Abstract
The G protein-coupled receptor GPR54 (AXOR12, OT7T175) is central to acquisition of reproductive competency in mammals. Peptide ligands (kisspeptins) for this receptor are encoded by the Kiss1 gene, and administration of exogenous kisspeptins stimulates hypothalamic gonadotropin-releasing hormone (GnRH) release in several species, including humans. To establish that kisspeptins are the authentic agonists of GPR54 in vivo and to determine whether these ligands have additional physiological functions we have generated mice with a targeted disruption of the Kiss1 gene. Kiss1-null mice are viable and healthy with no apparent abnormalities but fail to undergo sexual maturation. Mutant female mice do not progress through the estrous cycle, have thread-like uteri and small ovaries, and do not produce mature Graffian follicles. Mutant males have small testes, and spermatogenesis arrests mainly at the early haploid spermatid stage. Both sexes have low circulating gonadotropin (luteinizing hormone and follicle-stimulating hormone) and sex steroid (β-estradiol or testosterone) hormone levels. Migration of GnRH neurons into the hypothalamus appears normal with appropriate axonal connections to the median eminence and total GnRH content. The hypothalamic–pituitary axis is functional in these mice as shown by robust luteinizing hormone secretion after peripheral administration of kisspeptin. The virtually identical phenotype of Gpr54- and Kiss1-null mice provides direct proof that kisspeptins are the true physiological ligand for the GPR54 receptor in vivo. Kiss1 also does not seem to play a vital role in any other physiological processes other than activation of the hypothalamic–pituitary–gonadal axis, and loss of Kiss1 cannot be overcome by compensatory mechanisms.
Keywords: Gpr54, kisspeptin, mouse, puberty
Neuroendocrine events within the hypothalamus control sexual maturation and seasonal breeding in mammals (1). The gonadotropin-releasing hormone (GnRH)-induced secretion of the gonadotropic hormones luteinizing hormone (LH) and follicle-stimulating hormone (FSH) from the anterior pituitary is essential to invoke puberty and maintain reproductive function. The G protein-coupled receptor GPR54 (2) is a key protein involved in the pubertal activation of the hypothalamic pituitary gonadal axis because mice and humans with mutations in this receptor are sterile with hypogonadotropic hypogonadism (3–7).
A series of overlapping peptide ligands (kisspeptins) for the GPR54 receptor are produced by the Kiss1 gene (8–10). Kiss1 mRNA is expressed in hypothalamic regions that regulate gonadotropin secretion including the anteroventral periventricular nucleus (AVPV), the periventricular nucleus, and the arcuate nucleus (ARC) (11). Kiss1 expression increases at puberty in rodents (12–14) and primates (15) and fluctuates during the rat estrous cycle (12, 16). Kiss1 expression is also subject to differential regulation by sex steroids, providing a plausible mechanism for the feedback control of gonadotropin secretion by these hormones (17, 18). Administration of kisspeptins potently stimulates gonadotropin secretion in several mammalian species (11, 12, 19–24). These effects are likely mediated by a direct action on GnRH neurons, which express the GPR54 receptor (13, 21, 25). Kisspeptins can stimulate GnRH release from explanted rat hypothalamic fragments (20, 26) and after central injection in sheep (25). Kisspeptin immunoreactive neurons have been shown to project fibers directly onto GnRH neurons (14, 24). Finally, direct electrophysiological studies on GnRH-GFP neurons in mice revealed that kisspeptin stimulates a robust and long-lasting depolarization in most of the GnRH neurons (13).
Although the effects of acute administration of kisspeptins on gonadotropin secretion have been described, the role that endogenous kisspeptins play in this process has not been determined, and it is not known whether there are other physiological roles for this ligand. It is important to establish the function of endogenous neurotransmitters because exogenously administered neurotransmitters do not always indicate what happens at the physiological level in a whole animal. As a step toward understanding the physiological role of endogenous kisspeptins, we have generated and characterized Kiss1-null mutant mice.
Results
Generation of Kiss1 Knockout Mice.
The Kiss1 coding region is located within two exons on chromosome 1 and is part of an alternatively spliced transcript from the Golt1a gene (Ensembl Gene ID ENSMUSG00000041717). Both coding exons were deleted by gene targeting and replaced with an internal ribosome entry site (IRES)-LacZ reporter gene (Fig. 1A) so that expression of the Kiss1 allele (designated Kiss1tm1PTL) can be visualized by detection of β-galactosidase activity. Targeted ES clones were identified by PCR and confirmed by Southern blot analysis (data not shown). Correctly targeted clones were used to generate germ-line chimeras that transmitted the Kiss1tm1PTL allele at the expected Mendelian frequency. Heterozygous mice were fertile and gave rise to viable homozygotes at the expected frequency, demonstrating functional placentation by mutant fetuses and normal fetal development.
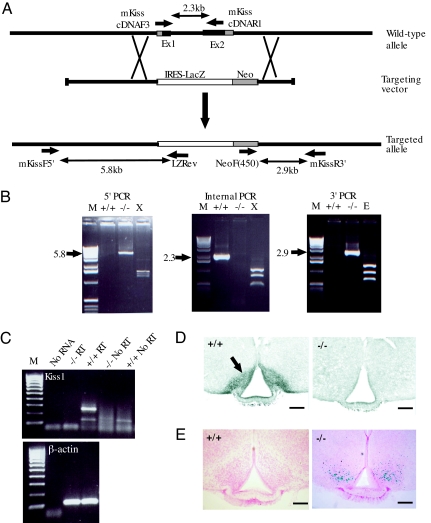
Fig. 1.
Kiss1 gene targeting. (A) Gene targeting strategy. Kiss1 exons are indicated as shaded boxes with the coding sequence in black. The targeting vector replaces the complete Kiss1 coding region with an IRES-LacZ sequence. The location of primers used to confirm the correct targeting event and loss of Kiss1 coding sequence in the mutant mice are shown along with the size of the PCR products. (B) PCR analysis of Kiss1 locus in mutant mice. The identity of the PCR products was confirmed by restriction enzyme digestion. X, XcmI; E, EcoRI; M, Invitrogen 1-kb DNA marker. (C) RT-PCR of Kiss1gene expression confirming the null allele. (D) Immunohistochemical detection of KISS1 neurons in the ARC of the hypothalamus. (Scale bars: 300 μm.) (E) β-galactosidase activity in the ARC of mutant mice. (Scale bars: 300 μm.)
PCR analysis of homozygous mutant mice confirmed the correct gene targeting event and absence of the Kiss1 coding region (Fig. 1B). The absence of Kiss1 mRNA was confirmed by RT-PCR because Kiss1 transcripts were detected only in wild-type animals (Fig. 1C). Immunohistochemistry of hypothalamic sections consistently failed to detect KISS1 protein-containing cell bodies in mutant mice as illustrated in the ARC (Fig. 1D), confirming a true null mutation. As predicted, the ARC showed β-galactosidase activity in the mutant mice consistent with the IRES-LacZ transgene being driven from the Kiss1 promoter. The β-galactosidase activity was confined to cell bodies and did not extend into axons (Fig. 1E). Low-level expression of β-galactosidase was also detected in the AVPV and periventricular nucleus of mutant mice but not in other regions of reported Kiss1 expression, such as the dorsomedial hypothalamic nucleus (14, 27, 28).
Anatomy of Kiss1tm1PTL-Null Mice.
Mutant mice were viable and born at the expected frequency. There were no gross anatomical or developmental abnormalities other than in the reproductive system. Thus, Kiss1 does not appear to have a major role in other physiological processes. Both male and female mutant mice, however, were significantly smaller than sex-matched littermates at 2 months old [supporting information (SI) Table 1]. When this weight difference was taken into account, no significant differences were found in the major organs of female mice apart from the ovaries and uterus, which were considerably smaller in the mutants (Fig. 2A and SI Table 1). The testes of Kiss1-null mice were almost 1/10th the size of wild-type testes (Fig. 3A), and the kidney, liver, and salivary glands were also smaller after correction for total body weight difference (SI Table 1).
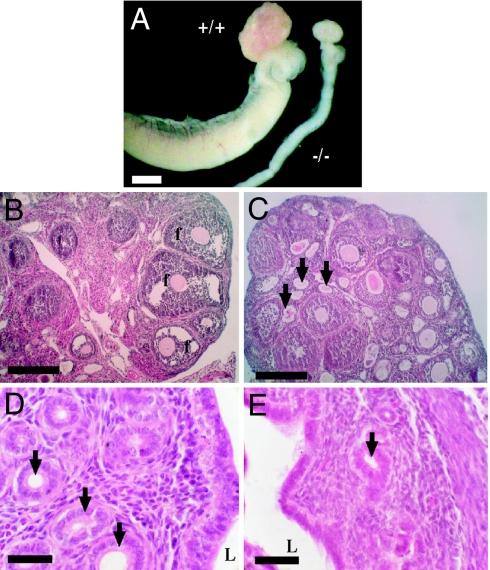
Fig. 2.
Reproductive system defects in female Kiss1tm1PTL-null mice. (A) Reduced ovary size and thread-like uterus in mutant mice. (Scale bar: 1 mm.) (B and C) Histology of ovaries showing larger number of atretic follicles (arrows) in mutant mice (C) and absence of late-stage follicle maturation (f) present in wild-type mice. (Scale bars: 100 μm.) (D and E) Histology of uterus showing full development of glands (arrows) in the endometrial layer in wild type (D) in contrast to mutant mice (E). L, uterine lumen. (Scale bars: 25 μm.)
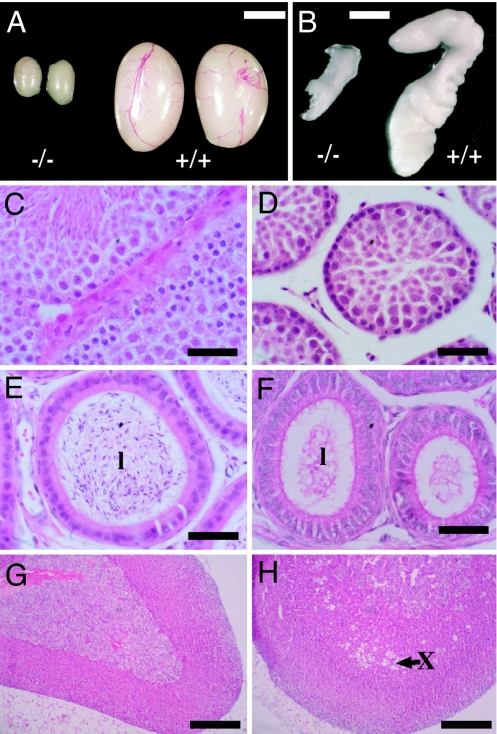
Fig. 3.
Reproductive system defects in male Kiss1tm1PTL-null mice. (A) Reduced testes size in mutant mice. (Scale bar: 0.25 cm.) (B) Reduced development of seminal vesicle in mutant mice. (Scale bar: 0.25 cm.) (C and D) Histology of seminiferous tubules showing intact spermatogenesis and mature sperm in wild-type mice (C) and impaired spermatogenesis in mutant mice (D). (Scale bars: 50 μm.) (E and F) Histology of epididymis showing sperm in the lumen (l) in wild-type mice (E) and absence of sperm in the lumen in mutant mice (F). (Scale bars: 50 μm.) (G and H) Histology of adrenal gland showing retention of fetal zone X (arrow) in mutant male mice (H) and absence of this zone in mature male wild-type mice (G). (Scale bars: 300 μm.)
Mutant mice of both sexes failed to undergo pubertal sexual maturation and suffered hypogonadism. Males had a microphallus and poor development of secondary sex glands such as the preputial gland and the seminal vesicles (Fig. 3B). Mutant females failed to undergo normal vaginal opening at pubertal age, and vaginal smears showed that they were not progressing through the estrous cycle (data not shown).
Histology of Kiss1tm1PTL-Null Mice.
Kiss1-null female mice showed histological abnormalities in the ovary and the uterus. The ovaries of mutant mice lacked late-stage antral follicles or corpora lutea, although early stage antral formation was observed (Fig. 2C). Mutant ovaries also had a large number of atretic follicles (Fig. 2C, arrows) compared with wild type (Fig. 2B). These changes are consistent with failure of folliculargenesis and ovulation in the Kiss1-null mice. The uteri of mutant mice appeared typical of an animal before puberty with a paucity of gland duct development in a narrow endometrial layer (Fig. 2E, arrow) compared with normal (Fig. 2D, arrows).
Kiss1-null male mice showed defective spermatogenesis with absence of spermatozoa in most seminiferous tubules (Fig. 3D). Spermatogenesis progressed through meiosis to the early haploid spermatid stage, but spermiogenesis was incomplete, and spermatozoa with condensed sperm heads were rarely observed compared with wild type (Fig. 3C). Occasionally, however, a few condensed sperm heads were observed in some seminiferous tubules, indicating a low level of spermiogenesis, but this appeared disorganized with far fewer spermatozoa than in wild-type testes. These limited numbers of spermatozoa did not exit into the lumen of the epididymis (Fig. 3F) compared with wild-type mice (Fig. 3E), perhaps because of lack of Sertoli cell fluid secretion, which requires testosterone. A reexamination of histological sections from Gpr54-null mice (5) has confirmed that they also show this low level of spermatozoa formation. The adrenal gland of mutant male mice retained the characteristic vacuolated fetal zone X (Fig. 3H), which was absent in age-matched wild-type animals that had progressed through puberty (Fig. 3G).
Hormonal Profile of Kiss1tm1PTL-Null Mice.
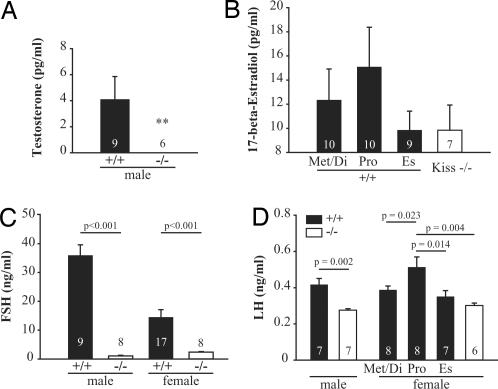
To understand the physiological mechanisms responsible for the defects in the reproductive organs, the endocrine profile of the mutant mice was assessed. Wild-type male mice had an average free plasma testosterone level of 4.1 ± 1.8 pg/ml (n = 9) whereas mutant male mice (n = 6) had undetectable plasma testosterone levels, i.e., <0.17 pg/ml (Fig. 4A). Female mice had lower but not statistically different circulating plasma levels of 17-β-estradiol (9.8 ± 2.1 pg/ml, n = 7) than wild-type mice at proestrus (15.1 ± 3.3 pg/ml, n = 10) (Fig. 4B). The Kiss1 mutant female mice also failed to show the cyclic fluctuations in 17-β-estradiol levels observed in the female wild-type mice. Kiss1-null mice of both sexes had significantly lower plasma FSH levels (1.0 ± 0.1 ng/ml, n = 8 for males; 2.4 ± 0.2 ng/ml, n = 8 for females) (Fig. 4C) compared with wild type (35.9 ± 3.7 ng/ml, n = 9 for males; 14.5 ± 2.6 ng/ml, n = 17 for females). Similarly, plasma LH levels were significantly lower in Kiss1-null male mice than wild type (0.28 ± 0.01 and 0.42 ± 0.03 ng/ml, respectively) (Fig. 4D). Mutant females' plasma LH levels (0.30 ± 0.01 ng/ml) were significantly lower compared with wild-type proestrus mice (0.46 ± 0.05 ng/ml, P = 0.004) but not compared with diestrus/metestrus and estrus mice (0.39 ± 0.02 and 0.41 ± 0.06 ng/ml, respectively).
Fig. 4.
Hormone profiles of Kiss1tm1PTL-null mice. (A) Plasma free testosterone levels in male mice. ∗∗, Testosterone concentration was below the limit of detection in the mutant mice. (B) Plasma 17-β-estradiol levels in female mice. Mutant mice had 17-β-estradiol levels close to the background limit of the assay (8 pg/ml). (C) Plasma FSH levels. Both male and female mutant mice showed significantly lower FSH levels than age-matched wild-type animals (P < 0.001, Student's t test). (D) Plasma LH levels. Mutant male mice showed significantly lower LH levels (P = 0.002) than age-matched wild-type males. In females, Kiss1tm1PTL-null mice had lower plasma LH levels than proestrus (Pro) wild-type animals (statistically significant differences are shown, one-way ANOVA with Student–Newman–Keuls test). The number of mice used for each analysis is indicated in each histogram. Met/Di, metestrus and diestrus; Es, estrous.
Kiss1tm1PTL-Null Mice Show Normal GnRH Neuronal Localization in the Hypothalamus and GnRH Content.
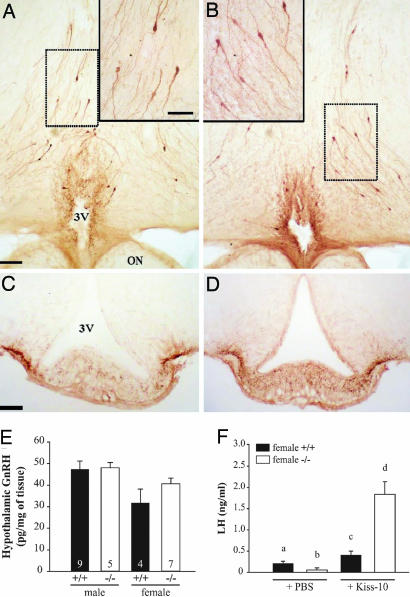
To determine whether the phenotype of the mutant mice was caused by the failure of GnRH neurons to migrate into the hypothalamus, we examined the presence of GnRH neurons by immunohistochemistry. GnRH immunoreactive neurons were found in the appropriate regions of the hypothalamus of mutant mice. Throughout the preoptic region, GnRH-positive neuronal cell bodies displayed the scattered distribution pattern typical of these neurons (Fig. 5 A and B). GnRH-positive neurons projected to the external zone of the median eminence (Fig. 5 C and D). In addition, measurement of hypothalamic GnRH content confirmed the immunohistochemistry data (Fig. 5E). There was no significant difference in hypothalamic GnRH content between wild-type and mutant mice of either sex.
Fig. 5.
GnRH neurons in the hypothalamus of Kiss1tm1PTL-null mice and responses to kisspeptin injection. Photomicrographs of 50-μm-thick coronal sections showing GnRH immunoreactive neurons in the hypothalamus of wild-type (A and C) and mutant (B and D) mice. (A and B) GnRH-positive cell bodies in the preoptic region at the level of the organum vasculosum laminae terminalis. (Scale bar: 100 μm.) Frames in top corners are higher magnifications of respective dotted line squared areas. (Scale bar: 50 μm.) (C and D) GnRH-positive axonal projections and nerve terminals in the median eminence. (Scale bar: 50 μm.) 3V, third ventricle; ON, optic nerve. (E) GnRH content in hypothalami from both sexes. (F) Stimulation of LH release by kisspeptin-10 in Kiss1tm1PTL-null mice. Kiss1-null (−/−) or wild-type (+/+) female mice at diestrus were injected i.p. with vehicle (PBS) or 1 nmol of kisspeptin-10 in PBS and killed after 30 min, and serum was measured for LH. a, b, c, and d indicate values significantly different from each other (P < 0.05, n = 6 per group, one-way ANOVA followed by Student–Newman–Keuls test).
Kiss1tm1PTL-Null Mice Respond to Injection of Kisspeptin by Secreting LH.
The ability of the hypothalamic–pituitary axis to respond to injection of kisspeptin-10 was tested in the mutant mice. Both wild-type (0.41 ± 0.09 ng/ml) and mutant (1.84 ± 0.30 ng/ml) females showed significantly higher levels of plasma LH after i.p. delivery of kisspeptin-10 compared with vehicle-treated animals (0.21 ± 0.06 and 0.06 ± 0.05 ng/ml, respectively) (Fig. 5F). Moreover, the Kiss1-null mice showed a higher LH release response to kisspeptin-10 injection than wild type. These data also demonstrate that the pituitary gonadotrophs are functional in the Kiss1-null mice.
Discussion
Since the initial discovery that the G protein-coupled receptor GPR54 has a key role in regulation of mammalian fertility, several studies have been performed to elucidate the way in which kisspeptin ligands activate this receptor in vivo. By necessity, these studies have relied on measuring physiological responses to exogenous delivery of kisspeptins. These experiments have shown that kisspeptins are potent agonists of GPR54, but direct proof that this is the true and only physiological role for kisspeptins requires phenotypic analysis of intact animals that lack kisspeptins. To examine the role of endogenous kisspeptins in activation of the hypothalamic–pituitary–gonadal axis and to identify other potential physiological actions of these ligands, we have generated mice that lack a functional Kiss1 gene. The major phenotype of the mutant mice is a lack of pubertal maturation, sterility, and hypogonadotropic hypogonadism. Mutant females do not progress through the estrous cycle, have thread-like uteri and small ovaries, and do not produce mature Graffian follicles. Mutant female mice could be induced to ovulate by injection of gonadotropic hormones, however, indicating that ovarian responses were intact (data not shown). Mutant male mice have atrophied testes and spermatogenic arrest mainly at the early haploid spermatid stage. Both sexes have low circulating gonadotropin (LH and FSH) and sex steroid (β-estradiol or testosterone) hormone levels. These phenotypes provide direct proof that the Kiss1 gene encodes the true physiological ligand for the GPR54 receptor in vivo.
The Kiss1-null mice also show that kisspeptins are not essential for other major physiological functions. Kiss1 is expressed in other tissues most notably the placenta, where expression is localized to syncytiotrophoblast cells (29). These cells are derived from the trophoblast of the developing fetus and invade the uterine wall to increase the surface area for nutrient exchange. Because Kiss1 expression can suppress metastasis in several different cancer cell types (30–33) it has been suggested that kisspeptins may regulate placental invasion (34). Our data, however, show that Kiss1 is not required for placenta formation per se because mutant null mice were born at the expected rate. Whether there are subtle differences between the placentae of normal mice and those of mutant fetuses remains to be established.
The Kiss1-null mice are unusual in having such a dramatic phenotype; many neuropeptide knockout mice do not display overt phenotypes without an experimental challenge. For example, neuropeptide Y null mice have the same weight and food intake as normal littermates (35) even though many studies have shown that exogenous injection of neuropeptide Y strongly stimulates hyperphagia and weight gain. It has been suggested that, in mice, there exist compensatory mechanisms that act to minimize the effects of a lack of some nonessential neuropeptides during development (36). Clearly, no such compensation takes place in the Kiss1-null mice, perhaps because the kisspeptin/GPR54 pathway is not required for any crucial developmental events during gestation.
The Kiss1 locus is tagged with an IRES-LacZ reporter gene, which provides a convenient tool to define the anatomical localization of cell bodies expressing Kiss1. The β-galactosidase expression we observe in the ARC, AVPV, and periventricular nuclei of mutant mice is consistent with the reported Kiss1 expression in these regions (14, 17, 18). The β-galactosidase staining also shows that Kiss1-expressing neurons are still present in the ARC of the knockout mice. The lower β-galactosidase expression in the AVPV compared with the ARC can be explained by the differential regulation of Kiss1 expression by estrogen in these regions (14, 17, 18), which is low in the mutant mice. In rodents, estrogen increases Kiss1 expression in the AVPV and deceases expression in the ARC (14, 17, 18).
The Kiss-1 gene was originally isolated as a human metastasis suppressor gene (30), and kisspeptins can inhibit cell migration in vitro and in vivo (10, 34, 37, 38). In addition, some forms of isolated hypogonadotropic hypogonadism are caused by failure of GnRH neurons to migrate from the olfactory placode to the hypothalamus (39). It was therefore possible that the reproductive defect in the Kiss1 mutant mice results from a GnRH migratory defect, perhaps resulting in failure of neurons to localize to the preoptic area or to target the median eminence. However, immunohistochemical localization of GnRH within the hypothalamus showed no major deficiency in neuronal migration. In addition, hypothalamic GnRH content was similar between wild-type and mutant mice. Moreover, the functionality of GnRH neurons in Kiss1-null mice was demonstrated by LH secretion in response to injection of exogenous kisspeptin-10. In fact, the responses in the mutant mice were significantly greater than those observed in wild-type females. This may be because of the slightly higher hypothalamic GnRH content found in Kiss1-null female mice, which may result in an exaggerated GnRH response after kisspeptin stimulation for the first time.
The phenotype of the Kiss1 mutant mice is consistent with the GPR54/kisspeptin receptor–ligand pair being necessary and required for progress through puberty and the acquisition of adult reproductive capabilities. Furthermore, this is consistent with the lack of ligand-stimulated GnRH release from the hypothalamus. Moreover, we demonstrate here that kisspeptins exert their major effects specifically on activation of the hypothalamic–pituitary–gonadal axis with little overt effect on other physiological pathways. The Gpr54-null mice raised the concept that this receptor is crucial for reproductive function (4, 5). By showing that the Kiss1-null and Gpr54-null mice are phenocopies, we have eliminated the possibility of an alternative ligand to GPR54. These data demonstrate the potential for a pivotal role for pharmacological modulators of the kisspeptin/GPR54 pathway in the treatment of inherited/congenital reproductive disorders and sex hormone-dependent cancers.
Materials and Methods
Gene Targeting and Generation of Mutant Mice.
The targeting vector was constructed by using homology arms amplified from 129S6/Sv/Ev mouse genomic DNA using the following primers: 5′armF, TTTGTCGACAGCTCACAGTACAGGAGCCACCTCTGG; 5′armR, TTTGCGGCCGCAGCCATTGAGATCATTCTGGGAGGAAG; 3′armF, AAAGGCGCGCCAAGGCAGGGAGCTTCTAGACTTGTGC; and 3′armR, AAAGGCCGGCCAAAACACCCCAGGGAGGAGGCATTGAG. The 5′armF/R primer pair amplified a 3.8-kb fragment, and the 3′armF/R primer pair amplified a 1.9-kb fragment. The arms were cloned on either side of a cassette containing an IRES-LacZ reporter gene and a promoted neomycin phosphoribosyltransferase selectable marker gene. Homologous recombination of this targeting construct results in the deletion of 2.4 kb of the Kiss1 locus, consisting of 88 bp of the first coding exon, all of the 2.0-kb downstream intron, and 319 bp of the second coding exon, covering all of the coding region of this exon including the key active region of the processed peptide.
ES cells (CCB; 129S6/Sv/Ev strain) were cultured, and gene targeting was performed as described previously (40). Targeted clones were identified by PCR and Southern blot analysis. Chimeras were generated by injection into C57/Bl6 blastocysts, and inbred mice were established by breeding germ-line chimeras with 129S6/Sv/Ev mice. All experiments were performed under the authority of a United Kingdom Home Office Project License and were approved by a local ethics committee.
Molecular Analysis.
Correct gene targeting in the mice was confirmed by long-range PCR using AccuTaq (Sigma-Aldrich, Dorset, U.K.) according to the manufacturer's instructions. The 5′ genomic region was amplified by using mKissF5′ (GAAGCAGAATCAAACATCTCCGAG) and LZRev (TTCTCCGTGGGAACAAACGG). The 3′ genomic region was amplified by using NeoF (450) (ATGGAAGCCGGTCTTGTCGATC) and mKissR3′ (CACCATGAGGATAATGGACTGAACC). Both mKissF5′ and mKissR3′ are located outside the genomic arms in the targeting vector. The Kiss1 gene was amplified by using the primers cDNAF3 (TGCTGCTTCTCCTCTGTGTCG) and cDNAR1(GCCGAAGGAGTTCCAGTTGTA). The 5.8-kb 5′ PCR product, which amplified only from the targeted locus, was digested with XcmI to give bands of 1,567 bp/1,546 bp (doublet), 1406 bp, and 1,278 bp. The 2.9-kb 3′ PCR product, which amplified only from the targeted allele, was digested with XcmI to give bands of 1,354 bp, 889 bp, and 657 bp. The 2.3-kb internal PCR product was amplified only from the wild-type allele and was digested with EcoRI to give band of 1,094 bp, 784 bp, and 422 bp.
RT-PCR was used to detect Kiss1 mRNA. RNA was made by using a Qiagen (Crawley, U.K.) RNAeasy kit, and cDNA was made by using SuperScript 2 from Invitrogen (Paisley, U.K.). Kiss1 cDNA was amplified by using cDNAF3 and cDNAR1, which span an intron and amplify a 285-bp fragment from cDNA. Primers specific for the mouse β-actin gene (CTGTATTCCCCTCCATCGTG and GGGTCAGGATACCTCTCTTGC) were used to confirm cDNA synthesis.
Hormone Assays.
Mice were killed by CO2 exposure between 1530 hours and 1630 hours. Vaginal smears were performed on wild-type female mice to determine their estrous cycle stage. Blood was collected in a heparinized syringe from the inferior vena cava and centrifuged at 1,000 × g for 10 min at 4°C. The plasma supernatant samples were collected and stored at −80°C until assayed. Testosterone was measured by using an ELISA kit (DB52181; IBL, Hamburg, Germany) with a sensitivity of 0.17 pg/ml (intraassay variation, 8.9%; interassay variation, 8.8%). 17-β-estradiol was measured by ELISA, the assay sensitivity was 8.0 pg/ml, and the intraassay and interassay coefficients of variations were both 10%. For the nontreated mouse plasma, LH was measured by using a RIA kit purchased from Biocode-Hycel (Liege, Belgium) (sensitivity, 0.14 ng/ml; intraassay variation, 10.5%; interassay variation, 12.1%). For the vehicle- or kisspeptin-treated female mouse plasma, LH was assessed by using an ELISA kit from Endocrine Technologies (Newark, CA) (sensitivity, 0.5 ng/ml; intraassay variation, 7%; interassay variation, 9.8%). FSH was measured by using an ELISA kit from Biocode-Hycel (sensitivity, 0.2 ng/ml; intraassay variation, 4.7%; interassay variation, 8.5%). Hypothalamic GnRH was measured by using a RIA kit from Phoenix Pharmaceuticals (Karlsruhe, Germany) (sensitivity, 4 pg per tube; intraassay variation, 4.7%; interassay variation, 8.3%).
Histology and Immunohistochemistry.
Mouse tissues were fixed for 16 h in 4% paraformaldehyde, washed in PBS, wax-embedded, and sectioned at 7 μm. Sections were stained with hematoxylin and eosin. Kisspeptin immunohistochemistry was performed on free-floating 30-μm frozen hypothalamic sections spanning the anterior preoptic area to the premammillary body as previously described (14) using a highly specific rabbit antibody raised against the highly conserved Kp10 sequence YNWNSFGLRY-NH2 (28). For GnRH immunohistochemistry, fixed brains were Vibratome-sectioned at 50 μm covering the preoptic area and the median eminence and then mounted on slides. Sections were blocked for 1 h at room temperature with PBS containing 5% goat serum (G-9023; Sigma) and then incubated overnight at 4°C in rabbit polyclonal anti-GnRH antibody [a generous gift from G. Tramu (41) provided by means of V. Prevot (Institut National de la Santé et de la Recherche Médicale, Lille, France)] diluted at 1:3,000, with 0.3% Triton X-100 and 5% goat serum in PBS. GnRH immunoreactivity was visualized by using a biotinylated goat anti-rabbit secondary antibody for 1 h at room temperature and avidin-biotin complex (ABC; Vector Laboratories, Peterborough, U.K.). An incubation step in 1% H2O2 was performed after the secondary antibody to quench endogenous peroxidase activity. The final staining was made with 3,3′-diaminobenzidine (DAB, SK-4100; Vector) as chromogen.
β-Galactosidase Staining.
To detect β-galactosidase activity in the mouse brain, floating 50-μm-thick sections were incubated overnight at 37°C in staining buffer containing 1 mM MgCl2, 0.5 mg/ml X-Gal (Melford Laboratories, Chelsworth, Ipswich, Suffolk, U.K.), 5 mM potassium ferrocyanide, and 5 mM potassium ferricyanide. The sections were rinsed in PBS, counterstained with 1% Neutral Red solution, dehydrated, and mounted on slides with DPX (Sigma–Aldrich).
Kisspeptin-10 injections in Wild-Type and Kiss1−/− Mice.
Wild-type 2- to 4-month-old female mice (n = 6) in diestrus stage and Kiss1tm1PTL-null 2- to 4-month-old female mice (n = 6) received one single i.p. injection of 100 μl of 10 μM mouse kisspeptin-10 in 0.1 M PBS [human Metastin (45–54) amide; Sigma–Aldrich) or PBS only (vehicle). The mice were killed by CO2 exposure 30 min after injection. Blood was collected as described above.
Statistical Analysis.
Data are presented as the mean ± SEM for each group. Differences among groups were assessed by one-way ANOVA with a Student–Newman–Keuls post hoc test. Student's t test was used when only two groups were being compared with a similar standard deviation. Differences were considered significant when P < 0.05.
Supplementary Material
Acknowledgments
This work was partly funded by a Biotechnology and Biological Sciences Research Council Industrial Partnership Award with Paradigm Therapeutics (BB/C003861/1). S.A.J.R.A. is supported by a Canada Research Chair in Molecular Oncology. W.H.C. is supported by the Ford Physiology Fund.
Abbreviations
- IRES
internal ribosome entry site
- FSH
follicle-stimulating hormone
- LH
luteinizing hormone
- AVPV
anteroventral periventricular nucleus
- ARC
arcuate nucleus
- GnRH
gonadotropin-releasing hormone.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704114104/DC1.
References
- 1.Ebling FJ. Reproduction. 2005;129:675–683. doi: 10.1530/rep.1.00367. [DOI] [PubMed] [Google Scholar]
- 2.Lee DK, Nguyen T, O'Neill GP, Cheng R, Liu Y, Howard AD, Coulombe N, Tan CP, Tang-Nguyen AT, George SR, O'Dowd BF. FEBS Lett. 1999;446:103–107. doi: 10.1016/s0014-5793(99)00009-5. [DOI] [PubMed] [Google Scholar]
- 3.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Proc Natl Acad Sci USA. 2003;100:10972–10976. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funes S, Hedrick JA, Vassileva G, Markowitz L, Abbondanzo S, Golovko A, Yang S, Monsma FJ, Gustafson EL. Biochem Biophys Res Commun. 2003;312:1357–1363. doi: 10.1016/j.bbrc.2003.11.066. [DOI] [PubMed] [Google Scholar]
- 5.Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS, Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, et al. N Engl J Med. 2003;349:1614–1627. doi: 10.1056/NEJMoa035322. [DOI] [PubMed] [Google Scholar]
- 6.Lanfranco F, Gromoll J, von Eckardstein S, Herding EM, Nieschlag E, Simoni M. Eur J Endocrinol. 2005;153:845–852. doi: 10.1530/eje.1.02031. [DOI] [PubMed] [Google Scholar]
- 7.Semple RK, Achermann JC, Ellery J, Farooqi IS, Karet FE, Stanhope RG, O'Rahilly S, Aparicio SA. J Clin Endocrinol Metab. 2005;90:1849–1855. doi: 10.1210/jc.2004-1418. [DOI] [PubMed] [Google Scholar]
- 8.Kotani M, Detheux M, Vandenbogaerde A, Communi D, Vanderwinden JM, Le Poul E, Brezillon S, Tyldesley R, Suarez-Huerta N, Vandeput F, et al. J Biol Chem. 2001;276:34631–34636. doi: 10.1074/jbc.M104847200. [DOI] [PubMed] [Google Scholar]
- 9.Muir AI, Chamberlain L, Elshourbagy NA, Michalovich D, Moore DJ, Calamari A, Szekeres PG, Sarau HM, Chambers JK, Murdock P, et al. J Biol Chem. 2001;276:28969–28975. doi: 10.1074/jbc.M102743200. [DOI] [PubMed] [Google Scholar]
- 10.Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, Kanehashi K, Terao Y, Kumano S, Takatsu Y, Masuda Y, et al. Nature. 2001;411:613–617. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 11.Gottsch ML, Cunningham MJ, Smith JT, Popa SM, Acohido BV, Crowley WF, Seminara S, Clifton DK, Steiner RA. Endocrinology. 2004;145:4073–4077. doi: 10.1210/en.2004-0431. [DOI] [PubMed] [Google Scholar]
- 12.Navarro VM, Castellano JM, Fernandez-Fernandez R, Barreiro ML, Roa J, Sanchez-Criado JE, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. Endocrinology. 2004;145:4565–4574. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- 13.Han SK, Gottsch ML, Lee KJ, Popa SM, Smith JT, Jakawich SK, Clifton DK, Steiner RA, Herbison AE. J Neurosci. 2005;25:11349–11356. doi: 10.1523/JNEUROSCI.3328-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarkson J, Herbison AE. Endocrinology. 2006;147:5817–5825. doi: 10.1210/en.2006-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shahab M, Mastronardi C, Seminara SB, Crowley WF, Ojeda SR, Plant TM. Proc Natl Acad Sci USA. 2005;102:2129–2134. doi: 10.1073/pnas.0409822102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JT, Popa SM, Clifton DK, Hoffman GE, Steiner RA. J Neurosci. 2006;26:6687–6694. doi: 10.1523/JNEUROSCI.1618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Endocrinology. 2005;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- 18.Smith JT, Dungan HM, Stoll EA, Gottsch ML, Braun RE, Eacker SM, Clifton DK, Steiner RA. Endocrinology. 2005;146:2976–2984. doi: 10.1210/en.2005-0323. [DOI] [PubMed] [Google Scholar]
- 19.Matsui H, Takatsu Y, Kumano S, Matsumoto H, Ohtaki T. Biochem Biophys Res Commun. 2004;320:383–388. doi: 10.1016/j.bbrc.2004.05.185. [DOI] [PubMed] [Google Scholar]
- 20.Thompson EL, Patterson M, Murphy KG, Smith KL, Dhillo WS, Todd JF, Ghatei MA, Bloom SR. J Neuroendocrinol. 2004;16:850–858. doi: 10.1111/j.1365-2826.2004.01240.x. [DOI] [PubMed] [Google Scholar]
- 21.Irwig MS, Fraley GS, Smith JT, Acohido BV, Popa SM, Cunningham MJ, Gottsch ML, Clifton DK, Steiner RA. Neuroendocrinology. 2004;80:264–272. doi: 10.1159/000083140. [DOI] [PubMed] [Google Scholar]
- 22.Dhillo WS, Chaudhri OB, Patterson M, Thompson EL, Murphy KG, Badman MK, McGowan BM, Amber V, Patel S, Ghatei MA, Bloom SR. J Clin Endocrinol Metab. 2005;90:6609–6615. doi: 10.1210/jc.2005-1468. [DOI] [PubMed] [Google Scholar]
- 23.Navarro VM, Fernandez-Fernandez R, Castellano JM, Roa J, Mayen A, Barreiro ML, Gaytan F, Aguilar E, Pinilla L, Dieguez C, Tena-Sempere M. J Physiol. 2004;561:379–386. doi: 10.1113/jphysiol.2004.072298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinoshita M, Tsukamura H, Adachi S, Matsui H, Uenoyama Y, Iwata K, Yamada S, Inoue K, Ohtaki T, Matsumoto H, Maeda K. Endocrinology. 2005;146:4431–4436. doi: 10.1210/en.2005-0195. [DOI] [PubMed] [Google Scholar]
- 25.Messager S, Chatzidaki EE, Ma D, Hendrick AG, Zahn D, Dixon J, Thresher RR, Malinge I, Lomet D, Carlton MB, et al. Proc Natl Acad Sci USA. 2005;102:1761–1766. doi: 10.1073/pnas.0409330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nazian SJ. J Androl. 2006;27:444–449. doi: 10.2164/jandrol.05144. [DOI] [PubMed] [Google Scholar]
- 27.Brailoiu GC, Dun SL, Ohsawa M, Yin D, Yang J, Chang JK, Brailoiu E, Dun NJ. J Comp Neurol. 2005;481:314–329. doi: 10.1002/cne.20350. [DOI] [PubMed] [Google Scholar]
- 28.Franceschini I, Lomet D, Cateau M, Delsol G, Tillet Y, Caraty A. Neurosci Lett. 2006;401:225–230. doi: 10.1016/j.neulet.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 29.Horikoshi Y, Matsumoto H, Takatsu Y, Ohtaki T, Kitada C, Usuki S, Fujino M. J Clin Endocrinol Metab. 2003;88:914–919. doi: 10.1210/jc.2002-021235. [DOI] [PubMed] [Google Scholar]
- 30.Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, Welch DR. J Nat Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Welch DR. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 32.Shirasaki F, Takata M, Hatta N, Takehara K. Cancer Res. 2001;61:7422–7425. [PubMed] [Google Scholar]
- 33.Sanchez-Carbayo M, Capodieci P, Cordon-Cardo C. Am J Pathol. 2003;162:609–617. doi: 10.1016/S0002-9440(10)63854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilban M, Ghaffari-Tabrizi N, Hintermann E, Bauer S, Molzer S, Zoratti C, Malli R, Sharabi A, Hiden U, Graier W, et al. J Cell Sci. 2004;117:1319–1328. doi: 10.1242/jcs.00971. [DOI] [PubMed] [Google Scholar]
- 35.Erickson JC, Hollopeter G, Palmiter RD. Science. 1996;274:1704–1707. doi: 10.1126/science.274.5293.1704. [DOI] [PubMed] [Google Scholar]
- 36.Gingrich JA, Hen R. Curr Opin Neurobiol. 2000;10:146–152. doi: 10.1016/s0959-4388(99)00061-6. [DOI] [PubMed] [Google Scholar]
- 37.Jiang Y, Berk M, Singh LS, Tan H, Yin L, Powell CT, Xu Y. Clin Exp Metastasis. 2005;22:369–376. doi: 10.1007/s10585-005-8186-4. [DOI] [PubMed] [Google Scholar]
- 38.Stafford LJ, Xia C, Ma W, Cai Y, Liu M. Cancer Res. 2002;62:5399–5404. [PubMed] [Google Scholar]
- 39.Lutz B, Rugarli EI, Eichele G, Ballabio A. FEBS Lett. 1993;325:128–134. doi: 10.1016/0014-5793(93)81428-3. [DOI] [PubMed] [Google Scholar]
- 40.van der Meer T, Chan WY, Palazon LS, Nieduszynski C, Murphy M, Sobczak-Thepot J, Carrington M, Colledge WH. Reproduction. 2004;127:503–511. doi: 10.1530/rep.1.00131. [DOI] [PubMed] [Google Scholar]
- 41.Beauvillain JC, Tramu G. J Histochem Cytochem. 1980;28:1014–1017. doi: 10.1177/28.9.6157712. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.