and 2 and data not shown) and strongly resistant to soaking-mediated RNAi of the same genes. However, similar to wild-type and in contrast to sid-1 mutants, sid-2(qt13) worms were fully sensitive to systemic RNAi initiated by injection or transgenic expression of dsRNA targeting somatic and germ-line-expressed genes (Tables 1 and 2; Fig. 1). Thus, sid-2 is required for the uptake of environmental dsRNA but, unlike sid-1, is not required for the subsequent spread of dsRNA between cells.
Table 1.
Characterization of sid-2 deficiency on RNAi of a germ-line-expressed gene
| dsRNA delivery | Hours after injection | Percent embryonic lethality (n)* |
||
|---|---|---|---|---|
| Wild-type N2 | sid-2(qt13) | sid-1(qt2) | ||
| Bacteria-mediated | NA | 100 (152) | 0 (113) | 1 (535) |
| Soaking-mediated | NA | 100 (435) | 8 (402) | ND |
| Anterior gonad | 7–22 | 80 (748) | 84 (254) | 49 (728) |
| Intestine cytoplasm | 7–22 | 80 (834) | 77 (650) | 2 (782) |
Progeny of worms exposed to mex-3 dsRNA by various methods were scored for embryonic lethality. Two or three anterior intestinal cells or the majority of a gonad arm were filled with dsRNA. Worms were maintained at 25°C.
*Data from ref. 4. NA, not applicable; n, number of progeny scored; ND, not determined.
Table 2.
Characterization of sid-2 deficiency on RNAi of an embryonically expressed gene
| dsRNA delivery | Hours after injection | Percent twitching progeny (n)* |
||
|---|---|---|---|---|
| Wild-type N2 | sid-2(qt13) | sid-1(qt2) | ||
| Bacteria-mediated | NA | 100 (299) | 1 (362) | 0 (363) |
| Anterior gonad | 11.5–23.5 | 74 (685) | 80 (834) | 0 (981) |
Progeny of worms exposed to unc-22 dsRNA by various methods were scored for the Unc-22 twitching phenotype. Two or three anterior intestinal cells or the majority of a gonad arm were filled with dsRNA. Worms were maintained at 25°C.
*Data from ref. 4. NA, not applicable; n, number of progeny scored.
Fig. 1.
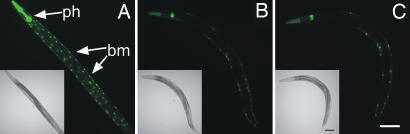
sid-2 worms are capable of spreading transgene-initiated RNAi. (A) Wild-type expression of GFP in the pharynx (ph) (myo-2::gfp) and body-wall muscle nuclei (bm) (myo-3::gfp) of an HC46 worm. (B) Expression of dsRNA (myo-2::gfp) in the pharynx causes incomplete silencing of pharynx and body-wall muscle GFP in the HC57 strain. (C) A sid-2(qt13) worm expressing the same transgenes as HC57 is capable of spreading transgene-initiated RNAi from the pharynx to the body-wall muscle. GFP images were taken at equal exposures. Anterior is upper left. (Scale bar, 0.1 mm.)
SID-2 Is an Intestinal Luminal Transmembrane Protein.
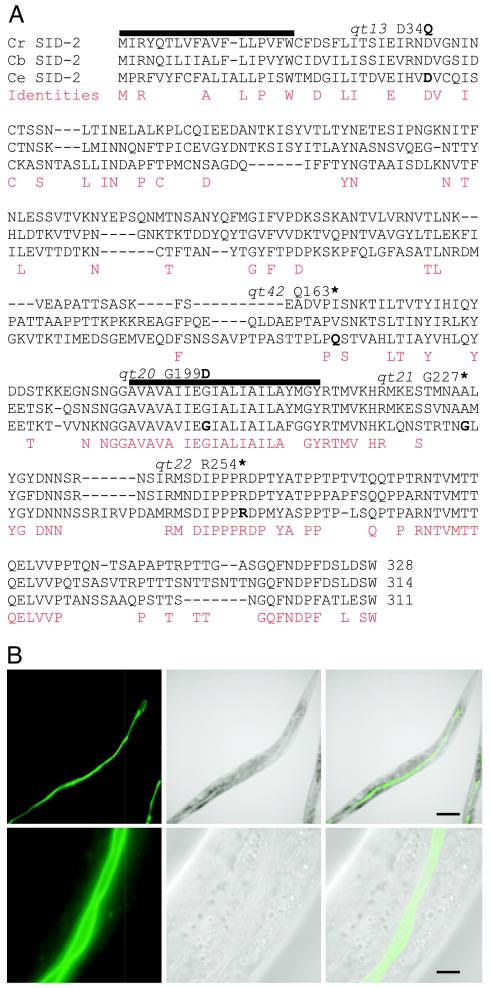
To analyze the molecular basis of dsRNA uptake from the environment, we identified and characterized the sid-2 gene. sid-2(qt13) was mapped to a small genetic interval on Linkage Group III and rescued by injection of amplified genomic DNA fragments [supporting information (SI) Fig. 5]. A fragment that contained the predicted gene ZK520.2 rescued the mutant phenotype, and injection of ZK520.2 dsRNA into wild-type worms produced worms resistant to bacteria-mediated RNAi (data not shown). Sequence analysis identified point mutations in each of five sid-2 alleles tested, confirming the identity of sid-2 as ZK520.2 (Fig. 2A). One of these (qt41) creates a stop codon before the predicted transmembrane domain, which is likely a null allele, and has a stronger environmental RNAi defect than sid-2(qt13) mutants, but was otherwise indistinguishable from wild type. sid-2 complementary DNA (cDNA) was amplified by RT-PCR, and one splice form was identified. sid-2 encodes a 311-aa transmembrane protein with sequence similarity only to a homologous protein in C. briggsae and in Caenorhabditis remanei (17, 18) (Fig. 2A).
Fig. 2.
SID-2 is a predicted membrane protein conserved in C. briggsae and C. remanei, and SID-2::GFP is expressed in the luminal intestinal membranes. (A) Translated sequence of C. elegans sid-2 cDNA aligned with a conceptual assembly of the homologous C. briggsae and C. remanei genes. Identical amino acids are marked with an asterisk and similar amino acids are marked with a colon or period. The predicted signal peptide and transmembrane domain are underlined. The positions and amino acid sequence changes for five sequenced alleles are shown above the sequences (∗, stop). (B) Localization of SID-2::GFP expression to intestinal lumen. Fluorescent, white light, and overlay respectively at low (Upper) and high (Lower) magnification. (Scale bars, 0.1 mm and 10 μm.)
To determine the spatial expression pattern of SID-2, we generated animals expressing a fusion of SID-2 to GFP (sid-2::gfp), which rescued the environmental RNAi defect of sid-2(qt13) animals, indicating that it was functional and therefore likely properly expressed and localized. SID-2::GFP localized to the intestinal lumen (Fig. 2B) and was also detected at much lower levels in excretory duct cells (data not shown), which are secretory cells of the excretory system (19). Consistent with the restricted expression, mosaic analysis of sid-2 confirmed that sid-2 activity is not required in muscle cells for uptake of silencing information into muscle cells (SI Fig. 6). These results suggest that sid-2 enables import of ingested dsRNA from the intestinal lumen.
SID-1 and SID-2 Are Both Required for the Import of Environmental dsRNA into Intestinal Cells.
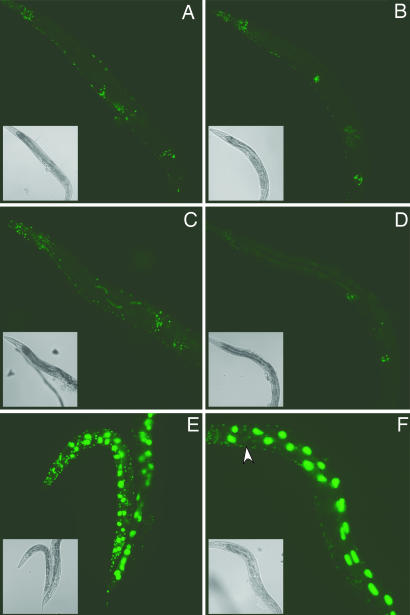
We previously showed that SID-1 functions as a channel for dsRNA, and that a SID-1::GFP reporter was expressed at high levels in cells directly exposed to the environment, including intestinal cells, suggesting that dsRNA may enter the animal via these sid-1-expressing cells (7, 16). To determine whether SID-1, in the absence of sid-2, is sufficient to import dsRNA into and to initiate RNAi in intestinal cells, we assayed GFP silencing in sur-5::gfp transgenic lines, which express GFP in all cells. In wild-type animals, this GFP expression is efficiently silenced in nonneuronal cells by gfp RNAi (Fig. 3 and data not shown). sid-2(qt13); sur-5::gfp worms fed or soaked in gfp dsRNA were fully resistant to GFP silencing in intestinal cells and all other cells, but when a single intestinal cell (or the body cavity) was injected with gfp dsRNA, the entire worm was fully sensitive to RNAi in all nonneuronal cells (Fig. 3 and data not shown). These results show that SID-1, in the absence of sid-2, can mediate uptake of dsRNA into intestinal cells from the body cavity, but not from the intestinal lumen. Similar to sid-2 mutants, sid-1(qt9); sur-5::gfp worms fed or soaked in gfp dsRNA were fully resistant to GFP silencing in all cells; however, unlike sid-2 mutants, sid-1 mutants injected with gfp dsRNA were fully resistant to GFP silencing, except in the injected intestinal cell (Fig. 3 E and F). These results suggest that both sid-1 and -2 are required for the initial import of dsRNA into intestinal cells from the lumen. However, the import of dsRNA into intestinal cells from the body cavity and the spread of dsRNA between intestinal cells as well as the subsequent dissemination of dsRNA throughout the animal does not require sid-2.
Fig. 3.
Injection of dsRNA bypasses sid-2 defects in nonneuronal cells. (A and B) Injection of gfp dsRNA (1 mg/ml) into the pseudocoelom (A) or anteriormost intestinal cell (B) of sid-2(qt13); sur-5::gfp adult hermaphrodites results in complete silencing of GFP in nonneuronal cells. (C and D) Injection into the pseudocoelom (C) or anterior intestine (D) of control sur-5::gfp worms yields comparable results. (E and F) sid-1(qt9); sur-5::gfp worms are completely resistant to RNAi after injection into the pseudocoelom (E) and show silencing in only the injected intestinal cell (arrow) (F). Animals were photographed 48 h after injection at 20°C. Insets are bright-field photomicrographs of the same worm. GFP images were taken at equal exposures.
SID-2 Function Is Not Conserved in C. briggsae.
To examine how the SID-2 protein mediates environmental RNAi, we first determined the topology of SID-2. We assayed transgenic lines expressing SID-2::β-Gal fusion proteins and found that SID-2 is a single-pass transmembrane protein with an intracellular C terminus (SI Fig. 7; Fig. 2A) (20, 21). SID-2 homologs are detected only in the closely related nematodes, C. briggsae and C. remanei. The degree of amino acid sequence conservation in the three domains of SID-2 is similar among the three species: 23% N-terminal (190 amino acids), 86% transmembrane (21 amino acids), and 53% C-terminal (100 amino acids), despite the relatively recent divergence of C. briggsae and C. remanei (Fig. 4). To determine whether the divergent C. briggsae SID-2 supports environmental RNAi, we targeted an essential gene by soaking C. briggsae worms in dsRNA. The C. briggsae animals were strongly resistant to silencing (Table 3). Injection of this same dsRNA caused efficient RNAi in C. briggsae that was indistinguishable from that observed in C. elegans (data not shown). A C. briggsae SID-2::GFP fusion protein expressed in C. briggsae showed intestinal expression localized to the luminal membrane, indicating that changes in gene expression and protein localization are not likely to account for the different sensitivity to environmental dsRNA (data not shown). Given the relative lack of sequence conservation between C. elegans and C. briggsae SID-2 (Fig. 2), we suspected that C. elegans SID-2 had gained or C. briggsae SID-2 had lost the ability to support environmental RNAi. To determine whether the sequence divergence between the SID-2 homologs explained the lack of environmental RNAi in C. briggsae, we transformed the C. elegans sid-2::gfp construct into C. briggsae. The transgene was expressed and localized as in C. elegans (data not shown) and conferred sensitivity to soaking initiated RNAi (Table 3 and SI Table 4). These results suggest that C. elegans sid-2 is sufficient to enable the uptake of environmental dsRNA by C. briggsae.
Fig. 4.
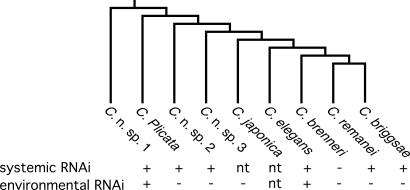
Phylogenetic relationship of select Caenorhabditis species and systemic and environmental RNAi proficiency. Phylogeny from ref. 30. Only C. brenneri (31) was conclusively deficient for systemic RNAi, whereas only C. elegans and C. sp. 1 (SB341) were proficient at environmental RNAi. nt, not tested.
Table 3.
C. elegans sid-2 confers sensitivity to soaking RNAi to C. briggsae
| Species and genotype | Percent embryonic lethality |
Percent affected adults |
||
|---|---|---|---|---|
| pal-1 dsRNA | Buffer only | pal-1 dsRNA | Buffer only | |
| C. elegans | ||||
| sid-2(qt13) | 55 (11,251) | 0 (455) | 69 (29) | 0 (9) |
| sid-2(qt13); sid-2::gfp | 99 (968) | 2 (122) | 100 (27) | 0 (5) |
| C. briggsae | ||||
| Wild type | 5 (716) | 1 (276) | 4 (27) | 0 (10) |
| Wild type; sid-2::gfp | 96 (775) | 5 (628) | 100 (22) | 0 (19) |
L4 hermaphrodites were soaked in species-specific pal-1 dsRNA for 24 hr and then transferred to culture plates. The fraction of progeny laid during the subsequent 24 hr that hatched (left two columns) and the fraction of strongly affected adults (right two columns) are shown. pal-1 is a more potent environmental RNAi trigger than the other genes we have tested, perhaps reflecting its dual maternal and zygotic functions (28, 29). Furthermore, two nonsense alleles, qt21 and qt32 (Fig. 2), and a deletion allele, gk505, show a stronger environmental RNAi deficiency than qt13 (M.S. C.P.H., and A.M.J., unpublished data). The majority of nonhatched embryos showed posterior defects characteristic of pal-1(RNAi).
*Percent of adults that produced >10 progeny and >30% embryonic lethality.
C. elegans Sensitivity to Environmental dsRNA Is Rare Among Caenorhabditis Nematodes.
To determine whether environmental RNAi sensitivity arose in C. elegans or had been lost in C. briggsae, we undertook a broad analysis of systemic RNAi among a selection of extant Caenorhabditis species (Fig. 4). We generated a set of species-specific RNA polymerase II subunit dsRNA that produced a similar early embryonic arrest phenotype among the progeny embryos (cell cycle arrest around gastrulation) when injected into the gonad of each species, showing that the dsRNA effectively initiated RNAi in each species. To test for systemic RNAi, we measured embryonic lethality among the progeny of intestine- or body-cavity-injected mothers, and to test for environmental RNAi, we measured embryonic lethality among the progeny of mothers soaked in dsRNA overnight. All but one species were sensitive to intestine- or body-cavity-injected dsRNA, showing that systemic RNAi is broadly conserved among Caenorhabditis (SI Table 4). In contrast, only C. elegans and one distantly related unnamed species were sensitive to environmental RNAi. This suggests that sensitivity to environmental RNAi may be rare, or that sensitivity may be regulated, perhaps in ways absent in the Bristol N2 strain. Notably, a wild C. elegans isolate shows reduced RNAi sensitivity (22). Nevertheless, our phylogenetic analysis shows that environmental RNAi has either arisen independently at least twice or been lost independently at least four times within the Caenorhabditis clade; thus this trait appears to have been subject to strong positive and/or negative selection.
It will be interesting, once DNA transformation techniques are developed for these more distant nematodes, to determine whether expressing C. elegans SID-2 in these species can similarly impart environmental RNAi. In an attempt to directly assess SID-2 transport activity, we measured dsRNA uptake in Drosophila S2 cells transfected with SID-2 expression constructs. Similar experiments with SID-1 enabled rapid and robust uptake of dsRNA (16). However, expression of induced SID-2 in S2 cells resulted in only a minimal increase in dsRNA internalization compared with noninduced cells and an indistinguishable increase compared with cells expressing a transport defective mutant version of SID-1 (D. de Jong, personal communication).
Conclusions
Our results delineate two steps in systemic RNAi, whereby sid-2 functions in the intestine to bring dsRNA into the animal, and sid-1 functions to distribute dsRNA into peripheral cells. However, sid-1 is also required for environmental RNAi-mediated silencing in intestinal cells. Therefore, it is probable that SID-2 does not directly mediate transport of dsRNA into the cytoplasm of intestinal cells. This may reflect SID-2-dependent endocytosis of intestinal dsRNA coupled to SID-1-mediated dsRNA efflux from endosomes into the intestinal cytosol, from which it can then be disseminated. Alternatively, SID-2 may mediate transcytosis of dsRNA from the lumen through the intestinal cell to the pseudocoelomic space for subsequent SID-1-mediated transport into pseudocoelom-exposed cells. SID-2 is unlikely to be required generally for endocytosis, because animals depleted for core endocytotic machinery are dead. sid-2 mutant animals are indistinguishable from wild type in development, morphology, and fertility and show no readily apparent defects in intestinal morphology.
C. elegans SID-2 functions in the intestine to mediate uptake of luminal dsRNA and the divergent extracellular portion of SID-2 is likely critical to this activity. The relative sequence divergence of the luminal domain compared with the transmembrane and cytoplasmic domains between the environmental RNAi enabling CeSID-2 and the nonenvironmental enabling homologs from C. briggsae and C. remanei suggest that this protein may function as an environmental sensor, perhaps sensing niche-specific information or acquisition of sequence-specific resistance to pathogens (23, 24). A dietary source of dsRNA could be dsRNA released from ingested and partially digested virions with dsRNA genomes. Alternatively, contact with tissues of infected animals (host, dead nematodes, or infected mother) may mediate sequence-specific immunity to viruses (25), because cells infected with viruses of many types produce abundant dsRNA (26).
Materials and Methods
Strains.
See SI Table 5 for details of strains used in this study. All mutants were generated and characterized in the N2 Bristol background.
sid-2::gfp.
The sid-2 promoter and coding region was amplified from N2 genomic DNA by PCR by using the primers A1 and A3 and digested with AvrII (see SI Table 6 for primer sequences). GFP coding and unc-54 3′ UTR sequences from pPD95.75 were amplified by PCR by using the primers A4 and A5 and digested with SpeI. The two fragments were then ligated in the presence of T4 DNA ligase, AvrII, and SpeI by using NEB buffer 2 (New England Biolabs, Ipswich, MA) and 10 mM ATP. Ligation was done by thermocycling: 20°C for 20 min and37°C for 10 min for 12 cycles. The desired 9.5-kb fragment was gel-purified (Zymoclean; Zymo Research, Orange, CA), reamplified by using primers A1 and A5, and injected at 20 mg/ml into N2 worms. Transformants were identified by GFP expression. A spontaneous integrated line (HC123) was identified.
C. briggsae Expressing C. elegans sid-2::gfp.
C. briggsae strain AF16 was injected with 10 ng/μl sid-2::gfp rescuing fragment along with 20 ng/μl pEON2 [rol-6(su1006)]. F2 lines were established by selecting Rol animals that expressed GFP in the gut lumen. All soaking experiments were performed with transgenic line HC189, which segregated >95% Rol in each generation. The C. elegans transgenic line HC188 (sid-2(qt13); sid-2::gfp) was constructed similarly but segregates 80% Rol in each generation.
Determination of SID-2 Membrane Topology.
sid-2::lacZ fusion constructs were generated, injected, and scored as described for sid-1 (16). Primers A1 and either L1 or L2 were used to amplify sid-2 fragments truncated following the signal sequence and predicted transmembrane, respectively. These were fused to one of two lacZ PCR products, one of which was preceded by a synthetic transmembrane domain. Diluted sid-2 fragments and lacZ products were mixed, reamplified by using nested primers L3 and L4, gel purified, and injected.
Mapping.
sid-2(qt13) was mapped to the right arm of linkage group III by using standard methods and phenotypic markers, chromosomal deficiency tDf10, and single-nucleotide polymorphisms (27) (SI Fig. 5).
Sequencing and Rescue.
The sid-2 rescue fragment (16 kb) was amplified from N2 genomic DNA with primers A1 and A2 and injected at 15 μg/ml with 25 μg/ml pRF4 [rol-6(su1006)] into HC105 worms. F1 worms were scored for sensitivity to bacteria-mediated RNAi of gfp. Successful rescue was also seen with a 7.8-kb fragment amplified with B2 and B6. To sequence mutant DNA, template was amplified from mutant genomic DNA by using primers CF1 and A6. Two independent amplifications were sequenced by using primers CF1, CF2, CF3, CF4, CF5, and CR1.
cDNA.
cDNA corresponding to the 5′ end of sid-2 was amplified from oligo dT primed first-strand cDNA by using primers SL1 and A6 and to the 3′ end by using primers A7 and dT (21). Amplified fragments were gel-purified and cloned into pCR4.1 (Invitrogen, Carlsbad, CA) for sequencing using Big Dye terminator ready reaction mix (Applied Biosystems, Foster City, CA), and M13 forward and reverse primers.
Genetic Mosaics.
sid-2 rescue fragment (amplified with primers B2 and B6) (15 μg/ml), pRF4 (15 μg/ml), and pHC183 (7) (50 μg/ml) were injected into HC105 worms and F2 lines recovered. To obtain sid-2 mosaics exposed to gfp hairpin RNA, five young adult rollers were picked to small drops of Escherichia coli strain OP50, eggs were laid overnight, and then bacteria expressing gfp hairpin RNA was added. The resulting adults were scored for cell autonomy of sid-2 function.
RNAi Methods.
Double-stranded mex-3 RNA was made by in vitro transcription with T7 RNA polymerase and PstI linearized pHC170 [mex-3 hairpin in pCR4.1 (Invitrogen)]. Double-stranded unc-22 hairpin RNA was made by in vitro transcription with T7 RNA polymerase and HindIII linearized pPD128.117. Double-stranded GFP hairpin was made by in vitro transcription with T7 polymerase, and PmeI-linearized pPD126.25. pPD128.117, and pPD126.25 were gifts from A. Fire (Stanford University, Palo Alto, CA). Double-stranded pal-1 RNA was made by in vitro transcription with a PCR product made with T3 and T7 chimeric primers PALT3 and PALT7 from early embryo cDNA, prepared as described in Hill et al. (32). ZK520.1 (primers B1, B2), ZK520.2 (B3, B4), ZK520.3 (B5, B6) RNAi templates were amplified with primers containing T7 RNA polymerase. C. briggsae pal-1 dsRNA corresponding to the 3′ UTR was made by in vitro transcription with T7 and SP6 polymerases and pHC97. All dsRNAs were injected at 1 mg/ml unless otherwise noted. Bacteria-mediated RNAi was performed as described in Winston et al. (7), and soaking-mediated RNAi was performed as described in Maeda et al. (33). mex-3 dsRNA was resuspended in soaking buffer at ≈4 mg/ml and pal-1 and gfp dsRNA at ≈5 mg/ml; 0.5 μl dsRNA solution was placed in the cap of a 0.2-ml PCR tube, and ≈20 young L4 animals were placed in the drop. Five microliters of soaking buffer was placed in the bottom of each tube to maintain humidity inside the tube during incubation. Tubes were sealed and incubated at 20°C for 21–24 h. After incubation, animals were placed on OP50-seeded plates, allowed to lay eggs for ≈24 h at 20°C, and assayed for embryonic lethality at 25°C and GFP expression at 20°C.
RNAi of body-wall-expressed GFP initiated by expression of gfp dsRNA in the pharynx is best viewed after starvation, likely because of the perdurance of GFP in normally growing worms, although starvation-delayed development may provide an additional time period for RNAi to spread. Five L4s were placed on an OP50 E. coli-seeded 60-mm NG plate at 20°C. Two days after bacteria were depleted, a chunk from the starved plate was transferred to a fresh plate at 20°C. The starved larvae (L1–L2) that became adults were observed.
Supplementary Material
Acknowledgments
We thank Andy Fire for plasmids, Celera Genomics for the sur-5::nls-gfp integrated transgene and Daniel Schott and Antony Jose for critical comments on the manuscripts. This work was supported by the National Science Foundation (MCB-01110452).
Abbreviation
- cDNA
complementary DNA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY466439).
This article contains supporting information online at www.pnas.org/cgi/content/full/0611282104/DC1.
References
- 1.Marsh M, Helenius A. Cell. 2006;124:729–740. doi: 10.1016/j.cell.2006.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen I, Christie PJ, Dubnau D. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider A, Marechal-Drouard L. Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 4.Lucas WJ, Lee JY. Nat Rev Mol Cell Biol. 2004;5:712–726. doi: 10.1038/nrm1470. [DOI] [PubMed] [Google Scholar]
- 5.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 7.Winston WM, Molodowitch C, Hunter CP. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- 8.Tabara H, Grishok A, Mello CC. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 9.Timmons L, Fire A. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 10.Orii H, Mochii M, Watanabe K. Dev Genes Evol. 2003;213:138–141. doi: 10.1007/s00427-003-0310-3. [DOI] [PubMed] [Google Scholar]
- 11.Chera S, de Rosa R, Miljkovic-Licina M, Dobretz K, Ghila L, Kaloulis K, Galliot B. J Cell Sci. 2006;119:846–857. doi: 10.1242/jcs.02807. [DOI] [PubMed] [Google Scholar]
- 12.Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, Newcomb RD. Insect Mol Biol. 2006;15:383–391. doi: 10.1111/j.1365-2583.2006.00656.x. [DOI] [PubMed] [Google Scholar]
- 13.Newmark PA, Reddien PW, Cebria F, Alvarado AS. Proc Natl Acad Sci USA. 2003;100:11861–11865. doi: 10.1073/pnas.1834205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakhetia M, Charlton WL, Urwin PE, McPherson MJ, Atkinson HJ. Trends Plants Sci. 2005;10:362–367. doi: 10.1016/j.tplants.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Soares CA, Lima CM, Dolan MC, Piesman J, Beard CB, Zeidner NS. Insect Mol Biol. 2005;14:443–452. doi: 10.1111/j.1365-2583.2005.00575.x. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg EH, Hunter CP. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz EM, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Canaran P, Chan J, Chen N, Chen WJ, Davis P, et al. Nucleic Acids Res. 2006;34:D475–D478. doi: 10.1093/nar/gkj061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson FK, Albert PS, Riddle DL. J Ultrastruct Res. 1983;82:156–171. doi: 10.1016/s0022-5320(83)90050-3. [DOI] [PubMed] [Google Scholar]
- 20.Li X, Greenwald I. Neuron. 1996;17:1015–1021. doi: 10.1016/s0896-6273(00)80231-7. [DOI] [PubMed] [Google Scholar]
- 21.Silhavy TJ, Beckwith JR. Microbiol Rev. 1985;49:398–418. doi: 10.1128/mr.49.4.398-418.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tijsterman M, Okihara KL, Thijssen K, Plasterk RH. Curr Biol. 2002;12:1535–1540. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- 23.Fitch DH. Curr Biol. 2005;15:R655–R658. doi: 10.1016/j.cub.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Hong RL, Sommer RJ. BioEssays. 2006;28:651–659. doi: 10.1002/bies.20404. [DOI] [PubMed] [Google Scholar]
- 25.Schott DH, Cureton DK, Whelan SP, Hunter CP. Proc Natl Acad Sci USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 28.Hunter CP, Kenyon CJ. Cell. 1996;87:217–226. doi: 10.1016/s0092-8674(00)81340-9. [DOI] [PubMed] [Google Scholar]
- 29.Edgar LG, Carr SH, Wang H, Wood WB. Dev Biol. 2001;229:71–88. doi: 10.1006/dbio.2000.9977. [DOI] [PubMed] [Google Scholar]
- 30.Kiontke K, Gavin NP, Raynes Y, Roehrig C, Piano F, Fitch DH. Proc Natl Acad Sci USA. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sudhaus W, Kiontke K. Zootaxa. 2007;1456:45–62. [Google Scholar]
- 32.Hill AA, Hunter CP, Tsung BT, Tucker-Kellogg G, Brown EL. Science. 2000;290:809–812. doi: 10.1126/science.290.5492.809. [DOI] [PubMed] [Google Scholar]
- 33.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.