Abstract
The outer dynein arm-docking complex (ODA-DC) is a microtubule-associated structure that targets the outer dynein arm to its binding site on the flagellar axoneme (Takada et al. 2002. Mol. Biol. Cell 13, 1015–1029). The ODA-DC of Chlamydomonas contains three proteins, referred to as DC1, DC2, and DC3. We here report the isolation and sequencing of genomic and full-length cDNA clones encoding DC3. The sequence predicts a 21,341 Da protein with four EF-hands that is a member of the CTER (calmodulin, troponin C, essential and regulatory myosin light chains) group and is most closely related to a predicted protein from Plasmodium. The DC3 gene, termed ODA14, is intronless. Chlamydomonas mutants that lack DC3 exhibit slow, jerky swimming because of loss of some but not all outer dynein arms. Some outer doublet microtubules without arms had a “partial” docking complex, indicating that DC1 and DC2 can assemble in the absence of DC3. In contrast, DC3 cannot assemble in the absence of DC1 or DC2. Transformation of a DC3-deletion strain with the wild-type DC3 gene rescued both the motility phenotype and the structural defect, whereas a mutated DC3 gene was incompetent to rescue. The results indicate that DC3 is important for both outer arm and ODA-DC assembly.
INTRODUCTION
The eukaryotic flagellar axoneme, characterized by the 9 + 2 arrangement of its microtubules, is an exquisite example of a highly ordered cytoskeletal structure. The perfect assembly of the axoneme, which contains some 250 proteins (Luck et al., 1977), is required for the efficient functioning of the flagellum. Inherent in the assembly process is the correct targeting of the outer dynein arms, which in Chlamydomonas reinhardtii have been estimated to generate as much as 4/5 of the force necessary for flagellar motility (Brokaw, 1994). As a result, the flagella of outer dynein arm-less (oda) strains beat with a greatly reduced beat frequency, leading to a distinct slow, jerky swimming phenotype (Kamiya, 1988). To date, 13 ODA genes (ODA1-ODA13) have been shown to be involved in outer arm assembly (Kamiya, 2002).
The Chlamydomonas outer dynein arm is a microtubule-associated ATPase composed of three heavy chains (α, β, and γ), two intermediate chains (IC1 and IC2, previously referred to as IC78 and IC69, respectively), and several light chains (LC1–) (King, 2000). The heavy chains form both the globular motor domains and the fibrous stems that extend to the base of the dynein complex. The base of the outer arm is permanently bound to the A-tubule of the outer doublet microtubule in an ATP-independent manner. The dynein motor domains transiently interact with the B-tubule of an adjacent outer doublet in an ATP-dependent manner to generate the interdoublet sliding that is converted to bending during flagellar beating (Satir, 1968; Summers and Gibbons, 1971). The generation and coordination of flagellar bending undoubtedly requires precise positioning of the arms relative to each other and to the other components of the axoneme. This positioning is brought about by the outer dynein arm-docking complex (ODA-DC), which is located at the outer-arm-binding site and is necessary for the arms to attach to the doublet microtubule (Takada and Kamiya, 1994; Wakabayashi et al., 2001; Takada et al., 2002).
The ODA-DC of Chlamydomonas contains equimolar amounts of three polypeptides: DC1, DC2, and DC3 (Takada et al., 2002). DC1 and DC2 are predicted to be coiled-coil proteins and are encoded by ODA3 and ODA1, respectively (Koutoulis et al., 1997; Takada et al., 2002). In contrast, nothing is known about DC3. To fully understand the function(s) of the ODA-DC, a detailed knowledge of each of its component parts is necessary. To that end, we describe the isolation and characterization of the gene and a full-length cDNA encoding DC3 and of mutants defective in DC3. The results show that DC3 is a novel member of the EF-hand superfamily of calcium-binding proteins and is important for the assembly of both the outer dynein arm and the ODA-DC.
MATERIALS AND METHODS
Strains
Chlamydomonas reinhardtii strains used in this study include: g1 (nit1, mt+) (Pazour et al., 1995); 1330.1 (nit1, mt–) (Pazour et al., 1999); 137c (mt+); H8–(arg7, mt–) (obtained from Dr. P. Lefebvre, University of Minnesota); CC124 (mt–); CC2664 (ida1, mt+) (Kamiya et al., 1991); CC2290 (mt–) (Gross et al., 1988); CC2229 (oda1, mt–), CC2233 (oda3-1, mt–), CC2236 (oda5, mt+), CC2239 (oda6, mt–), CC2240 (oda7, mt+), CC2242 (oda8, mt+), and CC2245 (oda9, mt–) (Kamiya, 1988); and CC2492 (pf13A, mt+) and CC1382 (pf22, mt+) (Huang et al., 1979). Strains with “CC” numbers and 137c are available from the Chlamydomonas Genetics Stock Center (Department of Botany, Duke University, Durham, NC). Other strains used were as follows: V06 (oda14-1::NIT1, nit1, mt+), V16 (oda14-3::NIT1, nit1, mt+), and V87.2 (oda10::NIT1, nit1, mt+) (Koutoulis et al., 1997), generated by insertional mutagenesis of g1; F28 (oda14-2::NIT1, nit1, mt–) (Koutoulis et al., 1997), generated by insertional mutagenesis of 1330.1; 2111.1 (oda14-2::NIT1, mt–; offspring of F28 × 137c cross); 026.2 (oda14-1::NIT1, arg7, mt+; offspring of V06 × H8–cross); and transformants t026.2-12, D76, and W215, obtained by transforming strain 026.2 with an ODA14 genomic clone and pARG7.8 (Debuchy et al., 1989).
Growth Media
The following media were used: M medium I (Sager and Granick, 1953) modified to contain 0.0022 M KH2PO4 and 0.00171 M K2HPO4 (referred to as M medium in this study); M–N (M medium without nitrogen); R (M medium supplemented with 0.0075 M sodium acetate); R + Arg (R medium supplemented with 50 μg/ml arginine); and TAP (Gorman and Levine, 1965).
DNA and RNA Isolation
For Southern blot analysis, Chlamydomonas genomic DNA was isolated as described by Pazour et al. (1995). In brief, 0.5-ml cell digestion buffer (20 mM Tris, pH 7.5, 20 mM EDTA, 5% SDS, and 1 mg/ml proteinase K) was added to ∼0.3 ml packed cells. The cells were digested overnight at 50°C. Ammonium acetate was added to 1.5 M, and the mixture was extracted once with an equal volume of phenol:chloroform, once with chloroform, precipitated with ethanol, and resuspended in 10 mM Tris-HCl, 1 mM EDTA, pH 8.0. Aliquots of the DNA were digested with BamHI, EcoRI, HindIII, PstI, or XhoI. For northern blot analysis, Chlamydomonas mRNA was isolated exactly as described by Koutoulis et al. (1997). Gel electrophoresis, Southern blotting, and northern hybridization were performed according to standard procedures (Sambrook et al., 1989).
Isolation of the Mr ∼25,000 ODA-DC Protein and Peptide Sequencing
Flagellar axonemes were isolated from CC2239 (oda6) cells using the method of Witman (1986). Axonemes were extracted with HMDE (30 mM HEPES, pH 7.5, 5 mM MgSO4, 1 mM DTT, 0.5 mM EGTA) containing 0.5 M potassium acetate (Nakamura et al., 1997) and then with 0.6 M KCl in HMDEK (HMDE containing 25 mM potassium acetate). The resultant extract was fractionated by 5–20% sucrose density gradient centrifugation under Mg2+-free conditions (Piperno and Luck, 1979; Pfister et al., 1982). The 7S fraction containing the ODA-DC (Takada and Kamiya, 1994) was collected, and the Mr ∼25,000 ODA-DC protein (DC3) was separated from other proteins on a 5–20% acrylamide gradient gel (Takada et al., 2002). The gel was blotted to polyvinylidene difluoride (PVDF) membrane (Immobilon-PSQ; Millipore, Bedford, MA) and stained with Ponceau S. DC3 was excised and partially hydrolyzed with trypsin. The resulting peptide fragments were separated by reverse-phase chromatography, and three were sequenced directly on an ABI/Perkin Elmer-Cetus 494 Procise Protein Sequencer (PE Biosystems, Foster City, CA).
Cloning of DC3 cDNAs
Chlamydomonas strain CC2664 (ida1) was deflagellated by pH shock (Witman et al., 1972). Cells were allowed to regenerate their flagella for 30 min to induce the synthesis of flagellar proteins. Total RNA was isolated from the cells as described in Wilkerson et al. (1994). First-strand cDNA was synthesized from the RNA using oligo-dT as primer and Superscript II Reverse Transcriptase (Life Technologies Inc., Rockville, MD).
To specifically amplify cDNAs encoding DC3, degenerate PCR primers were designed based on DC3 peptide sequence (see above) and the highly restricted codon usage of Chlamydomonas (Harris, 1989; Wilkerson et al., 1994). Primer 25E [5′-CGCGGAATTCAGGAGCT(CGT)AC(CGT)GAGCT(CGT)TT-3′] was designed from the sequence EELTELF in peptide 1, primer 25J [5′-CGCGGGATCCTT(ACG)AG(AG)TC(ACGT)GC(ACG)GT(ACG)CC(AG)AA-3′] was designed from the sequence FGTADLN in peptide 3, and primer 25K [5′-CGCGGGATCCAA(ACG)AC(CT)TGCTC(ACG)AG(CT)TGCAT-3′] was designed from the sequence MQLEQVF in peptide 3. To facilitate cloning of the PCR products, EcoRI or BamHI sites were engineered onto the 5′-ends of the forward (25E) and reverse (25J, 25K) primers, respectively.
cDNAs encoding DC3 were amplified by PCR using the 25E/25J primer pair. The resulting products were used as template for a second round of nested PCR using the 25E/25K primer pair. The resultant PCR products were extracted with phenol/chloroform (50:50), precipitated with ethanol, digested with both EcoRI and BamHI, and ligated into pBluescript II KS (Stratagene, La Jolla, CA) that had been digested with both EcoRI and BamHI and treated with alkaline phosphatase. A 180-base–pair insert (PCR clone 6–10) was sequenced using the Sequenase Version 2.0 DNA Sequencing Kit (USB Corporation, Cleveland, OH) and found to contain additional sequences from peptide 3 not used in primer design. This fragment was labeled with [α-32P]dCTP using the Prime-It II random primer labeling kit (Stratagene) and then used to screen a Lambda ZAP II cDNA library generated from wild-type Chlamydomonas cells (Wilkerson et al., 1995). Six phage clones that hybridized strongly with the probe were identified and the cDNA inserts were recovered by helper phage excision. Insert sizes were determined by digesting the resulting phagemids with both EcoRI and XhoI (Wilkerson et al., 1995). The ends of four of the cDNA clones were sequenced. These data were used to design internal sequencing primers, and one entire clone (p25K2–1) was sequenced using the Sequenase Version 2.0 DNA Sequencing Kit. The sequence of clone p25K2–1 was further confirmed by automated sequencing of both strands (DNA Sequencing Facility, Iowa State University, Ames, IA).
Cloning of DC3 Genomic DNA
The cDNA insert of p25K2-1 (see above) was amplified by PCR, electrophoresed in an agarose gel, and recovered by excising the gel band and extracting the DNA with NaI and GLASSMILK (Qbiogene, Inc., Carlsbad, CA). The DNA was released from the GLASSMILK with 10 mM Tris-HCl, 1 mM EDTA, pH 8.0, and used as template for probe synthesis using the Prime-It II random primer labeling kit. The labeled cDNA was used to screen a Chlamydomonas genomic BAC library (Incyte Genomics, Inc., Palo Alto, CA) and several positive clones were identified. To obtain smaller clones containing the DC3 gene, a positive clone was digested with a panel of restriction enzymes. Digestion with SmaI produced several bands, one of which hybridized with the cDNA probe. This 5.6-kb restriction fragment was cloned into the SmaI site of pBluescript II SK to create pDC3S-2.
Computational Analysis
The Lasergene99 SeqMan II program (DNAStar, Inc., Madison, WI) was used for sequence assembly. The BLASTP program (Altschul et al., 1997) was used to search the nonredundant GenBank CDS translations+PDB+SwissProt+PIR+PRF database for related protein sequences. To determine if DC3 was a member of an existing protein family, we used version 7.7b of Pfam (Bateman et al., 2002). Version 1.82 of ClustalW (Thompson et al., 1994) was used for multiple sequence alignment. Secondary structure was predicted using the PHDsec program (Rost and Sander, 1993; Rost and Sander, 1994; Rost, 1996). Regions of coiled-coil were predicted using the COILS program (Lupas, 1996). The Swiss-Model Automated Comparative Protein Modeling Server version 3.5 (Peitsch, 1995; Peitsch, 1996; Guex and Peitsch, 1997) was used to generate a theoretical three-dimensional model: the model was visualized with Protein Explorer version 1.901 Beta (http://molvis.sdsc.edu/protexpl/index.htm). A phylogenetic tree was constructed using the TreeTop Phylogenetic Tree Prediction service on the GeneBee Molecular Biology Server at Moscow State University, Russia (http://www.genebee.msu.su/services/phtree_reduced.html).
Site-directed Mutagenesis
The Chameleon Site-Directed Mutagenesis Kit (Stratagene) was used to create point mutations in the DC3 gene according to the manufacturer's instructions. A primer (5′-CTATGCCCGCAAGAATTAGCAGAACGAGCTAGAG-3′) was designed to change the codon 14 amino acids downstream of the start methionine from GAG to TAG, resulting in a premature STOP (construct pDC3T-7). The presence of the engineered STOP was confirmed by sequencing (DNA Sequencing Facility, Iowa State University, Ames, IA). Another primer (5′-CTATGCCCGCAAGAATGAGCAGAACGAGCTAGAG-3′) was used to restore the original sequence in a second round of mutagenesis. Restoration of the GAG codon was confirmed by sequencing and the repaired construct was named pDC3G-11.
Transformation
Using the silicon carbide fiber method of Dunahay (1993), we transformed strain 026.2, an arg7 derivative of oda14-1, with the pARG7.8 plasmid containing the cloned ARG7 gene (Debuchy et al., 1989) and plasmid clones containing the DC3 gene. ARG7 transformants were selected on solid R media. Individual transformants were picked into liquid R medium and screened by light microscopy for restored wild-type motility.
Genetic Analysis
Standard methods were used for mating and tetrad analysis (Levine and Ebersold, 1960; Harris, 1989). Briefly, gametes of opposite mating type were mated, and the resulting zygotes were transferred to solid R or R + Arg medium and hatched, and progeny were dissected using a glass needle. Once colonies were large enough, they were transferred to 5 ml of R or R + Arg liquid medium. Approximately 3 d after transfer to liquid medium, motility was scored by light microscopy using dim red illumination. The arg7 phenotype was scored by comparing cell growth on R vs. R + Arg media.
Electron Microscopy
Flagellar axonemes were isolated from cells grown in either M or TAP medium using the method of Witman (1986) and prepared for electron microscopy as described by Kamiya (1988).
SDS-PAGE and Western Blotting
Flagellar axonemes and the detergent-soluble membrane + matrix fraction were isolated using the method of Witman (1986). Proteins were separated by SDS-PAGE (Pfister et al., 1982) and electro-phoretically transferred to PVDF membrane (Immobilon-P, Millipore). Western blotting was performed according to standard procedures (Sambrook et al., 1989) using 5% nonfat dry milk and 1% gelatin from cold water fish skin (Sigma, St. Louis, MO) in 10 mM Tris, pH 7.5, 166 mM NaCl, 0.05% Tween 20 as blocking agent. Proteins of interest were visualized using antibodies against DC1 and DC2 (Wakabayashi et al., 2001), DC3 (see below), or IC140 (Yang and Sale, 1998; obtained from Dr. W. Sale, Emory University). Goat anti-rabbit IgG coupled to horseradish peroxidase (HRP; Pierce, Rockford, IL) was used as the secondary antibody. The HRP conjugate was detected using the LumiGLO Chemiluminescent Substrate Kit from Kirkegaard & Perry Laboratories (Gaithersburg, MD). The relative amount of protein in bands on western blots was quantitated using ImageQuant software (Amersham Biosciences, Piscataway, NJ); mutant values were calculated relative to wild type using either IC140 or RSP-1 as a loading control.
Immunoprecipitation of the ODA-DC
Immunoprecipitation and detection of the immunoprecipitated proteins were carried out exactly as described by Takada et al. (2002). In brief, 0.6 M KCl extracts of V06 and W215 cells were obtained as described above and dialyzed overnight against two changes of buffer in the absence of Mg2+ to dissociate the outer arm from the ODA-DC. The proteins were then biotinylated and immunoprecipitated using the anti-DC1 antibody. Proteins in the immunoprecipitate were separated by SDS-PAGE and transferred to nitrocellulose (Schleicher & Schull, Keene, NH); the biotinylated proteins were detected using streptavidin-HRP (Molecular Probes, Eugene, OR).
Analysis of Swimming Speed
The swimming speed of individual cells was determined using an ExpertVision (EV) motion analysis system (Motion Analysis Corp., Santa Rosa, CA) (Moss et al., 1995). To prevent light-induced behavior, cells were viewed with dim red illumination. The EV system “path” operator was used to track the movement of individual cells. As cells traversed the recording window, their position was recorded every 33 milliseconds (30 frames/s). Once paths were obtained, the EV system “speed” operator was used to calculate the speed of individual cells in micrometers per second. The mean swimming speed for each strain is the average of >100 cells.
Phototaxis and Photoshock
Cells that had been dark-adapted for 3 h were placed in a homemade phototaxis chamber (Moss et al., 1995) that was mounted on the stage of a Zeiss Universal microscope (Thornwood, NY). Randomly swimming cells were initially viewed using dark-field microscopy with dim red illumination. To analyze phototactic behavior, an argon ion laser light (488 nm) was passed through a fiber optic cable attached to one side of the phototaxis chamber. To analyze photoshock, the cells were exposed to a Vivitar 283 electronic camera flash, mounted ∼3 feet away from the microscope stage. The behavior of a population of cells in response to either the laser light stimulus or the camera flash was recorded on videotape by capturing images with a CCD camera mounted on the microscope.
DC3 Antibody Production
A rabbit polyclonal antibody against bacterially expressed DC3 was produced (Research Genetics, Huntsville, AL). Briefly, two New Zealand white rabbits were injected with 0.1 mg each of recombinant DC3 in Freund's complete adjuvant. Approximately every 2 weeks thereafter, the rabbits received another inoculation of recombinant DC3 in Freund's incomplete adjuvant. Week-10 bleeds were tested on whole axonemes from wild-type cells and one prominent band at Mr ∼25,000 was detected by immunoblotting. The rabbits received one more boost of recombinant DC3 before terminal bleeds were taken. For western blotting (see above), the antibody was used unpurified at 1:40,000 dilution.
RESULTS
Sequence and Characterization of cDNA and Genomic DNA Encoding DC3
As the first step toward isolation of a cDNA encoding DC3, we isolated axonemes from the mutant oda6, which lacks the outer dynein arms but retains the ODA-DC (Takada et al., 1994), so that the latter can be obtained free of the former. The ODA-DC was then extracted from the axonemes and partially purified by sucrose density gradient centrifugation (Takada et al., 2002). Proteins in the 7S fraction (containing the ODA-DC) were separated by SDS-PAGE and transferred to PVDF membrane, and the band corresponding to DC3 was excised and digested with trypsin. Peptides were separated by HPLC and selected peptides were sequenced. The following sequences were obtained: KIDAEELTELFLR (peptide 1), TYKAPPPPQKR (peptide 2), and AELDMQLEQVFGTADLNSGK (peptide 3). The sequences of peptide 1 and peptide 3 were then used to design the degenerate oligonucleotide primers 25E, 25J, and 25K (see MATERIALS AND METHODS).
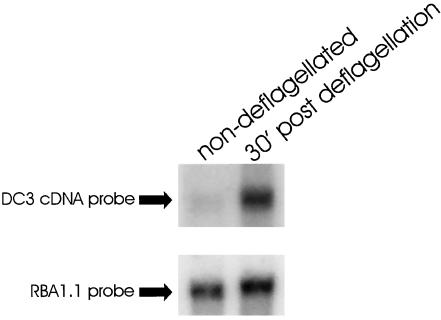
The 25E/25J primer pair and PCR were used to amplify reverse-transcribed cDNA from cells regenerating their flagella (Wilkerson et al., 1994). The reaction produced two major and several minor products. To amplify the product specific for DC3, the 25E/25J PCR products were reamplified with primer 25E and the nested primer 25K. This reaction produced just one prominent species of ∼300-base–pairs, which was subcloned. Sequencing of one of the subclones revealed that it encoded amino-acid sequence present in peptide 3 but not used to design the primers, indicating that the correct cDNA had been amplified. This subclone was then used to isolate 6 cDNA clones from a wild-type cDNA library (Wilkerson et al., 1995). Sequencing of the ends of these clones indicated that they all were derived from the same gene. To verify that the cDNAs encoded a flagellar protein, one of the clones was used to probe mRNA isolated from control and deflagellated wild-type cells. The cDNA hybridized to one prominent band of ∼1.4 kb. As is characteristic of genes encoding flagellar proteins, DC3 gene transcription was upregulated in wild-type cells that had been deflagellated and were actively regrowing their flagella (Figure 1, upper panel). On longer exposure, a very minor amount of a second message of ∼1.8 kb was also detected in the deflagellated samples.
Figure 1.
DC3 gene expression is induced by deflagellation. mRNA was isolated from wild-type cells that were either fully flagellated (non–deflagellated) or actively regrowing their flagella (30′ post deflagellation). mRNA was analyzed by northern hybridization using the DC3 cDNA as a probe (top panel). In non-deflagellated cells, a small amount of DC3 mRNA (∼1.4 kb) is visible; this message increases substantially upon deflagellation and growth of new flagella. With prolonged exposure, a second very minor message of ∼1.8 kb was also observed in the deflagellated samples. A probe (RBA1.1) to the fructose-bisphosphate aldolase gene, a gene involved in glycolysis, was used as a loading control; transcription of this gene is not induced by deflagellation (bottom panel).
The longest clone (p25K2–1) contained a single open reading frame with three in-frame stop codons located upstream of the first ATG (Figure 2A). With the putative translation initiation site at nucleotide position 148, the clone exactly predicts the sequence of the three peptides that were obtained by direct sequencing of tryptic fragments of DC3. Because peptide 2 was not used to obtain the cDNA clone, its presence in the predicted amino acid sequence unequivocally confirms that p25K2–1 encodes DC3. An in-frame stop codon (TGA) is located at nucleotide position 700. At nucleotide 1234 is a consensus polyadenylation signal and the poly A tail begins at nucleotide 1413. The clone predicts a protein product of 184 amino acids with a mass of 21,341 Da, in good agreement with the relative mobility of DC3 in SDS-polyacrylamide gels. We therefore conclude that the p25K2–1 cDNA clone contains the entire DC3 coding region.
Figure 2.
DC3 sequence analysis. (A) Nucleic acid and deduced amino-acid sequence of DC3. The three in-frame STOP codons upstream of the start ATG are in bold. The peptides obtained by direct sequencing of DC3 tryptic fragments are underlined. An asterisk indicates the STOP codon at the end of the DC3 open reading frame. The consensus Chlamydomonas polyadenylation signal is double underlined. Nucleotide and amino-acid numbers are shown on the right (amino-acid numbers are in bold). These sequence data are available from GenBank/EMBL/DDBJ under accession number AY294291. (B) Comparison of the DC3 sequence and its putative homologue from Plasmodium falciparum. The alignment was generated by version 1.82 of ClustalW. Consensus symbols: “*” identical sequence; “:” conserved substitution; “.” semiconserved substitution. (C) Predicted secondary structure of the DC3 protein. The predicted secondary structure is typical of many EF-hand protein family members (reviewed in Kawasaki et al., 1998). The EF-hands are double underlined. L, loop; H, helix; E, strand; “.” no prediction.
In contrast to the other two components of the ODA-DC (Koutoulis et al., 1997; Takada et al., 2002), DC3 is not predicted by the COILS program to form coiled coils. We used BLASTP 2.2.4 (Altschul et al., 1997) to search the database for related proteins. The best match (37% identity, 61% similarity) was to a protein predicted from the genomic sequence of both Plasmodium yoelii yoelii and Plasmodium falciparum. An alignment of DC3 and the protein from P. falciparum, the human malaria parasite, is shown in Figure 2B. Although the parasite proteins have an N-terminal extension making them longer than the Chlamydomonas protein, the identities extend throughout the sequences, suggesting they are true homologues. Secondary structure analysis using the PHDsec program (Rost and Sander, 1993; Rost and Sander, 1994; Rost, 1996) predicted four helix-loop-helix motifs (double underlined in Figure 2C). Analysis of the sequence at the Pfam protein families (Bateman et al., 2002) site (http://pfam.wustl.edu) confirmed that these are EF-hands and that DC3 is a novel member of the EF-hand superfamily of calcium-binding proteins (see DISCUSSION).
The DC3 cDNA was used to select a BAC clone containing the DC3 gene. A 5.6-kb subfragment of this BAC was sequenced and found to contain the entire DC3 gene. Unusual for a Chlamydomonas gene, the DC3 gene contains no introns.
Identification of Mutants with Defects in the DC3 Gene
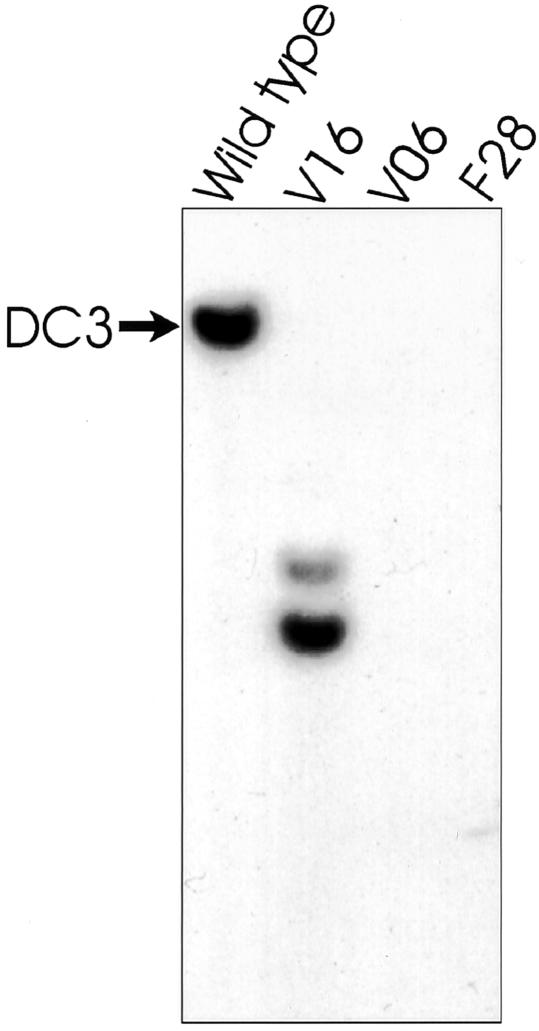
In Chlamydomonas, reverse genetics has been very useful for deducing the function of cloned genes encoding flagellar proteins (Pazour and Witman, 2000). To find a mutant defective in DC3, a collection of insertional mutants having flagellar/cytoskeleton-related phenotypes (Pazour et al., 1995; Koutoulis et al., 1997; Pazour and Witman, 2000) was screened by Southern blot using the DC3 cDNA as probe. The cDNA hybridized to a single band in wild-type genomic DNA digested with PstI (Figure 3, arrow), BamHI, EcoRI, HindIII, and XhoI. These data indicate that only one DC3 gene is present in the Chlamydomonas genome. Two insertional mutants (F28 and V06) lacked hybridizing bands, indicating that the gene was completely deleted (Figure 3). A third strain (V16) gave two hybridizing bands, suggesting that the gene had been disrupted by an insertion (Figure 3). When the DC3 mutants were analyzed by light microscopy, they all had slow, jerky swimming, a hallmark of strains lacking the outer dynein arm (Kamiya, 1988). One of the deletion mutants, V06 (hereafter referred to as the DC3-deletion strain), was selected for further studies. The DC3-deletion strain grew and divided normally, indicating that DC3 is not vital to cell survival. Conclusive evidence that deletion of the DC3 gene specifically is responsible for the mutant phenotype is presented later in this article.
Figure 3.
Identification of insertional mutants with defects in the DC3 gene. Genomic DNA was isolated from insertional mutants having flagellar/cytoskeletal defects, cut with PstI, and analyzed by Southern blot using the DC3 cDNA as a probe. The probe hybridized to a single band (arrow) in wild type, but did not hybridize to any bands in strains V06 or F28. In strain V16, the probe hybridized to two bands, indicating the DC3 gene had been disrupted by an insertion.
DC3-null Axonemes Contain a Partial Complement of Outer Arms and ODA-DCs
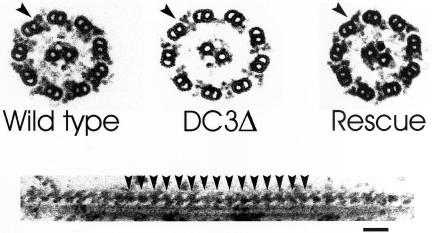
To determine if axonemes of the DC3-deletion strain had an abnormal ultrastructure, we examined them by EM. In contrast to null mutants for DC1 (Koutoulis et al., 1997) and DC2 (Takada et al., 2002), which lack outer arms, some DC3-null outer doublets had outer dynein arms (Figure 4, upper panel, center). Quantitative analysis of 59 DC3-null axonemal cross sections revealed an average of 3.1 outer dynein arms per axonemal cross section. Arms were more abundant in proximal than distal regions of the axoneme: an average of 4.73 outer arms was observed in 22 proximal axonemes (i.e., axonemes with beak structures; Hoops and Witman, 1983), whereas an average of 2.14 outer arms was observed in 37 distal axonemes (i.e., axonemes without beak structures). The arms appeared to be attached to the correct location on the A-tubules; no ectopic location (e.g., on the B-tubules) of outer arms was observed. Although the outer arms were attached at the correct location, a gap was often seen between the outer arm and the A-tubule to which it was attached (Figure 4, upper panel, center, arrowhead), suggesting the arms may not be as tightly associated with the outer doublet microtubule in the absence of DC3. This gap was not observed in wild-type axonemes prepared under identical conditions (Figure 4, upper panel, left). In longitudinal sections, the outer arms appeared to be evenly spaced at 24-nm intervals along the DC3-null axoneme (Figure 4, lower panel, arrowheads), indicating that, when present, they assemble in a cooperative manner.
Figure 4.
Loss of DC3 results in partial loss of outer dynein arms. Top panel: Axonemal cross sections from a wild-type strain (left), the DC3-deletion strain (center), and the DC3-deletion strain rescued by transformation with the DC3 gene (right). DC3-null axonemes do not assemble a full complement of outer dynein arms. A gap was often seen between the A-tubule and the arm (arrowhead), suggesting that the arms are not as tightly associated with the outer doublet in the absence of DC3. Outer dynein arms (arrowhead) are completely restored in the null strain transformed with the cloned DC3 gene. Bottom panel: Longitudinal section showing the 24-nm periodicity of outer dynein arms (arrowheads) along a portion of a DC3-null outer doublet. The normal periodicity indicates that when outer arms are present, they assemble cooperatively. Bar, 50 nm.
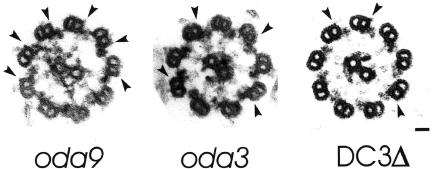
Because the ODA-DC is essential for the correct positioning of outer arms on the axoneme (Koutoulis et al., 1997; Takada et al., 2002), our observation that DC3-null axonemes contain some correctly positioned arms suggested a docking complex was present in these axonemes. If so, it would indicate that a docking complex lacking DC3 is competent to assemble on the axoneme and correctly position some outer dynein arms. To investigate this possibility, we compared axonemal cross sections from a strain lacking outer dynein arms but retaining the ODA-DC (oda9), a strain lacking outer dynein arms and the ODA-DC (oda3), and the DC3-deletion strain. The ODA-DC is visible on oda9 axonemes as a small projection on the A-tubules at the sites where the outer dynein arms would normally attach (Figure 5, left, arrowheads). These projections are missing from the A-tubules of oda3 (Figure 5, center, arrowheads). Because outer arms normally obstruct the docking complex from view, we searched for docking complexes on those A-tubules lacking arms in the DC3-deletion strain. A small projection (Figure 5, right, arrowheads), positioned at sites where the outer arms would normally attach, was indeed present on some but not all doublets of the DC3-deletion strain. We refer to this structure as a “partial” ODA-DC, i.e., one missing DC3.
Figure 5.
The DC3-deletion strain assembles a “partial” docking complex. Axonemal cross sections from a strain lacking outer arms but retaining the ODA-DC (oda9), a strain lacking outer arms and the ODA-DC (oda3), and the DC3-deletion strain (DC3Δ). The ODA-DC is visible on oda9 axonemes as a small projection on the A-tubules at the sites where the outer dynein arms would normally attach (left, arrowheads). These projections are missing from the A-tubules of oda3 (center, arrowheads). DC3-null A-tubules that lacked the outer dynein arm frequently had a structure resembling the docking complex at sites where outer arms would normally attach (right, arrowheads). We refer to this structure as a “partial” docking complex. Bar, 25 nm.
A Complex of DC1 and DC2 Assembles onto DC3-null Axonemes
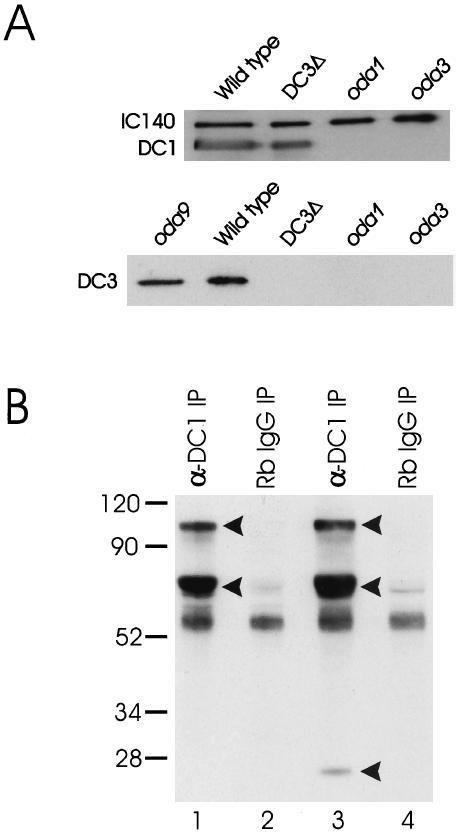
To confirm that DC1 and DC2 assemble onto axonemes of the DC3-deletion strain, we isolated and subfractionated DC3-null flagella to obtain membrane-free axonemes and the detergent-soluble membrane + matrix fraction. We then analyzed the fractions by western blot using antibodies specific for DC1 and DC2. Although there was no signal in the membrane + matrix fraction, DC3-null axonemes contained both DC1 (Figure 6A, upper panel) and DC2 (our unpublished results).
Figure 6.
(A) DC1 and DC2 assemble on the axoneme in the absence of DC3, but not vice versa. Top panel: Axonemes from wild type, the DC3-deletion strain (DC3Δ), oda1, and oda3 were isolated and analyzed by western blotting. oda1 is null for DC2; oda3 is null for DC1. The blot was probed with antibodies to DC1, DC2, and the inner arm IC, IC140 (used as a loading control). As expected, antibodies to DC1 detected protein in wild-type axonemes, but not in oda1 or oda3 axonemes, which lack the ODA-DC. Importantly, the antibody also detected protein in axonemes of the DC3-deletion strain. Essentially identical results were obtained with antibodies to DC2 (our unpublished results). These data indicate that DC1 and DC2 can assemble on the axoneme in the complete absence of DC3. Bottom panel: Axonemes from wild type, the DC3-deletion strain (DC3Δ), oda1, oda3, and oda9 were prepared as above (oda9 has a defect in IC1 and lacks outer dynein arms but retains the ODA-DC). The blot was probed with a polyclonal antibody to DC3. The antibody detects a single protein of Mr ∼25,000 in both oda9 and wild-type axonemes but detects no protein in DC3-null axonemes, indicating that it is specific for DC3. Axonemes from oda1 and oda3 do not contain DC3, indicating that DC3 assembly is dependent on the presence of DC1 and DC2. (B) Immunoprecipitation of the ODA-DC in the absence of Mg2+. The DC1 antibody was used to immunoprecipitate the ODA-DC from biotinylated 0.6 M KCl extracts of DC3-transformant (strain W215) and DC3-null axonemes. The immunoprecipitated proteins were then resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and detected using streptavidin-HRP. The anti-DC1 antibody immuno-precipitated three proteins of Mr ∼105,000, ∼70,000, and ∼25,000 (arrowheads) from DC3-transformant axonemal extracts (lane 3). These proteins, corresponding to DC1, DC2, and DC3, respectively, were not immunoprecipitated from DC3-transformant axonemal extracts using normal rabbit IgG (lane 4). In contrast, the anti-DC1 antibody immunoprecipitated only DC1 and DC2 (arrowheads) from the DC3-null axonemal extracts (lane 1). These proteins were not immunoprecipitated from DC3-null axonemal extracts using normal rabbit IgG (lane 2). These data confirm that a “partial” docking complex composed of DC1 and DC2 assembles on the axoneme when DC3 is missing and that transformation of the DC3-deletion strain with the DC3 gene restores DC3 to the ODA-DC. Numbers on left indicate molecular weight markers.
To determine whether DC1 and DC2 occur together as a complex and to investigate whether any additional polypeptides are contained in the ODA-DC when DC3 is missing, we immunoprecipitated the ODA-DC from biotinylated 0.6 M KCl extracts of DC3-null axonemes using an antibody specific for DC1. Immunoprecipitated proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and detected using streptavidin-HRP. Two major bands corresponding to DC1 and DC2 were detected in the DC3-null immunoprecipitate (Figure 6B, lane 1). Except for bands present in the normal IgG control (Figure 6B, lane 2), no other bands were observed, indicating that no other protein substitutes for DC3 when DC3 is absent. These data confirm that a docking complex containing only DC1 and DC2 is competent to assemble on the axoneme in the complete absence of DC3. However, “partial” ODA-DCs apparently do not bind outer arms as well as wild-type ODA-DCs, because not all “partial” ODA-DCs are occupied by arms (Figure 5, right panel).
As an independent test of our ultrastructural observation that the number of ODA-DCs is reduced in the DC3-deletion strain, we quantitatively analyzed gels such as that shown in Figure 6A (upper panel) to determine the relative amounts of DC1 and DC2 in wild-type vs. DC3-null axonemes. The data were normalized to those for the inner arm intermediate chain IC140 and the radial spoke protein RSP-1 (unpublished data), both of which are expected to be present in normal amounts in the mutant strain. The results indicated that the amounts of DC1 and DC2 in the mutant axonemes were ∼83% and ∼81% (average of two experiments) of their levels in the wild-type axonemes, confirming that the mutant does not assemble a full complement of ODA-DCs. Similar analyses revealed that DC3-null axonemes contain less than one-half the normal levels of the outer dynein arm subunits IC1 and IC2, confirming that the loss of outer arms is greater than the loss of ODA-DCs in the mutant strain.
DC3 Cannot Assemble in the Absence of DC1 and DC2
Our finding that a “partial” docking complex composed of only DC1 and DC2 could assemble in the DC3-deletion strain raised the question of whether DC3 could assemble onto the axoneme independently of DC1 and DC2. To investigate this, we probed western blots of DC1-null (oda3) and DC2-null (oda1) axonemes with our antibody to DC3 (Figure 6A, lower panel). The antibody detected one prominent axonemal protein of Mr ∼25,000 in both wild type and oda9 (which lacks outer arms but has the ODA-DC). No protein was detected in axonemes from the DC3-deletion strain, indicating that the antibody is specific for DC3. The DC3 antibody did not react with any proteins in axonemes isolated from the DC1- or DC2-null strains. Therefore, although DC1 and DC2 can assemble in the absence of DC3, DC3 cannot assemble in the absence of the other two docking complex components. Moreover, because the only known structural difference between oda1, oda3, and oda9 is that oda9 has the ODA-DC, whereas oda1 and oda3 do not, these results provide additional evidence that DC3 is part of the ODA-DC.
DC3-null Outer Arms Can Generate Force
oda mutants completely lacking outer dynein arms typically swim in a jerky manner at about one-third the speed of wild type (Kamiya, 1988). Although the DC3 mutants are slow, jerky swimmers, it was important to measure their swimming speed to see if the arms that were present were functional. If they were not functional, the swimming speed should be comparable to that of an oda mutant completely lacking the outer arms; if they were functional, the swimming speed should be intermediate between that of such a mutant and wild type. Therefore, we compared the swimming speed of the DC3-deletion strain to that of oda9 (completely lacking outer dynein arms) and g1 (wild type). As expected, the mean swimming speed of oda9 (39 μm/s) was about one-third the mean swimming speed of wild type (116 μm/s). In contrast, the mean swimming speed of the DC3-deletion strain (61 μm/s) was about one-half that of wild type, indicating that the outer arms that assemble in the absence of DC3 are force-generating and functional.
DC3-null Mutants Have an Altered Photoshock Response
Chlamydomonas displays two distinct behaviors in response to light: phototaxis and photoshock (Witman, 1993). Phototaxis is directed forward swimming toward or away from a light source. Photoshock, which occurs in response to an intense flash of light, is a sudden stop in forward swimming, brief backward swimming, and then resumed forward swimming. An essential feature of photoshock is a change in outer arm activity: although still able to phototax, outer armless strains have an altered photoshock response. They stop in response to a flash of bright light but either cannot generate the symmetrical waveform needed for backward swimming (Kamiya and Okamoto, 1985) or produce small bend amplitudes with their flagella improperly aligned (Mitchell and Rosenbaum, 1985). These data indicate that the outer arm is important for normal backward swimming during photoshock (Kamiya and Okamoto, 1985).
We tested the DC3-deletion strain's ability to phototax and photoshock (see MATERIALS AND METHODS). Although phototaxis was normal, the photoshock response was altered; the cells stopped in response to an intense flash of light, but little or no backward swimming was evident. Thus, although the outer dynein arms in the DC3-deletion strain can generate force for forward swimming (see above), either the absence of DC3 or the incomplete number of arms precludes normal backward swimming during the photoshock response.
The DC3-null Phenotype Can Be Rescued by Transformation with DNA Containing DC3
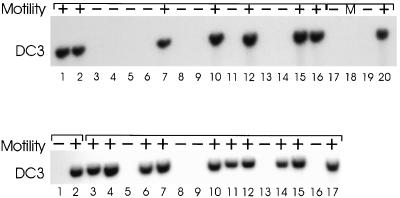
To determine if the slow, jerky swimming was directly linked to the DC3 gene deletion, we crossed the DC3-deletion strain to a wild-type strain of opposite mating type. Offspring from the cross were scored for motility by light microscopy. DNA was isolated from one product each of 16 different tetrads, cut with PstI, blotted, and probed with the DC3 cDNA. As shown in Figure 7 (upper panel), all progeny lacking the DC3 gene had mutant motility, whereas those inheriting a DC3 gene had normal motility. These results strongly suggest that the DC3 gene deletion is responsible for the mutant phenotype.
Figure 7.
Top panel: The slow-swimming motility phenotype and the DC3 gene deletion cosegregate. The DC3-deletion strain was crossed to a wild-type strain, tetrads were dissected, and progeny from 16 different tetrads (lanes 1–16) were scored for motility by light microscopy and for the presence of the DC3 gene by Southern blotting. Whenever a cell line lacked the DC3 gene, it also had mutant motility (–). Lanes 17 and 19 contain DNA from the two DC3-deletion strains, V06 and F28, respectively. Lane 20 contains DNA from a wild-type strain. Bottom panel: Wild-type motility (+) segregates with the DC3 transgene. A DC3-deletion strain that had been rescued by transformation with the DC3 gene was crossed to a DC3-deletion strain of opposite mating type. Tetrads from the cross were dissected and the progeny scored for motility. DNA was isolated from the DC3-deletion strain (lane 1), the rescued strain (lane 2), and from randomly selected progeny representing 15 independent tetrads (lanes 3–17). Samples were analyzed by Southern blotting using the DC3 cDNA as a probe. Wild-type motility was observed only in progeny containing the inserted DC3 gene. +, wild-type motility; –, mutant motility; M, marker.
A frequently used procedure for determining whether a disruption or deletion of a particular gene is responsible for a mutant phenotype is to transform the mutant with a wild-type copy of the gene and see if it rescues the phenotype. Therefore, we cotransformed the DC3-deletion strain with a 5.6-kb fragment of genomic DNA (pDC3S-2) containing the DC3 gene and a separate DNA containing a selectable marker (see MATERIALS AND METHODS). Wild-type motility was restored in ∼21% (53/253) of the cell lines that also took up the selectable marker. Genomic DNA was isolated from randomly picked transformants and probed for integration of the DC3 DNA by Southern blotting. Whenever a transformant had wild-type motility, it also had a DC3-positive genomic insertion (our unpublished results). Moreover, when one of the rescued strains was crossed to one of the DC3-deletion strains and the progeny from 15 different tetrads analyzed, wild-type motility always correlated with the presence of a DC3-positive insertion (Figure 7, lower panel). These results conclusively demonstrate that the restored motility was due to the presence of the integrated DC3 DNA. The results also indicate that the DC3 DNA used for transformation contains all the 5′ and 3′ sequences necessary for DC3 expression.
We measured the swimming velocity of a DC3-transformant to determine if its speed had been restored to wild-type levels. The DC3-transformant swam at a speed of 92 μm/s, a number comparable to that of wild type (see above). We also examined the rescued strain by EM to determine whether all the outer arms had been restored. Axonemal cross sections from the rescued strain showed the complete restoration of outer dynein arms (Figure 4, upper panel, right; one of the nine doublets normally does not have outer arms [Hoops and Witman, 1983]). In addition, the rescued strain displayed normal backward swimming during the photoshock response. To see if DC3 had been restored to the ODA-DC, we immunoprecipitated the ODA-DC from biotinylated 0.6 M KCl extracts of DC3-transformant axonemes using an antibody specific for DC1. The DC3-transformant extract was treated exactly as the DC3-null extract (see above). Three major bands corresponding to DC1, DC2, and DC3 were detected in the DC3-transformant immunoprecipitate (Figure 6B, lane 3). These data indicate that transformation of the DC3-deletion strain with DNA containing DC3 completely rescues the mutant phenotype.
The DC3 Gene Is Responsible for the Rescued Phenotype
The above results demonstrated that DNA containing the DC3 gene could rescue the DC3-null phenotype. However, because the intronless DC3 gene is only ∼1.4 kb, there was a possibility that the 5.6-kb rescuing clone contained one or more additional genes. If the insertional event that deleted DC3 also deleted a gene tightly linked to DC3 and if this gene were included in the 5.6-kb clone, it, rather than the DC3 gene, may have been responsible for rescuing the mutant phenotype. Therefore, to ascertain if the rescue was specifically due to DC3, we used site-directed mutagenesis to create a premature STOP codon in the DC3 gene and then transformed the mutagenized DNA into the DC3-deletion strain. Out of 495 transformants screened, none were rescued. To confirm that the modified gene's failure to rescue was due to the introduction of the STOP codon and not to some other mutation introduced during the mutagenesis procedure, we corrected the premature STOP by recreating the original sequence in a second round of mutagenesis. Transformation with the repaired DC3 gene restored wild-type swimming in 32 of 157 transformants screened, an efficiency of rescue comparable to that obtained with the wild-type gene (see above). These data demonstrate that DC3 is the only gene within the genomic clone that can rescue the DC3-deletion strain, thereby confirming that loss of DC3 is responsible for the DC3-null phenotype.
The DC3 Gene Is a Novel ODA Gene
To determine whether the DC3 gene was a novel gene or one of the known but still uncharacterized ODA genes, we crossed 137c-derived lab strains carrying mutations in the latter genes to the wild-type Chlamydomonas field isolate CC2290 (Gross et al., 1988). The CC2290 and 137c strains are interfertile yet polymorphic such that when genomic DNA from the two strains is probed with the DC3 cDNA, an RFLP is detected (our unpublished results). The presence of a DC3 RFLP and the ability to score motility by light microscopy makes the identification of parental and nonparental offspring from these crosses straightforward. For instance, if the DC3 gene is either identical to or tightly linked to one of the mutant genes, all progeny with the mutant phenotype will have the 137c version of the DC3 RFLP (because the mutants are in the 137c background). Alternatively, if the DC3 gene is not linked to the previously identified genes, then the mutant phenotype and the 137c version of the DC3 RFLP will segregate independently. Using the cDNA insert of p25K2–1 as a probe, we screened the progeny of crosses between CC2290 and oda5, oda7, oda8, pf13a, and pf22 by Southern blot and scored motility by light microscopy. In all cases, the mutant phenotype and the 137c version of the DC3 RFLP segregated independently (Table 1).
Table 1.
Segregation of the DC3 RFLP
| Crossa | Resultsb |
|---|---|
| CC2290 (mt-, ODA5, DC3G) × CC2236 (mt+, oda5, DC3R) | 4P:6NP |
| CC2290 (mt-, ODA7, DC3G) × CC2240 (mt+, oda7, DC3R) | 1P:9NP |
| CC2290 (mt-, ODA8, DC3G) × CC2242 (mt+, oda8, DC3R) | 6P:4NP |
| CC2290 (mt-, PF13A, DC3G) × CC2492 (mt+, pf13A, DC3R) | 4P:4NP |
| CC2290 (mt-, PF22, DC3G) × CC1382 (mt+, pf22, DC3R) | 4P:6NP |
Genotypes are shown in parentheses; DC3G and DC3R are the CC2290 and 137c versions of the PstI RFLP detected by the DC3 cDNA clone.
P, parental; NP, nonparental.
Lastly, to determine whether the DC3 gene was identical to the uncloned ODA10 gene, we screened an oda10 insertional mutant (Koutoulis et al., 1997) by Southern blot using the DC3 cDNA as a probe. No RFLPs were detected. Taken together, these data suggest that the DC3 gene is a previously unidentified gene that affects outer dynein arm assembly. We have named this new gene ODA14; the alleles of the two deletion strains V06 and F28 are termed oda14-1 and oda14-2, respectively, and that of the V16 strain is termed oda14-3. RFLP analysis (courtesy of Dr. C. Silflow, University of Minnesota) was used to map the ODA14 locus to Chlamydomonas Linkage group XV (http://www.biology.duke-.edu/chlamy_genome/nuclear_maps.html).
DISCUSSION
Different ODA-DC Subunits Have Distinct Functions
It has been proposed that DC1 and DC2 interact via their coiled-coil domains to form a heterodimeric rod-shaped structure, and further, that the heterodimers polymerize to form a filament running longitudinally along the A-tubule (Koutoulis et al., 1997; Takada et al., 2002). Recent cross-linking experiments (Takada et al., 2002) have demonstrated that these proteins indeed interact with one another, supporting the heterodimer hypothesis. By analogy to the arrangement of tropomyosin and troponin on actin filaments, these results are consistent with a model in which DC1 and DC2 form a filament with DC3 attached peripherally (see Figure 6 of Takada et al., 2002).
In addition to attaching to specific sites on the A-tubules, the outer dynein arms occur at 24-nm intervals along their length. How this periodicity is established is unclear; however, using immuno-EM, Wakabayashi et al. (2001) showed that in the absence of outer arms DC1 itself has a 24-nm periodicity. These data suggest that the ODA-DC specifies outer arm periodicity (Wakabayashi et al., 2001). Our data indicate that when outer arms are present on doublet microtubules of the DC3-deletion strain, their periodicity is preserved (see Figure 4, lower panel). Because DC1 and DC2 are present on DC3-null axonemes, it seems likely that these two subunits alone are sufficient to determine outer arm periodicity, suggesting that different ODA-DC subunits have distinct functions. Indeed, our results show that DC1 and DC2 can assemble onto the axoneme independently of DC3, but not vice versa. Although outer dynein arms can attach to sites that have “partial” ODA-DCs, not all “partial” ODA-DCs are occupied by arms. In addition, not all ODA-DC sites are occupied by “partial” ODA-DCs. We therefore conclude that DC3 enhances attachment of the ODA-DC to its site and enhances attachment of the outer arms to the ODA-DC.
DC3 as a Potential Calcium Sensor
The identification of DC3 as a novel member of the EF-hand superfamily of calcium-binding proteins raises the intriguing possibility that it plays a role in calcium-regulated outer dynein arm activity. Backward swimming during the photoshock response of Chlamydomonas is triggered by a sudden increase in the intraflagellar calcium concentration (for reviews see Witman, 1993 and Pazour et al., 1995). Although outer dynein arms are dispensable for forward swimming, outer dynein armless mutants show little or no backward movement during photoshock (Kamiya and Okamoto, 1985; Mitchell and Rosenbaum, 1985). These data imply there is a protein that senses the rise in intraflagellar calcium and relays the information to the outer arms, which respond by altering their activity in such a way that the cell swims backwards. One candidate for this Ca2+-sensor is LC4, a Ca2+-binding outer arm dynein light chain (King and Patel-King, 1995). However, inasmuch as the ODA-DC is in direct contact with the outer arm (Takada et al., 2002), DC3 also must be considered a candidate for the Ca2+ sensor. The behavior of the DC3-deletion strain lends support to this hypothesis: although the DC3-deletion strain retains some outer arms, it behaves like an outer-armless strain during photoshock.
The Human Malaria Parasite Has a DC3 Homologue
DC3 is 37% identical (61% similar) to a protein predicted from the genomic sequence of P. falciparum, the human malaria parasite. The high degree of sequence conservation between these proteins suggests that the Plasmodium protein is the functional homologue of Chlamydomonas DC3. The protein probably functions in assembly of the outer dynein arms in male sexual stage parasites, which have a typical “9 + 2” flagellar axoneme (Garnham et al., 1967). Interestingly, a similarity search using Chlamydomonas DC1 and DC2 sequences revealed no Plasmodium homologues. This result was surprising in light of our finding that DC3 requires DC1 and DC2 for assembly onto the axoneme of Chlamydomonas. However, the parasite protein has an N-terminal extension that is lacking in DC3. Perhaps this domain, through interactions with other axonemal proteins, assembles DC3 onto the axoneme in the absence of DC1 and DC2. Alternatively, Plasmodium may have DC1 and DC2 homologues that were missed by the similarity searches because of lack of sequence conservation. Another, not mutually exclusive, possibility is that the Plasmodium DC3 gene was acquired by lateral gene transfer from a Chlamydomonas-like endosymbiont, as appears to have been the case for the parasite's COXII genes (Funes et al., 2002). In this case, the Plasmodium DC3 homologue would be closely related to Chlamydomonas DC3, whereas its DC1 and DC2 homologues might not be closely related to algal DC1 and DC2.
DC3 Is a New CTER Member
A comparison of DC3's EF-hand sequences to a database of EF-hand proteins (courtesy of Dr. H. Kawasaki, Yokohama City University, Japan) indicates DC3 is a new member of CTER, a group of congruent EF-hand subfamilies that includes calmodulin, troponin C, and the essential and regulatory light chains of myosin. The second and third EF-hands of DC3 resemble other EF-hands that are known to bind calcium (∼30% of known EF-hands do not bind calcium; Kawasaki et al., 1998). The sequence of the second EF-hand appears more canonical (see Figure 4 of Kawasaki et al., 1998, for a description of the canonical EF-hand). It is of interest that the calcium-binding loop of the second EF-hand contains two cysteine residues; this is the only known member of the CTER group to have this feature and raises the possibility that DC3's function is regulated on two different levels—through redox state and calcium binding.
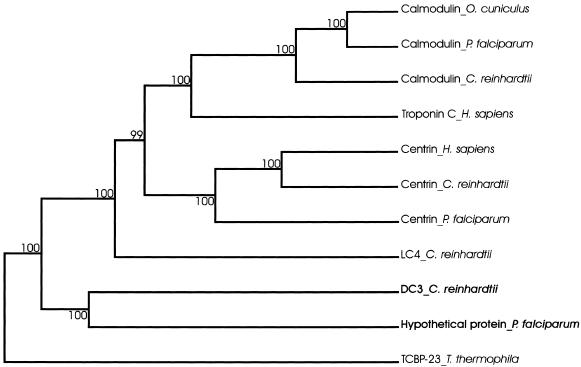
In a phylogenetic analysis of CTER family proteins (Figure 8), Chlamydomonas DC3 and the Plasmodium protein grouped together, supporting the prediction that they are true homologues. DC3 grouped independently of calmodulin, troponin C, centrin, and the calcium-binding light chain of Chlamydomonas outer arm dynein (King and Patel-King, 1995), indicating DC3 represents a distinct subfamily within CTER.
Figure 8.
Phylogenetic tree showing the relatedness of DC3 to other CTER members. A cluster algorithm was used to build the tree. Branch lengths represent evolutionary relatedness. Numbers at branch points are bootstrap values. The 23-kDa T. thermophila calcium-binding protein (TCBP-23), which is an EF-hand protein but not a member of the CTER group, was used to root the tree. DC3 and its putative P. falciparum homologue (in bold) group independently of other CTER members, indicating they form a distinct subfamily within CTER. Protein names are followed by organism names; LC4 is the 18-kDa calcium-binding outer dynein arm light chain, O. cuniculus is rabbit. Accession numbers (in parentheses) are as follows: Calmodulin_O. cuniculus (1003191A); Calmodulin_P. falciparum (P24044); Calmodulin_C. reinhardtii (P04352); Troponin C_H. sapiens (P02590); Centrin_H. sapiens (NP_004335); Centrin_C. reinhardtii (P05434); Centrin_P. falciparum (NP_702332); LC4_C. reinhardtii (Q39584); DC3_C. reinhardtii (AY294291); Hypothetical protein_P. falciparum (NP_702309); and TCBP-23_T. thermophila (P20473).
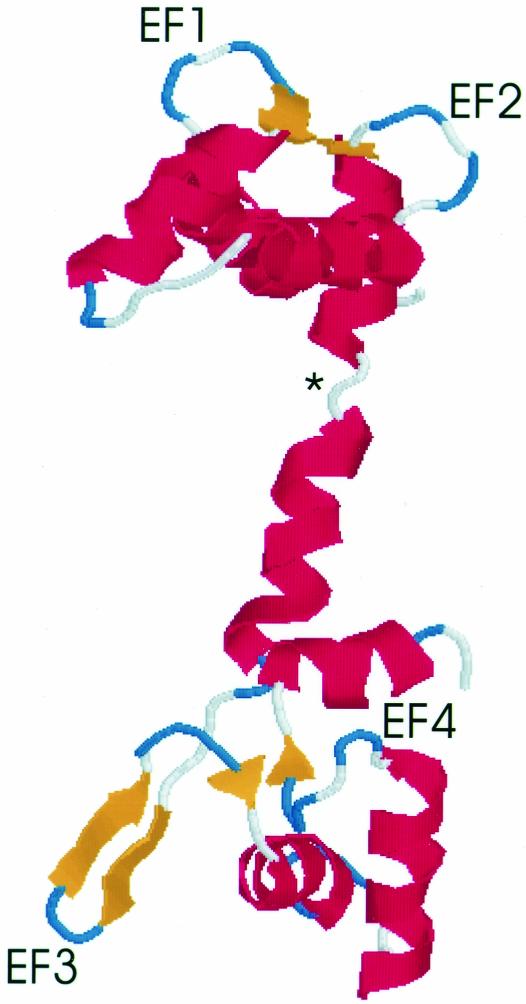
A molecular model of DC3 was generated using Swiss-Model (Peitsch, 1995; Peitsch, 1996; Guex and Peitsch, 1997). The homology model is based on the crystal structure of recombinant calmodulin from Drosophila melanogaster (Taylor et al., 1991). Eight amino acids from the NH2-terminus and 24 amino acids from the COOH-terminus were not included in the DC3 model because they could not be matched with corresponding sequence from the smaller calmodulin protein. The theoretical model was rendered in Protein Explorer and is shown in Figure 9. The predicted overall structure of DC3 is very similar to the solved calmodulin structure (Taylor et al., 1991) with a pair of EF-hands located at each end of the molecule. However, the third EF-hand is predicted to form a β-hairpin (two adjacent antiparallel β-strands connected by a loop); a β-hairpin is similarly predicted for the third EF-hand of the Plasmodium protein (unpublished data). The extended geometry of the β-strands would most likely preclude the coordination of a calcium ion by this EF-hand. As in calmodulin (Barbato et al., 1992; Ikura et al., 1992; Meador et al., 1992), a loop region in the middle of the molecule (Figure 9, asterisk) breaks into two helices what otherwise would be a long alpha helix.
Figure 9.
Homology model of DC3. The atomic coordinates of DC3 were assigned by Swiss-Model using calmodulin (Taylor et al., 1991) as a template. The theoretical model was rendered in Protein Explorer; the amino terminus is at the top. The four EF-hands are labeled EF1–EF4. Short antiparallel β-sheets are predicted to link adjacent loop regions, perhaps stabilizing the EF-hands (Taylor et al., 1991). An asterisk marks the predicted nonhelical region that breaks what would be a long central helix into two shorter helices. Red: alpha-helices; blue and white: loops; yellow: beta strands.
Use of Site-directed Mutagenesis To Identify Gene Responsible for Mutant Phenotype
We used site-directed mutagenesis to demonstrate conclusively that the DC3 gene was the only gene within the genomic clone that could rescue the DC3-deletion strain and therefore that the mutant phenotype was caused by loss of DC3. These results demonstrate the general applicability of site-directed mutagenesis (and correction) to determine whether a particular gene is responsible for a mutant phenotype. This approach provides an alternative to the more traditional but time-consuming method of cutting back the genomic clone until the smallest piece of rescuing DNA is found. The new approach saves time because it obviates the need to subclone progressively smaller genomic fragments and to perform many transformations with each. In addition, only a small amount of sequence is needed to create a point mutation encoding a premature STOP; this should be particularly advantageous for large genes.
The Intronless DC3 Gene May Be Useful as a cDNA Expression Vector
A conspicuous feature of most Chlamydomonas nuclear genes is the presence of several small intronic sequences (Silflow, 1998). Although some intronless genes have been described (Fabry et al., 1995; Fischer and Rochaix, 2001), introns are believed to play an important role in augmenting gene expression (Lumbreras et al., 1998). This view is supported by the limited success in expressing cDNAs in Chlamydomonas (reviewed in Fischer and Rochaix, 2001). The finding that the DC3 coding sequence lacks introns indicates that the 5′- and 3′-genomic regions must contain all the information necessary for efficient gene expression. Therefore, the DC3 flanking regions could potentially be used to generate a vector for the efficient expression of any Chlamydomonas cDNA. Recently, an expression vector incorporating the flanking regions of the intronless PsaD gene (PsaD is a nuclear gene encoding a chloroplast protein) was used to express the nuclear-encoded PsaF chloroplast protein and argininosuccinate lyase from their cDNAs (Fischer and Rochaix, 2001). An expression vector using the DC3 5′- and 3′-genomic regions may be particularly useful for expressing flagellar proteins because the DC3 promoter is sensitive to deflagellation (see Figure 1).
Acknowledgments
The authors thank Dr. J. Leszyk of the UMMS Proteomic Mass Spectrometry lab for peptide sequencing and Dr. A. Koutoulis for the initial identification of V06, V16, and F28 as “partial” oda mutants. This work was supported by National Institutes of Health (NIH) grant GM30626 to G.W., by the Robert W. Booth Fund at the Greater Worcester Community Foundation (G.W.), by an NIH Individual National Research Service Award (Predoctoral) to D.C., by a Summer Program in Japan fellowship from the National Science Foundation and the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT) to D.C., by a graduate research fellowship from the Worcester Foundation for Biomedical Research Auxiliary to D.C., by a fellowship from the Japan Society for the Promotion of Science to S.T., by a postdoctoral fellowship from The Lalor Foundation to K.W., and by a grant from MEXT to R.K. We also gratefully acknowledge the Diabetes Endocrinology Research Center grant DK32520 for support of core facilities used in this research.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0057. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0057.
Abbreviations used: BAC, bacterial artificial chromosome; EM, electron microscopy; EV, ExpertVision; HRP, horseradish peroxidase; IC, dynein intermediate chain; LC, dynein light chain; ODA-DC, outer dynein arm-docking complex; PCR, polymerase chain reaction; PVDF, polyvinylidene difluoride; RFLP, restriction fragment length polymorphism.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato, G., Ikura, M., Kay, L.E., Pastor, R.W., and Bax, A. (1992). Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry 31, 5269–5278. [DOI] [PubMed] [Google Scholar]
- Bateman, A. et al. (2002). The Pfam protein families database. Nucleic Acids Res. 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw, C.J. (1994). Control of flagellar bending: a new agenda based on dynein diversity. Cell Motil. Cytoskel. 28, 199–204. [DOI] [PubMed] [Google Scholar]
- Debuchy, R., Purton, S., and Rochaix, J.D. (1989). The arginosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8, 2803–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunahay, T.G. (1993). Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. BioTechniques 15, 452–454. [PubMed] [Google Scholar]
- Fabry, S., Muller, K., Lindauer, A., Park, P.B., Cornelius, T., and Schmitt, R. (1995). The organization structure and regulatory elements of Chlamydomonas histone genes reveal features linking plant and animal genes. Curr. Genet. 28, 333–345. [DOI] [PubMed] [Google Scholar]
- Fischer, N., and Rochaix, J.-D. (2001). The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genom. 265, 888–894. [DOI] [PubMed] [Google Scholar]
- Funes, S., Davidson, E., Reyes-Prieto, A., Magallon, S., Herion, P., King, M.P., and Gonzalez-Halphen, D. (2002). A green algal apico-plast ancestor. Science 298, 2155. [DOI] [PubMed] [Google Scholar]
- Garnham, P.C.C., Bird, R.G., and Baker, J.R. (1967). Electron microscope studies of motile stages of malaria parasites. V. Exflagellation in Plasmodium, Hepatocystis and Leucocytozoon. Trans. R. Soc. Trop. Med. Hyg. 61, 58–68. [DOI] [PubMed] [Google Scholar]
- Gorman, D.S., and Levine, R.P. (1965). Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54, 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, C.H., Ranum, L.P.W., and Lefebvre, P.A. (1988). Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13, 503–508. [DOI] [PubMed] [Google Scholar]
- Guex, N., and Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press. [DOI] [PubMed]
- Hoops, H.J., and Witman, G.B. (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97, 902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Piperno, G., and Luck, D.J.L. (1979). Paralyzed flagella mutants of Chlamydomonas reinhardtii defective for axonemal doublet microtubule arms. J. Biol. Chem. 254, 3091–3099. [PubMed] [Google Scholar]
- Ikura, M., Clore, G.M., Gronenborn, A.M., Zhu, G., Klee, C.B., and Bax, A. (1992). Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science 256, 632–638. [DOI] [PubMed] [Google Scholar]
- Kamiya, R. (1988). Mutations at twelve independent loci result in absence of outer dynein arms in Chlamydomonas reinhardtii. J. Cell Biol. 107, 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R. (2002). Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 219, 115–155. [DOI] [PubMed] [Google Scholar]
- Kamiya, R., Kurimoto, E., and Muto, E. (1991). Two types of Chlamydomonas flagellar mutants missing different components of inner arm dynein. J. Cell Biol. 12, 441–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya, R., and Okamoto, M. (1985). A mutant of Chlamydomonas that lacks the flagellar outer dynein arm but can swim. J. Cell Sci. 74, 181–191. [DOI] [PubMed] [Google Scholar]
- Kawasaki, H., Nakayama, S., and Kretsinger, R.H. (1998). Classification and evolution of EF-hand proteins. BioMetals 11, 277–295. [DOI] [PubMed] [Google Scholar]
- King, S.M. (2000). The dynein microtubule motor. Biochim. Biophys. Acta 1496, 60–75. [DOI] [PubMed] [Google Scholar]
- King, S.M., and Patel-King, R.S. (1995). Identification of a Ca2+-binding light chain within Chlamydomonas outer arm dynein. J. Cell Sci. 108, 3757–3764. [DOI] [PubMed] [Google Scholar]
- Koutoulis, A., Pazour, G.J., Wilkerson, C.G., Inaba, K., Sheng, H., Takada, S., and Witman, G.B. (1997). The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J. Cell Biol. 137, 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, R.P., and Ebersold, W.T. (1960). The genetics and cytology of Chlamydomonas. Annu. Rev. Microbiol. 14, 197–216. [DOI] [PubMed] [Google Scholar]
- Luck, D., Piperno, G., Ramanis, Z., and Huang, B. (1977). Flagellar mutants of Chlamydomonas: studies of radial spoke-defective strains by dikaryon and revertant analysis. Proc. Natl. Acad. Sci. USA 74, 3456–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumbreras, V., Stevens, D.R., and Purton, S. (1998). Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14, 441–447. [Google Scholar]
- Lupas, A. (1996). Prediction and analysis of coiled-coil structures. Methods Enzymol. 266, 513–525. [DOI] [PubMed] [Google Scholar]
- Meador, W.E., Means, A.R., and Quiocho, F.A. (1992). Target enzyme recognition by calmodulin: 2.4 Å structure of a calmodulin-peptide complex. Science 257, 1251–1255. [DOI] [PubMed] [Google Scholar]
- Mitchell, D.R., and Rosenbaum, J.L. (1985). A motile Chlamydomonas flagellar mutant that lacks outer dynein arms. J. Cell Biol. 100, 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, A.G., Pazour, G.J., and Witman, G.B. (1995). Assay of Chlamydomonas phototaxis. Methods Cell Biol. 47, 281–287. [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Wilkerson, C.G., and Witman, G.B. (1997). Functional interaction between Chlamydomonas outer arm dynein subunits: the γ subunit suppresses the ATPase activity of the αβ dimer. Cell Motil. Cytoskel. 37, 338–345. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., and Witman, G.B. (2000). Forward and reverse genetic analysis of microtubule motors in Chlamydomonas. Methods 22, 285–298. [DOI] [PubMed] [Google Scholar]
- Pazour, G.J., Koutoulis, A., Benashski, S.E., Dickert, B.L., Sheng, H., Patel-King, R.S., King, S.M., and Witman, G.B. (1999). LC2, the Chlamydomonas homologue of the t complex-encoded protein tctex2, is essential for outer dynein arm assembly. Mol. Biol. Cell 10, 3507–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G.J., Sineshchekov, O.A., and Witman, G.B. (1995). Mutational analysis of the phototransduction pathway of Chlamydomonas reinhardtii. J. Cell Biol. 131, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peitsch, M.C. (1996). ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24, 274–279. [DOI] [PubMed] [Google Scholar]
- Peitsch, M.C. (1995). Protein modeling by E-mail. Bio/Technology 13, 658–660. [Google Scholar]
- Pfister, K.K., Fay, R.B., and Witman, G.B. (1982). Purification and polypeptide composition of dynein ATPases from Chlamydomonas flagella. Cell Motil. Cytoskel. 2, 525–547. [DOI] [PubMed] [Google Scholar]
- Piperno, G., and Luck, D.J.L. (1979). Axonemal adenosine triphosphates from flagella of Chlamydomonas reinhardtii: purification of two dyneins. J. Biol. Chem. 254, 3084–3090. [PubMed] [Google Scholar]
- Rost, B. (1996). PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Rost, B., and Sander, C. (1993). Prediction of protein structure at better than 70% accuracy. J. Mol. Biol. 232, 584–599. [DOI] [PubMed] [Google Scholar]
- Rost, B., and Sander, C. (1994). Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19, 55–72. [DOI] [PubMed] [Google Scholar]
- Sager, R., and Granick, S. (1953). Nutritional studies with Chlamydomonas reinhardtii. Ann. NY Acad. Sci. 56, 831–838. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
- Satir, P. (1968). Studies on cilia III. Further studies on the cilium tip and a “sliding filament”model of ciliary motility. J. Cell Biol. 39, 77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow, C.D. (1998). Organization of the nuclear genome. In: The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, ed. J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant, Dordrecht: Kluwer Academic Publishers, 25–40.
- Summers, K.E., and Gibbons, I.R. (1971). Adenosine-triphosphate induced sliding of tubules in trypsin-treated flagella of sea-urchin sperm. Proc. Natl. Acad. Sci. USA. 68, 3092–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., and Kamiya, R. (1994). Functional reconstitution of Chlamydomonas outer dynein arms from α-β and γ subunits: requirement of a third factor. J. Cell Biol. 126, 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada, S., Wilkerson, C.G., Wakabayashi, K., Kamiya, R., and Witman, G.B. (2002). The outer dynein arm-docking complex: composition and characterization of a subunit (Oda1) necessary for outer arm assembly. Mol. Biol. Cell 13, 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, D.A., Sack, J.S., Maune, J.F., Beckingham, K., and Quiocho, F.A. (1991). Structure of a recombinant calmodulin from Drosophila melanogaster refined at 2.2-Å resolution. J. Biol. Chem. 266, 21375–21380. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi, K., Takada, S., Witman, G.B., and Kamiya, R. (2001). Transport and arrangement of the outer-dynein-arm docking complex in the flagella of Chlamydomonas mutants that lack outer dynein arms. Cell Motil. Cytoskel. 48, 277–286. [DOI] [PubMed] [Google Scholar]
- Wilkerson, C.G., King, S.M., Koutoulis, A., Pazour, G.J., and Witman, G.B. (1995). The 78, 000 Mr intermediate chain of Chlamydomonas outer arm dynein is a WD-repeat protein required for arm assembly. J. Cell Biol. 129, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson, C.G., King, S.M., and Witman, G.B. (1994). Molecular analysis of the γ heavy chain of Chlamydomonas flagellar outer-arm dynein. J. Cell Sci. 107, 497–506. [DOI] [PubMed] [Google Scholar]
- Witman, G.B. (1993). Chlamydomonas phototaxis. Trends Cell Biol. 3, 403–408. [DOI] [PubMed] [Google Scholar]
- Witman, G.B. (1986). Isolation of Chlamydomonas flagella and axonemes. Methods Enzymol. 134, 280–290. [DOI] [PubMed] [Google Scholar]
- Witman, G.B., Carlson, K., Berliner, J., and Rosenbaum, J.L. (1972). Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, membranes, matrix and mastigonemes. J. Cell Biol. 54, 507–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., and Sale, W.S. (1998). The Mr 140, 000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol. Biol. Cell 9, 3335–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]