Abstract
Conjugation with ubiquitin acts as a sorting signal for proteins in the endocytic and biosynthetic pathways at the endosome. Signal-transducing adaptor molecule (STAM) proteins, STAM1 and STAM2, are associated with hepatocyte growth factor-regulated substrate (Hrs) but their function remains unknown. Herein, we show that STAM proteins bind ubiquitin and ubiquitinated proteins and that the tandemly located VHS (Vps27/Hrs/STAM) domain and ubiquitin-interacting motif serve as the binding site(s). STAM proteins colocalize with Hrs on the early endosome. Overexpression of STAM proteins, but not their mutants lacking the ubiquitin-binding activity, causes the accumulation of ubiquitinated proteins and ligand-activated epidermal growth factor receptor on the early endosome. These results suggest that through interaction with ubiquitinated cargo proteins on the early endosome via the VHS domain and ubiquitin-interacting motif, STAM proteins participate in the sorting of cargo proteins for trafficking to the lysosome.
INTRODUCTION
Conjugation with ubiquitin serves as a sorting signal that determines the destination of various proteins at different subcellular sites such as the plasma membrane, the trans-Golgi network (TGN), and the endosome (Dupre et al., 2001; Hicke, 2001). The early endosome is an organelle where the sorting of endocytosed cell surface receptors that are to be recycled to the cell surface or destined for degradation in the lysosome occurs (Gruenberg and Maxfield, 1995). Newly synthesized lysosomal hydrolases are also delivered to the lysosome from the TGN via the endosome. At this organelle, ubiquitination is a sorting signal for endocytosed and newly synthesized lysosomal proteins to be incorporated into the luminal vesicles of the late endosome/multivesicular body (MVB) that bud inward from its limiting membrane (Dupre et al., 2001; Hicke, 2001). The late endosome/MVB then fuses with the lysosome, delivering the sorted proteins within the inner vesicles into the lumen of the lysosome. The molecular machinery of this trafficking route, the MVB pathway, has just begun to be elucidated in yeast (Conibear, 2002).
Hepatocyte growth factor-regulated substrate (Hrs) has been implicated in vesicular trafficking via the early endosome in higher eukaryotes. First, Hrs is mostly localized to the early endosome (Komada et al., 1997), and its yeast ortholog Vps27 belongs to the class E vacuolar protein sorting (Vps) proteins, mutations of which cause defective vesicular trafficking via the endosome (Raymond et al., 1992; Piper et al., 1995). Second, mice and flies lacking Hrs exhibit defective morphology and endocytic function of the endosome (Komada and Soriano, 1999; Lloyd et al., 2002). On the other hand, overexpression of Hrs also affects the morphology of the early endosome (Komada et al., 1997). Finally, Hrs can bind several proteins that are involved in vesicular trafficking such as SNAP-25 (Bean et al., 1997; Kwong et al., 2000), eps15 (Bean et al., 2000), sorting nexin 1 (Chin et al., 2001), and clathrin heavy chain (Raiborg et al., 2001). Recently, the ubiquitin-interacting motif (UIM), composed of ∼15 amino acid residues and originally identified as a polyubiquitin-binding site in the S5a subunit of the 26S proteasome (Young et al., 1998), was found in Hrs and its yeast ortholog Vps27 (Hofmann and Falquet, 2001). Later, it was demonstrated that the UIMs in these proteins also bind ubiquitin and that Vps27 is required for sorting ubiquitinated proteins for the MVB pathway (Bilodeau et al., 2002; Bishop et al., 2002; Polo et al., 2002; Raiborg et al., 2002; Shih et al., 2002). In addition, overexpression of Hrs, but not its mutant lacking the UIM, inhibits recycling of the transferrin receptor that is fused to ubiquitin, suggesting the possibility that Hrs is involved in the same sorting pathway in higher eukaryotes (Raiborg et al., 2002).
The signal-transducing adaptor molecule (STAM) family consists of two proteins, STAM1 and STAM2, which are 53% identical in amino acid sequences to each other (Takeshita et al., 1996; Lohi et al., 1998; Takata et al., 2000; Endo et al., 2000; Pandey et al., 2000). STAM2 is also known as Hrs-binding protein and EAST, and both STAM1 and STAM2 are tightly associated with Hrs (Asao et al., 1997; Takata et al., 2000). These proteins also possess the UIM (Hofmann and Falquet, 2001), suggesting that they cooperate with Hrs in vesicular trafficking of ubiquitinated proteins. Recently, it was shown that Hse1, a single yeast ortholog of STAM proteins, is associated with Vps27 and plays a role in sorting ubiquitinated proteins for the MVB pathway in a UIM-dependent manner (Bilodeau et al., 2002). However, the molecular function of STAM proteins in this process remains unknown.
STAM proteins possess the VHS (Vps27/Hrs/STAM) domain at the N terminus (Lohi and Lehto, 2001). This domain comprises ∼150 amino acids and is present in proteins involved in vesicular trafficking in eukaryotic cells (Lohi et al., 2002). In mammals, the VHS domain is present in eight proteins, including Golgi-localizing/γ-adaptin ear homology domain/ADP-ribosylation factor-binding (GGA) proteins, Hrs, STAM proteins, target of Myb 1 (Tom1), and Src-activating signaling molecule (Srcasm) (Lohi et al., 2002). The VHS domains in GGA proteins bind sortilin and mannose 6-phosphate receptors at the TGN, whereby GGAs mediate the trafficking of the sorting receptors to the lysosome (Nielsen et al., 2001; Puertollano et al., 2001; Takatsu et al., 2001; Zhu et al., 2001). However, the VHS domains in other proteins do not bind the sorting receptors (Puertollano et al., 2001; Takatsu et al., 2001), suggesting that they have distinct ligands yet to be identified. Herein, we demonstrate that STAM proteins bind ubiquitin via the tandemly located VHS domain and UIM and implicate the proteins in the sorting of ubiquitinated proteins in the MVB pathway.
MATERIALS AND METHODS
Far Western Screening
A λZAP mouse liver cDNA expression library was screened with 32P-labeled STAM2 by a far Western method as described previously (Kato et al., 2000).
Expression Constructs and Transfection
cDNAs for mouse STAM2 and Hrs were cloned as described previously (Komada et al., 1995; Takata et al., 2000). cDNAs for STAM1 (full-length) and Tom1 (VHS domain; amino acids 1–160) were amplified by polymerase chain reaction from a Marathon-Ready mouse brain cDNA library (BD Biosciences Clontech, Palo Alto, CA). cDNA for GGA1 (VHS domain; amino acids 1–159) was amplified similarly from a Marathon-Ready human brain cDNA library (BD Biosciences Clontech). Their sequences were subsequently verified by DNA sequencing.
Deletion constructs of STAM proteins were obtained by in vitro mutagenesis by using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA), and their sequences were verified by DNA sequencing. Deleted regions in STAM2ΔVHS, STAM2ΔUIM, STAM2-VHS, and STAM2ΔSH3 are amino acids 8–151, 163–180, 152–523, and 207–258, respectively. Deleted regions in STAM1ΔVHS, STAM1ΔUIM, and STAM1-VHS are amino acids 8–154, 170–186, and 155–548, respectively. The VHS domain of Hrs comprises amino acids 1–164. cDNA constructs were cloned into FLAG epitope- and hemag-glutinin (HA) epitope-tagged mammalian expression vectors pME-FLAG and pME-HA (Kato et al., 2000), respectively, and transfected into HeLa and COS-7 cells by using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN).
Glutathione S-Transferase (GST) Fusion Proteins
The VHS domain (amino acids 1–151), UIM (amino acids 152–192), and N-terminal region (amino acids 1–192) of STAM2, as well as the UIM of Hrs (amino acids 216–289), were cloned into the pGEX vectors (Amersham Biosciences, Piscataway, NJ) to generate the GST fusion constructs. GST fusion proteins were purified from the Escherichia coli strain BL21 transformed with the GST fusion constructs by using glutathione-Sepharose affinity beads (Amersham Biosciences).
Pull-Down Assay with Ubiquitin Beads
Two days after transfection of the FLAG-tagged expression constructs, HeLa cells in 100-mm dishes were lysed in 0.8 ml of lysis buffer (20 mM Tris-HCl pH 7.4, 100 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A). After centrifugation at 12,000 × g for 15 min at 4°C, the supernatants were incubated with 10 μl of ubiquitin-agarose beads (Boston Biochem, Cambridge, MA) for 16 h at 4°C. The beads were then washed three times with washing buffer (10 mM Tris-HCl pH 7.4, 100 mM NaCl, and 0.1% Nonidet P-40), and the bound proteins eluted by boiling with 1× Laemmli sample buffer. Eluted proteins were separated by SDS-PAGE, and FLAG-tagged proteins were detected by Western blotting by using anti-FLAG antibody (4 μg/ml; Sigma-Aldrich, St. Louis, MO), peroxidase-conjugated anti-mouse IgG antibody (1:20,000; Amersham Biosciences), and the enhanced chemiluminescence reagent (Amersham Biosciences).
To examine the binding of purified GST fusion proteins, ∼3 μg of the proteins was incubated with the ubiquitin beads in phosphate-buffered saline containing 1% Triton X-100, and bound GST fusion proteins were detected by Western blotting with anti-GST antibody (1 μg/ml; Santa Cruz Biotechnology, Santa Cruz, CA) after SDS-PAGE.
Pull-Down Assay with GST Fusion Proteins
The VHS domain, UIM, and N-terminal region (1–192) of STAM2 (3 μg) were coupled to 10 μl of glutathione beads as GST fusion proteins. To examine the binding of purified ubiquitin, 1 μg of a mixture of polyubiquitin chains (Ub2–7; Affiniti Research Products, Exeter, UK) was incubated with the beads in phosphate-buffered saline containing 1% Triton X-100 for 16 h at 4°C. The beads were washed with the washing buffer, and bound ubiquitins were detected by Western blotting with anti-ubiquitin antibody (1 μg/ml; MBL, Nagoya, Japan).
To examine the binding of ubiquitinated proteins, COS-7 cells in a 60-mm dish were transfected with a FLAG-tagged monoubiquitin expression vector (pcDNA3.1-FLAG-Ub, a kind gift of Dr. T. Suzuki, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). Two days after transfection, the cells were lysed with 0.3 ml of the lysis buffer, and the supernatant after centrifugation at 12,000 × g was used for the pull-down assay. The bound ubiquitinated proteins were detected with anti-FLAG antibody as described above. The amount of GST fusion proteins used for the pull-down assay was assessed by Western blotting with anti-GST antibody.
Detection of Intracellular Ubiquitinated Proteins
COS-7 cells in 60-mm dishes were cotransfected with HA-tagged STAM constructs and FLAG-tagged monoubiquitin. Two days after transfection, the cells were lysed with 0.3 ml of the lysis buffer. The lysates were then immunoprecipitated with anti-FLAG antibody (1 μg), and the immunoprecipitates were analyzed by Western blotting by using the same antibody. Anti-HA antibody (0.5 μg/ml; Sigma-Aldrich) was used in Western blotting of total cell lysates to detect expression of the STAM constructs.
Immunofluorescence Staining
Two days after transfection, cells were fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, and stained with mouse monoclonal anti-HA (4 μg/ml; Sigma-Aldrich), mouse monoclonal anti-FLAG (4 μg/ml; Sigma-Aldrich), rabbit polyclonal anti-FLAG (2 μg/ml; Affinity BioReagents, Golden, CO), rabbit polyclonal anti-Hrs (1:2000 dilution; Komada and Kitamura, 1995), mouse monoclonal FK2 (10 μg/ml; Affiniti Research Products), or mouse monoclonal anti-epidermal growth factor (EGF) receptor (10 μg/ml; MBL) by standard procedures. The secondary antibodies used were Alexa488- and Alexa594-conjugated anti-mouse and -rabbit IgG antibodies (Molecular Probes, Eugene, OR). Fluorescent images were captured using a confocal microscope.
To quantify the proportion of cells exhibiting FK2-positive early endosomes among those overexpressing each FLAG-tagged STAM2 construct, ∼100 cells strongly positive for anti-FLAG staining were randomly chosen and examined for FK2 staining. The experiment was repeated three times and the mean ± SD of the proportion were determined. The proportion of cells exhibiting enlarged early endosomes among those overexpressing the STAM2 constructs was determined in a similar manner.
To stain the internalized EGF receptor, COS-7 cells transfected with FLAG-tagged STAM2 constructs for 2 d were grown in the presence of 0.5% fetal bovine serum for another 24 h, and treated with 100 ng/ml EGF (PeproTech, Rocky Hill, NJ) and 10 μg/ml cycloheximide for 3 h. The cells were then subjected to immunofluorescence staining as described above.
RESULTS
STAM2 Binds Ubiquitin
To understand the function of STAM proteins, we have tried to identify proteins that directly bind STAM2 by far Western screening. We previously reported an interaction of the deubiquitinating enzyme UBPY with the Src homology 3 (SH3) domain of STAM2 (Kato et al., 2000). In the screening, we also isolated a partial cDNA encoding polyubiquitin. Polyubiquitin consists of several tandem repeats of the 76-aminoacid monoubiquitin and is cleaved to monoubiquitins to supply the intracellular free ubiquitin pool (Finley and Chau, 1991). To detect an interaction between STAM2 and ubiquitin in vitro, HeLa cells were transfected with a FLAG epitope-tagged STAM2 expression vector, and the cell lysate incubated with agarose beads to which monoubiquitin was conjugated (ubiquitin beads). After washing the beads, bound proteins were eluted with SDS and detected by Western blotting with anti-FLAG antibody (ubiquitin pull-down assay). As shown in Figure 1B, STAM2 bound to the ubiquitin beads but not to control beads to which no ubiquitin was conjugated, confirming the result of the far Western screening.
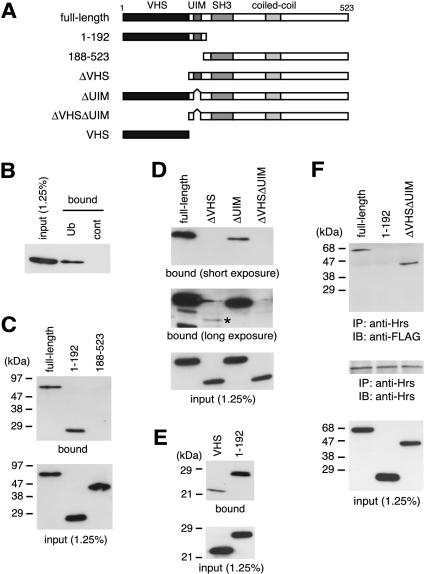
Figure 1.
Binding of STAM2 to ubiquitin. (A) Domain structure of STAM2 and its mutants used in this study. VHS domain, UIM, SH3 domain, and coiled-coil region are indicated. (B) FLAG-tagged STAM2 was expressed in HeLa cells and incubated with ubiquitin beads (Ub) or control beads (cont) to which no ubiquitin was conjugated. Bound proteins were separated by SDS-PAGE and detected by Western blotting with anti-FLAG antibody. The input of STAM2 (1.25%) was detected by Western blotting of the same lysate as used in the pull-down assay. (C–E) Ubiquitin pull-down assay of the N-terminal region (amino acids 1–192) and the C-terminal region (amino acids 188–523) of STAM2 (C), STAM2 lacking the VHS domain (ΔVHS), UIM (ΔUIM), or both (ΔVHSΔUIM) (D), and the VHS domain alone (VHS) compared with the N-terminal region (1–192) of STAM2 (E) is shown (top). In D, short exposure (top) and long exposure (middle) of the Western blot membrane are shown. An asterisk in the middle of D indicates STAM2ΔVHS, which was only detected after long exposure. The input of the constructs was assessed by Western blotting of the same lysates as used in the pull-down assay and is shown in bottom panels. (F) Binding of Hrs to STAM2 and its mutants. Lysates of HeLa cells transfected with FLAG-tagged full-length STAM2, N-terminal region (1–192), or STAM2ΔVHSΔUIM, were immunoprecipitated (IP) with anti-Hrs antibody and examined by Western blotting (IB) with anti-FLAG (top) or anti-Hrs (middle) antibodies. The expression level (input) of the STAM2 constructs was assessed by Western blotting of the same lysates as used in the immunoprecipitation (bottom).
STAM2 Binds Ubiquitin via the VHS Domain and UIM
To determine the ubiquitin-binding site of STAM2, we first examined the ability of the N-terminal region containing the VHS domain and UIM (amino acids 1–192) as well as the C-terminal region containing the SH3 domain and the coiled-coil region (amino acids 188–523) to bind ubiquitin. As shown in Figure 1C, the N-terminal region bound to the ubiquitin beads as efficiently as the full-length protein, whereas the C-terminal region exhibited no binding, indicating that the ubiquitin-binding site is located in the N-terminal region. Deletion of the VHS domain from the full-length protein dramatically reduced the ubiquitin binding (Figure 1D, ΔVHS). The binding, however, was detectable after longer exposure of the Western blot membrane, indicating that it was not completely abolished (Figure 1D, middle). Deletion of the UIM also reduced the binding activity to a large extent (Figure 1D, ΔUIM), and a mutant lacking both the VHS domain and UIM did not bind ubiquitin at all (Figure 1D, ΔVHSΔUIM). These results suggest that the VHS domain plays an essential role in ubiquitin binding and that the UIM is also required for maximal binding.
Next, we examined whether the VHS domain is sufficient to bind ubiquitin. The VHS domain alone was able to bind ubiquitin (Figure 1E). However, the binding activity was much lower than that of the N-terminal region, which contains both the VHS domain and UIM (Figure 1E, 1–192). We were not able to express the UIM alone in cells. However, very weak ubiquitin-binding activity of STAM2ΔVHS, which contains the UIM (Figure 1D), suggests that the UIM alone does not bind ubiquitin efficiently compared with the VHS domain. These results suggest that the inability of the mutants lacking the VHS domain and/or UIM to bind ubiquitin is due to the absence of these domains and not to the perturbation of its conformation and that these domains play a synergistic role on ubiquitin binding. Similar results were obtained when glutathione beads to which the GST-tagged monoubiquitin was coupled were used for the pull-down assay (our unpublished data).
The interaction in far Western screening suggested that the binding between STAM proteins and ubiquitin is direct. However, as STAM proteins are associated with Hrs, which binds ubiquitin, the ubiquitin binding of the STAM2 constructs in the pull-down assay might be mediated by endogenous Hrs. We therefore examined the association of full-length STAM2, the N-terminal region (1–192), and STAM2ΔVHSΔUIM with Hrs by coimmunoprecipitation experiments. Consistent with previous reports showing that Hrs and STAM proteins interact via their coiled-coil regions (Asao et al., 1997; Takata et al., 2000), anti-Hrs coprecipitated full-length STAM2, and STAM2ΔVHSΔUIM but not the N-terminal region (1–192) (Figure 1F). These results indicate that the ubiquitin-binding activity of the STAM2 constructs is not Hrs dependent.
VHS Domain and UIM of STAM2 Bind Ubiquitin Directly
To further confirm that STAM proteins bind ubiquitin directly, we examined the ubiquitin-binding activity of purified, GST-tagged VHS domain, UIM, and N-terminal region (1–192) of STAM2 by the pull-down assay. Whereas the binding of the control GST protein was hardly detectable, the VHS domain and UIM bound to the ubiquitin beads independently (Figure 2A, left). In this assay, the VHS domain and UIM bound ubiquitin in a similar efficiency. Consistent with the results shown in Figure 1, D and E, the N-terminal region containing both the VHS domain and UIM (1–192) bound it much more efficiently than either individual domain. The UIM of Hrs bound ubiquitin to a similar extent to the N-terminal region of STAM2 (1–192) (Figure 2A).
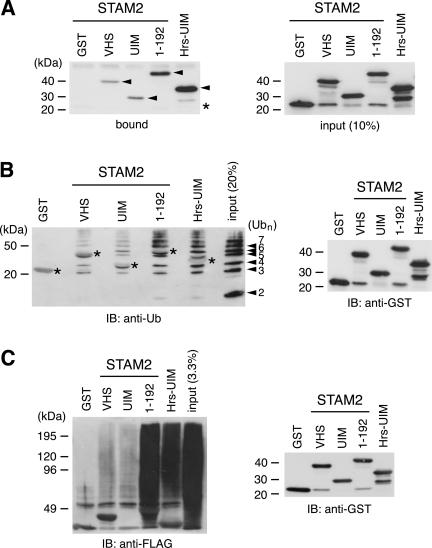
Figure 2.
Direct binding of the STAM2 VHS domain and UIM to ubiquitin and ubiquitinated proteins. (A) The purified GST-tagged VHS domain, UIM, and N-terminal region (1–192) of STAM2, as well as the UIM of Hrs, were incubated with ubiquitin beads, and bound proteins were analyzed by Western blotting with anti-GST antibody (left). The bound GST fusion proteins are indicated by arrowheads. The position of GST is indicated by an asterisk. The input (10%) of the GST fusion proteins detected by anti-GST antibody is shown (right). (B) A mixture of polyubiquitin chains was incubated with glutathione beads to which the VHS domain, UIM, and N-terminal region (1–192) of STAM2, as well as the UIM of Hrs, were coupled. Bound ubiquitin chains were detected by Western blotting with anti-ubiquitin antibody (left). The numbers on the right of the left panel indicate the n values of the polyubiquitin chain (Ubn). Larger bands represent multimeric complexes of ubiquitin with unknown nature. Asterisks indicate background staining of the GST-fusion proteins, which were faintly detected due to their large amount. (C) Lysate of cells transfected with FLAG-ubiquitin was incubated with glutathione beads to which the VHS domain, UIM, and N-terminal region (1–192) of STAM2, as well as the UIM of Hrs, were coupled. Bound proteins were separated by SDS-PAGE and ubiquitinated proteins were detected by Western blotting by using anti-FLAG antibody (left). The amount of the GST fusion proteins used for the pull-down assay was assessed by Western blotting with anti-GST antibody (B and C, right).
Reciprocally, binding of ubiquitin in solution to immobilized VHS domain and/or UIM of STAM2 was also examined. A mixture of polyubiquitin chains (dimer to heptamer) was incubated with the VHS domain and/or UIM, which were fused to GST and coupled to glutathione beads, and bound ubiquitin chains were detected by Western blotting with anti-ubiquitin antibody. Also in this assay, the ubiquitin chains bound to the VHS domain and UIM to a similar extent, and much more efficiently to the N-terminal region (1–192) (Figure 2B). The control GST protein did not bind ubiquitin, and the UIM of Hrs bound it as efficiently as the N-terminal region of STAM2 (1–192) (Figure 2B).
These results indicate that both the VHS domain and UIM in STAM2 can serve as a ubiquitin-binding site, and suggest a synergistic effect of these domains on the binding when they are tandemly located. It is, however, not known whether the VHS domain and UIM cooperatively bind a single ubiquitin molecule or bind two ubiquitin molecules independently in the context of the full-length protein.
STAM2 Binds Intracellular Ubiquitinated Proteins
To examine whether STAM proteins bind ubiquitinated proteins, lysate of COS-7 cells transfected with FLAG-tagged monoubiquitin was used for the pull-down assay with the STAM2 VHS domain and/or UIM, which were coupled to glutathione beads. Ubiquitinated proteins that bound to the beads were detected by Western blotting with anti-FLAG antibody. Although ubiquitinated proteins did not bind the control GST protein, they bound both the VHS domain and UIM (Figure 2C). Again, the binding activity was drastically increased when these domains were tandemly located (Figure 2C, 1–192). The UIM of Hrs bound the ubiquitinated proteins more efficiently than that of STAM2, but not as efficiently as the N-terminal region (1–192) (Figure 2C). These results suggest that STAM proteins bind ubiquitinated proteins within cells via the VHS domain and UIM.
STAM1 Binds Ubiquitin in a Similar Manner to STAM2
We next examined whether STAM1, the close homolog of STAM2, binds ubiquitin in a similar manner. In the pull-down assay, STAM1 also bound ubiquitin (Figure 3A, full-length). In addition, deletion of the VHS domain from STAM1 severely affected its ubiquitin-binding activity and deletion of the UIM also reduced the binding to some extent (Figure 3A, ΔVHS and ΔUIM). The VHS domain of STAM1 was also capable of binding ubiquitin, and the N-terminal region containing both the VHS domain and UIM (1–213) bound it more efficiently (Figure 3B). These results indicate that STAM2 and STAM1 are functionally redundant in binding ubiquitin.
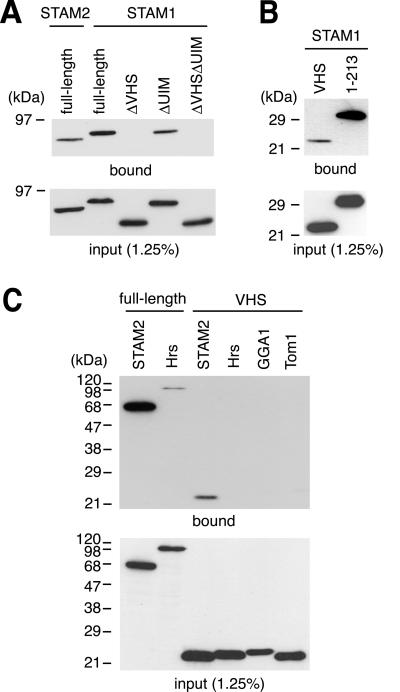
Figure 3.
Binding of STAM1 and other VHS domains to ubiquitin. FLAG-tagged STAM1 constructs (A and B) and VHS domains from other proteins (C) were expressed in HeLa cells and examined by the pull-down assay with ubiquitin beads. (A and B) Pull-down assay of full-length STAM1 and its mutants lacking the VHS domain (ΔVHS), UIM (ΔUIM), or both (ΔVHSΔUIM) (A, top), as well as the VHS domain alone (VHS) and an N-terminal region containing the VHS domain and UIM (amino acids 1–213) (B, top). (C) Pull-down assay of full-length Hrs and the VHS domains from Hrs, GGA1, and Tom1, together with STAM2 and its VHS domain (top). The input of the constructs was assessed by Western blotting of the same lysates as used in the pull-down assay (bottom).
Ubiquitin Binding of VHS Domains of Other Proteins
The VHS domain is found in several proteins including Hrs. We next examined whether VHS domains in other proteins are also capable of binding ubiquitin. As shown in Figure 3C, none of the VHS domains tested (Hrs, GGA1, and Tom1) exhibited detectable ubiquitin binding in a condition in which the STAM2 VHS domain did. Although these results do not completely exclude the possibility that other VHS domains bind ubiquitin, they suggest that the VHS domains of STAM proteins have higher ubiquitin-binding affinity than, if any, that of other VHS domains. As reported previously (Bishop et al., 2002; Lloyd et al., 2002; Polo et al., 2002; Raiborg et al., 2002), full-length Hrs bound ubiquitin although less efficiently than STAM2 (Figure 3C).
STAM Proteins Colocalize with Hrs on the Early Endosome
To elucidate the intracellular site where STAM proteins interact with ubiquitinated proteins, the subcellular localization of STAM proteins was examined. HeLa cells were transiently transfected with HA-tagged STAM2 or STAM1, and then stained with anti-HA antibody. In cells expressing a low level of HA-STAM proteins, the proteins were mainly localized to punctate structures in the cytoplasm (Figure 4, A and B). These structures were also positive for endogenous Hrs (Figure 4, A′, A″, B′, and B″), indicating that they are the early endosomes. When HA-STAM2 was expressed at a higher level, it was localized throughout the cytoplasm (Figure 4C). Staining with anti-Hrs antibody showed that overexpression of STAM2 causes the enlargement of the Hrs-positive early endosomes (Figure 4, C′ and C″, arrows), similar to the effect of Hrs overexpression (Komada et al., 1997). STAM1 overexpression caused the same effect (our unpublished data). Together with the previous observation that Hrs and STAM proteins are tightly associated (Asao et al., 1997; Takata et al., 2000), these results suggest that STAM proteins function as a complex with Hrs on the early endosome.
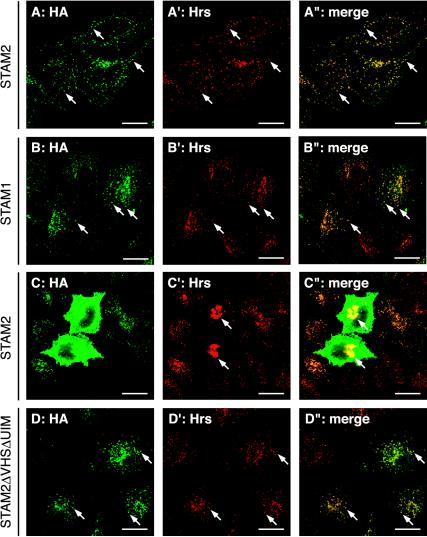
Figure 4.
Localization of STAM proteins on the early endosome. HeLa cells were transiently transfected with HA-tagged STAM2 (A–A″ and C–C″), STAM1 (B–B″), or STAM2ΔVHSΔUIM (D–D″), and double-stained with anti-HA (A–D) and anti-Hrs (A′. B′, C′, and D′) antibodies. A″, B″, C″, and D″ are merged images. Cells expressing a low level of HA-STAM2 (A–A″) and those expressing a high level (C–C″) are shown. Arrows in A–A″, B–B″, and D–D″ indicate typical early endosomes. Arrows in C′ and C″ indicate enlarged early endosomes in STAM2-overexpressing cells. Bars, 20 μm.
Next, the subcellular localization of STAM2 mutants lacking the VHS domain, UIM, or both, was examined. When expressed at a low level, all the mutants exhibited a similar localization pattern to that of the full-length protein (Figure 4, D–D″; our unpublished data). In contrast, the VHS domain alone and the N-terminal region containing the VHS domain and UIM (1–192) were localized throughout the cytoplasm (our unpublished data). These results indicate that the ubiquitin-binding activity is not required for the early endosomal localization of STAM proteins.
STAM Overexpression Causes the Accumulation of Ubiquitinated Proteins on the Early Endosome
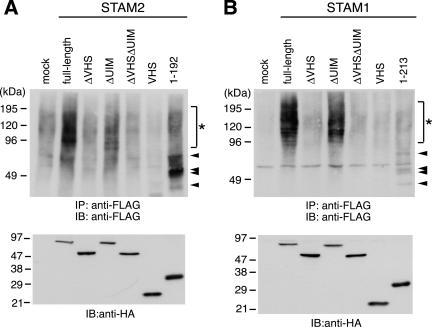
If STAM proteins interact with ubiquitinated proteins on the early endosome, their overexpression may affect the amount of ubiquitinated proteins associated with this organelle. To test this possibility, we examined the effect of overexpressing STAM proteins and their deletion mutants on the ubiquitination level of intracellular proteins. COS-7 cells were transfected with HA-tagged STAM2 or STAM1 constructs, together with FLAG-tagged ubiquitin. Ubiquitinated proteins were immunoprecipitated with anti-FLAG antibody and then detected by Western blotting by using the same antibody. Overexpression of full-length STAM1 and STAM2 significantly increased the ubiquitination level of intracellular proteins, mostly ranging from 100 to 200 kDa, compared with the mock transfection control (Figure 5, A and B, asterisks). This effect was not observed when mutants lacking the VHS domain alone (ΔVHS) or both the VHS domain and UIM (ΔVHSΔUIM) were overexpressed (Figure 5, A and B). In contrast, deletion of the UIM alone gave only a moderate reduction in the ubiquitination level observed with the full-length proteins (Figure 5, A and B, ΔUIM). This was consistent with the residual ubiquitin-binding activity of these mutants (Figures 1D and 3A). These effects are not simply due to overexpression of a ubiquitin-binding site because the VHS domains alone (Figure 5, A and B, VHS) and the N-terminal regions containing the VHS domain and UIM (Figure 5A, 1–192 for STAM2; B, 1–213 for STAM1) had no such effect. Instead, however, specific ubiquitinated proteins were detected when the N-terminal regions were overexpressed (Figure 5, A and B, arrowheads). This effect must be related to the ubiquitin-binding activity of these regions and require the UIM, because these ubiquitinated proteins were not detected when the VHS domains alone were overexpressed (Figure 5, A and B). The UIM was recently shown to play a role in monoubiquitination of UIM-containing proteins including eps15, epsin, Hrs, as well as STAM1 (Katz et al., 2002; Polo et al., 2002). However, most of the ubiquitinated proteins detected in Figure 5 were not the monoubiquitinated forms of the STAM constructs as judged from their size.
Figure 5.
Accumulation of ubiquitinated proteins in cells overexpressing STAM proteins. COS-7 cells were transfected with FLAG-tagged ubiquitin together with HA-tagged STAM2 (A) or STAM1 (B) constructs. The cells were lysed and ubiquitinated proteins were detected by immunoprecipitation (IP) followed by Western blotting (IB) with anti-FLAG antibody (top). Expression of STAM2 (A) and STAM1 (B) constructs as assessed by Western blotting of the total cell lysates with anti-HA antibody (bottom). Ubiquitinated proteins which are specific in cells overexpressing full-length STAM2 or STAM1, and those specific in cells overexpressing the N-terminal regions (1–192 for STAM2, 1–213 for STAM1), are indicated by asterisks and arrowheads, respectively.
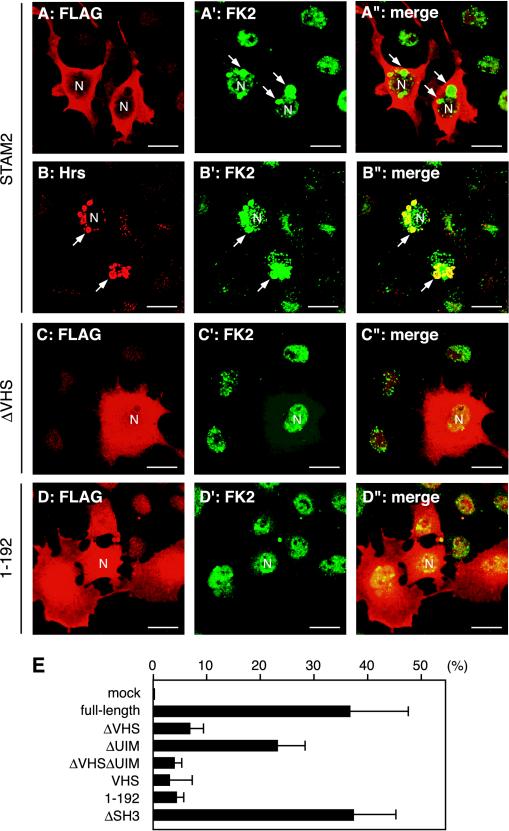
To reveal where these ubiquitinated proteins were accumulated within the cells, cells transfected with FLAG-tagged STAM2 or its deletion mutants were immunostained with FK2, an antibody that recognizes the ubiquitin moiety of ubiquitinated proteins (Fujimuro et al., 1994). In cells overexpressing full-length STAM2, perinuclear vacuolar structures were often stained (Figure 6A′, arrows). Double staining with anti-Hrs showed that these structures are also positive for Hrs, indicating that they are enlarged early endosomes (Figure 6, B–B″). The proportion of cells exhibiting such FK2-positive enlarged endosomes was reduced when mutants lacking the ubiquitin-binding activity (ΔVHS, ΔUIM, and ΔVHSΔUIM) or consisting only of the ubiquitin-binding site (VHS and 1–192) were overexpressed. Figure 6, C–C″ and D–D″ show cells overexpressing STAM2ΔVHS and the N-terminal region (1–192), respectively, and exhibiting no FK2-positive endosomes. The effects of the deletion constructs on the accumulation of ubiqutinated proteins were quantified as a proportion of the FK2-positive cells among those overexpressing each construct. Consistent with the results of Western blot analysis (Figure 5), only a low proportion of STAM2ΔVHS- and STAM2ΔVHSΔUIM-overexpressing cells exhibited FK2-positive enlarged endosomes (Figure 6E). Overexpression of the VHS domain alone or the N-terminal region (1–192) had similar effects (Figure 6E). STAM2ΔUIM, in contrast, retained some activity to generate FK2-positive early endosomes, although it was not as high as that of the full-length protein (Figure 6E, ΔUIM). STAM2 lacking the SH3 domain had the same effect as the full-length protein (Figure 6E, ΔSH3). Similar results were obtained with STAM1 (our unpublished data).
Figure 6.
Accumulation of ubiquitinated proteins on the early endosome in STAM2-overexpressing cells. (A–D″) COS-7 cells were transfected with FLAG-tagged full-length STAM2 (A–A″ and B–B″), STAM2ΔVHS (C–C″), or N-terminal region (1–192) (D–D″). The cells were stained with FK2 (A′, B′, C′, and D′), together with anti-FLAG (A, C, and D) or anti-Hrs (B) antibodies. A″, B″, C″, and D″ are merged images. Arrows indicate enlarged early eodosomes in STAM2-overexpressing cells. The nuclei (N) are also indicated in some cells. Bars, 20 μm. (E) Proportion of cells exhibiting FK2-positive early endosomes among those overexpressing the indicated STAM2 deletion mutants. Mean ± SD of three independent experiments are shown.
These results suggest that STAM proteins interact with ubiquitinated proteins on the early endosome and that the VHS domain as well as the UIM play important roles in this interaction.
Enlargement of the Early Endosome by STAM Requires the VHS Domain and UIM
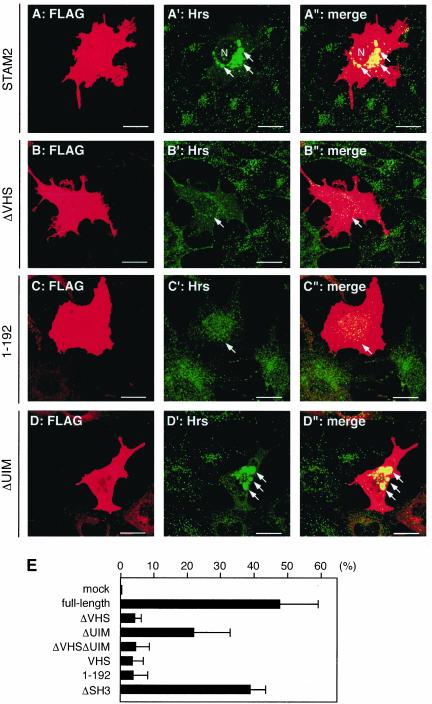
Overexpression of STAM2 causes enlargement of the early endosome in HeLa cells (Figure 4, C–C″). To examine whether the domains required for ubiquitin binding (i.e., the VHS domain and UIM) are also required for this effect, we transfected COS-7 cells with various STAM2 deletion mutants and visualized the early endosomes by staining for endogenous Hrs. Compared with full-length STAM2, which caused enlargement of the organelle also in COS-7 cells (Figure 7, A–A″), overexpression of STAM2ΔVHS, STAM2ΔVHSΔUIM, the VHS domain, or the N-terminal region (1–192) had no effect or only slightly increased the size of the early endosome in most of the cells (Figure 7, B–B″ and C–C″; our unpublished data). In contrast, STAM2ΔUIM overexpression caused enlargement of the early endosome (Figure 7, D–D″). Again, however, the effect was not as efficient as that of the full-length protein. Figure 7E shows the quantification of the proportion of cells exhibiting enlarged early endosomes among those overexpressing each deletion construct. STAM2ΔSH3 had a similar effect to that of the full-length protein also in this assay. Similar results were obtained with STAM1 (our unpublished data). These results suggest that the ability of STAM proteins to bind ubiquitin and to affect the early endosome morphology are closely related.
Figure 7.
Enlargement of the early endosome in STAM2-overexpressing cells. (A–D″) COS-7 cells were transfected with FLAG-tagged full-length STAM2 (A–A″), STAM2ΔVHS (B–B″), N-terminal region (1–192) (C–C″), or STAM2ΔUIM (D–D″). The cells were double stained with anti-FLAG (A, B, C, and D) and anti-Hrs (A′, B′, C′, and D′) antibodies. A″, B″, C″, and D″ are merged images. Arrows in A′, A″, D′, and D″ indicate enlarged early endosomes in STAM2- and STAM2ΔUIM-overexpressing cells. Arrows in B′, B″, C′, and C″ indicate STAM2ΔVHS- and N-terminal region (1–192)-overexpressing cells with early endosomes of normal size. The nucleus (N) is also indicated (A′ and A″). Bars, 20 μm. (E) Proportion of cells exhibiting enlarged early endosomes among those overexpressing the indicated STAM2 deletion mutants. Mean ± SD of three independent experiments are shown.
Effect of STAM2 Overexpression on the EGF Receptor Trafficking
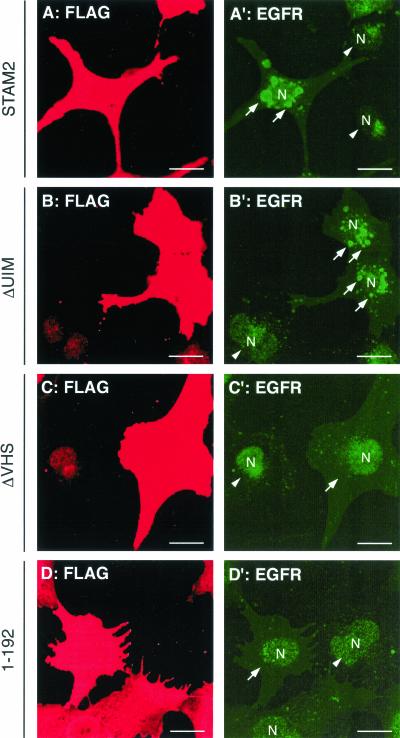
On EGF stimulation, the EGF receptor is internalized and delivered to the lysosome for degradation via the MVB pathway (Felder et al., 1990). We next examined the effect of STAM2 overexpression on the trafficking of the EGF receptor. COS-7 cells were transfected with FLAG-tagged STAM2 or its deletion mutants and exposed to EGF for 3 h in the presence of cycloheximide. The receptor localization was then visualized by immunofluorescence staining with anti-EGF receptor antibody. Untransfected cells exhibited weak EGF receptor staining, which is indicative of degradation of the receptor in the lysosome (Figure 8, A′, B′, C′, and D′, arrowheads). In cells overexpressing full-length STAM2, the EGF receptor was localized to vacuolar structures around the nucleus (Figure 8A′, arrows). These structures were positive for Hrs (our unpublished results), suggesting that inhibition of the EGF receptor trafficking to the lysosome led to its accumulation on the early endosome. Although not as effective as the full-length protein, STAM2ΔUIM retained some activity to accumulate the EGF receptor on the organelle (Figure 8B′, arrows). In contrast, cells overexpressing STAM2ΔVHS or the N-terminal region (1–192) were only slightly stained by anti-EGF receptor antibody, similar to untransfected cells (Figure 8, C′ and D′, arrows). STAM2ΔSH3 had a similar effect to that of the full-length protein (our unpublished data). These results suggest that STAM proteins play a role in the trafficking of ligand-activated EGF receptor to the lysosome and that the VHS domain is essential for the function.
Figure 8.
Effect of STAM2 overexpression on the EGF receptor trafficking. COS-7 cells were transfected with FLAG-tagged STAM2 (A and A′), STAM2ΔUIM (B and B′), STAM2ΔVHS (C and C′), or N-terminal region (1–192) (D and D′), and treated with EGF for 3 h in the presence of cycloheximide. The cells were double-stained with anti-FLAG (A, B, C, and D) and anti-EGF receptor (EGFR; A′, B′, C′, and D′) antibodies. Arrows in A′ and B′ indicate EGF receptor-positive early endosomes in STAM2- and STAM2ΔUIM-overexpressing cells. Arrows in C′ and D′ indicate cells overexpressing STAM2ΔVHS and the N-terminal region (1–192). Arrowheads indicate untransfected cells. The nuclei (N) are also indicated (A′, B′, C′, and D′). Bars, 20 μm.
DISCUSSION
In this study, we demonstrate that STAM proteins bind ubiquitin in a unique manner that involves the tandemly located VHS domain and UIM. Accumulation of ubiquitinated proteins and ligand-activated EGF receptor on the early endosome in STAM-overexpressing cells suggests that STAM proteins interact with ubiquitinated cargo proteins on this organelle and participate in sorting these proteins for trafficking to the lysosome.
VHS Domain as a Ubiquitin-binding Site
The VHS domain is present in eight mammalian proteins (GGA1/2/3, Hrs, STAM1/2, Tom1, and Srcasm). Crystal structures of the VHS domains in Hrs and Tom1 revealed that the domain forms a super helix with eight α helices (Mao et al., 2000; Misra et al., 2000). Because superhelical domains of several other proteins serve as a site of protein–protein interaction, the VHS domain has been suggested to be a protein-binding site. Indeed, the VHS domains of GGA proteins have been shown to bind the acidic-cluster dileucine (ACLL) signal of sorting receptors such as the mannose 6-phosphate receptors and sortilin (Nielsen et al., 2001; Puertollano et al., 2001; Takatsu et al., 2001; Zhu et al., 2001). In this study, we show that the VHS domains of STAM proteins bind another protein, ubiquitin. This is the second molecule identified as a ligand for the VHS domain, and thus we herein demonstrate a novel function of the VHS domain.
VHS domains other than those of GGA proteins do not bind the ACLL signal (Puertollano et al., 2001; Takatsu et al., 2001). The reason for this specificity was revealed from the crystal structures of the GGA VHS domains in complex with the ACLL signal of cation-independent mannose 6-phosphate receptor (Misra et al., 2002; Shiba et al., 2002; Kato et al., 2002). This analysis demonstrated that the amino acid residues of the VHS domains critical for the ACLL signal binding are located in helices 6 and 8 and are conserved only in GGA proteins. Similarly, the VHS domains from proteins other than STAM (Hrs, GGA1, and Tom1) did not exhibit detectable ubiquitin-binding activity (Figure 3C). Because the sequence homology of the VHS domains between STAM1 and STAM2 (74% amino acid identity) is much higher than that between STAM proteins and others (<40%), the amino acid residues conserved only in STAM proteins are likely to play important roles in ubiquitin binding. However, as the VHS domains of Hrs, GGA1, and Tom1 used in the pull-down assay were expressed in HeLa cells and not purified, a possibility that cellular protein(s) bound to these VHS domains and masked their ubiquitin-binding activity cannot be excluded. To confirm the specificity of the STAM VHS domain, therefore, it is necessary to compare their ubiquitin-binding activity using purified domains.
Regions in ubiquitin that are essential for its function as a sorting signal have been identified. A region surrounding isoleucine44 is required to serve as both proteasomal degradation and endocytosis signals, whereas a region surrounding phenylalanine4 is only required for endocytosis (Nakatsu et al., 2000; Shih et al., 2000; Sloper-Mould et al., 2001). In addition, isoleucine44 but not phenylalanine4 is required for ubiquitin to bind the UIMs of Vps27 and Ent1 (Shih et al., 2002). Further studies are necessary to determine whether these regions are involved in the interaction with the STAM VHS domain.
Manner of Ubiquitin Binding by STAM Proteins
The UIM was originally identified in the S5a subunit of the 19S regulatory complex of the 26S proteasome as a polyubiquitin-binding site (Young et al., 1998). Similar sequence motifs were later found in a number of proteins involved in the proteasomal and endocytic machinery (Hofmann and Falquet, 2001). Recently, the UIMs in endocytic proteins such as eps15, epsin, and Hrs were also shown to be critical for ubiquitin binding of these proteins (Bilodeau et al., 2002; Bishop et al., 2002; Polo et al., 2002; Raiborg et al., 2002; Shih et al., 2002). STAM proteins also possess the UIM. However, the manner of ubiquitin binding by this family of proteins is unique compared with other UIM-containing proteins because their ubiquitin-binding activity was drastically reduced by deleting the VHS domain (Figures 1D and 3A). In addition, the N-terminal region containing both the VHS domain and UIM bound ubiquitin much more efficiently than the individual domains (Figures 1E, 2, and 3B). These results support the possibility that these domains play a cooperative, rather than an additive, role on ubiquitin binding. It is not clear, however, whether the VHS domain and UIM bind a single ubiquitin molecule by recognizing distinct regions of ubiquitin or if the domains individually bind distinct ubiquitin molecules when tandemly located in the full-length STAM proteins. A crystal structural analysis of the VHS domain and UIM in complex with ubiquitin should answer this question.
Hrs also binds ubiquitin and possesses the VHS domain and UIM, raising the possibility that its manner of ubiquitin binding is similar to that of STAM proteins. This is, however, unlikely because first, deletion of or point mutations in the UIM of Hrs and its yeast ortholog Vps27 very significantly reduce their ubiquitin-binding activity (Raiborg et al., 2002; Shih et al., 2002; Bilodeau et al., 2002). Second, the Hrs VHS domain exhibited undetectable ubiquitin-binding activity in this study (Figure 3C). Finally, because the VHS domain and UIM are not tandemly located but are divided by the phosphatidylinositol (3)-phosphate-binding FYVE domain in Hrs, it is unlikely that these domains function cooperatively.
In the ubiquitin pull-down assay with the STAM constructs expressed in HeLa cells, deletion of the VHS domain affected the binding much more severely than deletion of the UIM (Figures 1D and 3A). However, the binding activity of the UIM was not significantly different from that of the VHS domain when the individual domains were purified as GST fusion proteins (Figure 2). Although the reason for this discrepancy is unclear, it may suggest that the UIM behaves differently depending on whether it is embedded in the full-length protein or it is isolated.
Role for STAM Proteins in Endosomal Protein Sorting
Three phenotypes were observed in the early endosome of cells overexpressing STAM proteins: accumulation of ubiquitinated proteins (Figures 5 and 6), accumulation of ligand-activated EGF receptor (Figure 8), and change in morphology (enlargement) (Figures 4 and 7). As reported previously (Komada et al., 1997; Chin et al., 2001; Raiborg et al., 2001; Bishop et al., 2002), these effects were also caused by Hrs overexpression, further suggesting that STAM proteins and Hrs act cooperatively in the same cellular process. All these effects were drastically diminished by deleting the VHS domain and also diminished to some extent by deleting the UIM from STAM proteins (Figures 5, 6, 7, 8). The mutant lacking the SH3 domain, on the other hand, was as effective as the full-length protein (Figures 6 and 7; our unpublished data). These results were in good correlation with the in vitro ubiquitin-binding activity of the deletion mutants (Figures 1, 2, 3), strongly suggesting that interaction with ubiquitin is required for STAM proteins to cause these cellular phenotypes. We cannot, however, rule out the other possibility that the VHS domain has a function other than binding ubiquitin, which is responsible for the phenotypes. The N-terminal region of STAM2 containing only the VHS domain and UIM was also hardly effective in causing the phenotypes (Figures 5, 6, 7, 8, 1–192), although it was sufficient for ubiquitin binding (Figure 1C). Because this region does not bind Hrs (Figure 1F), these results suggest that association with Hrs is also required for STAM proteins to exert their function.
The class E Vps proteins are a category of proteins in which gene disruption causes defective vesicular trafficking via the endosome in yeast (Raymond et al., 1992). Recently, three protein complexes composed of the class E Vps proteins, endosomal sorting complex required for transport (ESCRT)-I (Vps23, Vps28, and Vps37), ESCRT-II (Vps22, Vps25, and Vps36), and ESCRT-III (Vps2, Vps20, Vps24, and Snf7/Vps32), were shown to be involved in sorting ubiquitinated proteins in the MVB pathway (Katzmann et al., 2001, Babst et al., 2002a,b). Mammalian orthologs of the components in the ESCRT complexes have been identified, suggesting that the MVB sorting machinery is conserved among eukaryotes (Babst et al., 2000; Bishop and Woodman, 2001; Kamura et al., 2001). Vps27, the yeast ortholog of mammalian Hrs, also belongs to the class E Vps proteins and is required for sorting ubiquitinated proteins in the MVB pathway (Shih et al., 2002; Bilodeau et al., 2002). As is the case for Hrs and STAM proteins in mammalian cells, it was recently shown that Vps27 is associated with Hse1, a single yeast ortholog of STAM proteins (Bilodeau et al., 2002). In addition, disruption of the hse1 gene also leads to the class E vps mutant phenotype, including defects in 1) recycling of the sorting receptor Vps10 to the TGN from the endosome, 2) formation of the luminal vesicles of the MVB, and 3) sorting of ubiquitinated proteins into the MVB vesicles (Bilodeau et al., 2002). An Hse1 mutant protein lacking the UIM complements the former two defects but not the third (i.e., sorting ubiquitinated proteins), suggesting that the UIM of Hse1 plays a specific role in sorting ubiquitinated proteins for the MVB pathway. Together with these observations, our results presented in this study suggest that a complex of Hrs and STAM sorts ubiquitinated proteins for the MVB pathway in higher eukaryotes by directly binding to cargo proteins. The redundant activity of Hrs and STAM proteins to bind ubiquitin may increase the capacity or efficiency of the complex to sort ubiquitinated proteins on the early endosome. Another possibility is that one of the proteins first recognizes ubiquitinated proteins, which are then presented to the other during the sequential processes of the sorting pathway.
In this study, we were not able to determine whether the VHS domain and UIM of STAM proteins are required for the lysosomal degradation of known ubiquitinated cargos (e.g., EGF receptor) in a quantitative manner. This was due to a difficulty in examining the dominant-negative effects of overexpressing mutants lacking these domains, because the overexpression of the full-length STAM proteins exhibited an inhibitory effect on the degradation (Figure 8). The same effect has been demonstrated for Hrs. Although Hrs is believed to be required for the lysosomal trafficking of ubiquitinated cargos, its overexpression inhibits that of the EGF ligand/receptor complex (Chin et al., 2001; Raiborg et al., 2001; Bishop et al., 2002). In these cases, alternative approaches such as knocking out/down an endogenous protein and exogenously expressing a moderate level of mutant proteins are probably more useful. Further studies with such approaches are necessary to determine the significance of the VHS domain and UIM in the STAM function in the MVB sorting of ubiquitinated proteins.
Although similar roles have been suggested for the Hrs/STAM complex in higher eukaryotes and the Vps27/Hse1 complex in yeast, there might be some differences in the mechanisms of action between them. First, binding of Hse1 to ubiquitin is undetectable in an experimental condition in which Vps27 binds ubiquitin (Bilodeau et al., 2002), whereas STAM proteins directly bind ubiquitin as demonstrated in this study. In addition, Hrs has just one UIM whereas Vps27 has two tandemly located functional UIMs (Hofmann and Falquet, 2001; Shih et al., 2002). Therefore, the two complexes may interact with ubiquitinated proteins in a distinct manner. Second, both Hrs and STAM proteins become tyrosine phosphorylated in response to stimulation with growth factors and cytokines (Komada and Kitamura, 1995; Takeshita et al., 1996; Asao et al., 1997; Lohi et al., 1998; Takata et al., 2000), suggesting that the function of the Hrs/STAM complex in sorting endocytosed, ubiquitinated receptors is regulated by tyrosine phosphorylation. This will not be the case for the Vps27/Hse1 complex because yeast has no tyrosine-specific protein kinases. In addition, it is not yet known whether ubiquitination serves as an MVB sorting signal for newly synthesized lysosomal hydrolases in mammals as no mammalian protein has been shown to be ubiquitinated in the biosynthetic trafficking pathway to the lysosome. Finally, the deubiquitinating enzyme UBPY is associated with STAM proteins via their SH3 domains (Kato et al., 2000). UBPY shares several conserved domains such as the Cys and His boxes as well as the rhodanese homology domain with Doa4, a yeast deubiquitinating enzyme that releases ubiquitin molecules from ubiquitinated cargoes in the MVB pathway (Naviglio et al., 1998; Amerik et al., 2000). Doa4 also interacts with Vps27 genetically (Amerik et al., 2000). Therefore, in higher eukaryotes, UBPY may be recruited to the endosome by STAM proteins and play a similar role as Doa4 in yeast. However, Doa4 is possibly recruited to the endosome by ESCRT-III because its component Vps24 is required for this recruitment (Amerik et al., 2000). Therefore, the molecular mechanisms of recruiting deubiquitinating enzymes might also be distinct.
Elucidating how the interaction of the VHS domain and UIM of STAM proteins with ubiquitinated proteins is involved in the MVB sorting, as well as how the Hrs/STAM complex interacts with other components of the sorting machinery such as the ESCRT complexes, is important to understand the precise molecular mechanisms of the MVB sorting pathway in mammalian cells.
Acknowledgments
We thank Dr. T. Suzuki for providing the FLAG-tagged ubiquitin expression vector pcDNA3.1-FLAG-Ub.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–12–0823. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-12-0823.
Abbreviations used: ACLL, acidic-cluster dileucine; EGF, epidermal growth factor; ESCRT, endosomal sorting complex required for transport; GGA, Golgi-localizing/γ-adaptin ear homology domain/ADP-ribosylation factor-binding; GST, glutathione S-transferase; Hrs, hepatocyte growth factor-regulated substrate; MVB, multivesicular body; SH3, Src homology 3; STAM, signal-transducing adaptor molecule; TGN, trans-Golgi network; UIM, ubiquitin-interacting motif; VHS, Vps27/Hrs/STAM; Vps, vacuolar protein sorting.
References
- Amerik, A.Y., Nowak, J., Swaminathan, S., and Hochstrasser, M. (2000). The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell 11, 3365–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asao, H., Sasaki, Y., Arita, T., Tanaka, N., Endo, K., Kasai, H., Takeshita, T., Endo, Y., Fujita, T., and Sugamura, K. (1997). Hrs is associated with STAM, a signal-transducing adaptor molecule. J. Biol. Chem. 272, 32785–32791. [DOI] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D.J., Estepa-Sabal, E.J., Meerloo, T., and Emr, S.D. (2002a). ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D.J., Snyder, W.B., Wendland, B., and Emr, S.D. (2002b). Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283–289. [DOI] [PubMed] [Google Scholar]
- Babst, M., Odorizzi, G., Estepa, E.J., and Emr, S.D. (2000). Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1, 248–258. [DOI] [PubMed] [Google Scholar]
- Bean, A.J., Davanger, S., Chou, M.F., Gerhardt, B., Tsujimoto, S., and Chang, Y. (2000). Hrs-2 regulates receptor-mediated endocytosis via interactions with Eps15. J. Biol. Chem. 275, 15271–15278. [DOI] [PubMed] [Google Scholar]
- Bean, A.J., Seifert, R., Chen, Y.A., Sacks, R., and Scheller, R.H. (1997). Hrs-2 is an ATPase implicated in calcium-regulated secretion. Nature 385, 826–829. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., Urbanowski, J.L., Winistorfer, S.C., and Piper, R.C. (2002). The Vps27p-Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4, 534–539. [DOI] [PubMed] [Google Scholar]
- Bishop, N., and Woodman, P. (2001). TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J. Biol. Chem. 276, 11735–11742. [DOI] [PubMed] [Google Scholar]
- Bishop, N., Horman, A., and Woodman, P. (2002). Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J. Cell Biol. 157, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin, L.-S., Raynor, M.C., Wei, X., Chen, H.-Q., and Li, L. (2001). Hrs interacts with sorting nexin 1 and regulates degradation of epidermal growth factor receptor. J. Biol. Chem. 276, 7069–7078. [DOI] [PubMed] [Google Scholar]
- Conibear, E. (2002). An ESCRT into the endosome. Mol. Cell 10, 215–216. [DOI] [PubMed] [Google Scholar]
- Dupre, S., Volland, C., and Haguenauer-Tsapis, R. (2001). Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11, R932–R934. [DOI] [PubMed] [Google Scholar]
- Endo, K., et al. (2000). STAM2, a new member of the STAM family, binding to the Janus kinases. FEBS Lett. 477, 55–61. [DOI] [PubMed] [Google Scholar]
- Felder, S., Miller, K., Moehren, G., Ullrich, A., Schlessinger, J., and Hopkins, C.R. (1990). Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell 61, 623–634. [DOI] [PubMed] [Google Scholar]
- Finley, D., and Chau, V. (1991). Ubiquitination. Annu. Rev. Cell Biol. 7, 25–69. [DOI] [PubMed] [Google Scholar]
- Fujimuro, M., Sawada, H., and Yokosawa, H. (1994). Production and characterization of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349, 173–180. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J., and Maxfield, F.R. (1995). Membrane transport in the endocytic pathway. Curr. Opin. Cell Biol. 7, 552–563. [DOI] [PubMed] [Google Scholar]
- Hicke, L. (2001). A new ticket for entry into budding vesicles-ubiquitin. Cell. 106, 527–530. [DOI] [PubMed] [Google Scholar]
- Hofmann, K., and Falquet, L. (2001). A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26, 347–350. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Burian, D., Khalili, H., Schmidt, S.L., Sato, S., Liu, W.-J., Conrad, M.N., Conaway, R.C., Conaway, J.W., and Shilatifard, A. (2001). Cloning and characterization of ELL-associated proteins EAP45 and EAP20. J. Biol. Chem. 276, 16528–16533. [DOI] [PubMed] [Google Scholar]
- Kato, Y., Misra, S., Puertollano, R., Hurley, J.H., and Bonifacino, J.S. (2002). Phosphoregulation of sorting signal-VHS domain interactions by a direct electrostatic mechanism. Nat. Struct. Biol. 9, 532–536. [DOI] [PubMed] [Google Scholar]
- Kato, M., Miyazawa, K., and Kitamura, N. (2000). A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. J. Biol. Chem. 275, 37481–37487. [DOI] [PubMed] [Google Scholar]
- Katz, et al. (2002). Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3, 740–751. [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Babst, M., and Emr, S.D. (2001). Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Komada, M., and Kitamura, N. (1995). Growth factor-induced tyrosine phosphorylation of Hrs, a novel 115-kilodalton protein with a structurally conserved putative zinc finger domain. Mol. Cell. Biol. 15, 6213–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada, M., Masaki, R., Yamamoto, A., and Kitamura, N. (1997). Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J. Biol. Chem. 272, 20538–20544. [DOI] [PubMed] [Google Scholar]
- Komada, M., and Soriano, P. (1999). Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 13, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong, J., Roudabush, F.L., Moore, P.H., Montague, M., Oldham, W., Li, Y., Chin, L.-S., and Li, L. (2000). Hrs interacts with SNAP-25 and regulates Ca2+-dependent exocytosis. J. Cell Sci. 113, 2273–2284. [DOI] [PubMed] [Google Scholar]
- Lloyd, T.E., Atkinson, R., Wu, M.N., Zhou, Y., Pennetta, G., and Bellen, H.J. (2002). Hrs regulates endosome membrane invagination and tyrosine kinase receptor signaling in Drosophila. Cell 108, 261–269. [DOI] [PubMed] [Google Scholar]
- Lohi, O., and Lehto, V.-P. (2001). STAM/EAST/Hbp adapter proteins - integrators of signalling pathways. FEBS Lett. 508, 287–290. [DOI] [PubMed] [Google Scholar]
- Lohi, O., Poussu, A., Mao, Y., Quiocho, F., and Lehto, V.-P. (2002). VHS domain - a longshoreman of vesicle lines. FEBS Lett. 513, 19–23. [DOI] [PubMed] [Google Scholar]
- Lohi, O., Poussu, A., Merilainen, J., Kellokumpu, S., Wasenius, V.-M., and Lehto, V.-P. (1998). EAST, an epidermal growth factor receptor- and eps15-associated protein with Src homology 3 and tyrosine-based activation motif domains. J. Biol. Chem. 273, 21408–21415. [DOI] [PubMed] [Google Scholar]
- Mao, Y., Nickitenko, A., Duan, X., Lloyd, T.E., Wu, M.N., Bellen, H., and Quiocho, F.A. (2000). Crystal structure of the VHS and FYVE tandem domains of Hrs, a protein involved in membrane trafficking and signal transduction. Cell 100, 447–456. [DOI] [PubMed] [Google Scholar]
- Misra, S., Beach, B.M., and Hurley, J.H. (2000). Structure of the VHS domain of human Tom1 (target of Myb 1): insights into interaction with proteins and membranes. Biochemistry 39, 11282–11290. [DOI] [PubMed] [Google Scholar]
- Misra, S., Puertollano, R., Kato, Y., Bonifacino, J.S., and Hurley, J.H. (2002). Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933–937. [DOI] [PubMed] [Google Scholar]
- Nakatsu, F., Sakuma, M., Matsuo, Y., Arase, H., Yamasaki, S., Nakamura, N., Saito, T., and Ohno, H. (2000). A di-leucine signal in the ubiquitin moiety. J. Biol. Chem. 275, 26213–26219. [DOI] [PubMed] [Google Scholar]
- Naviglio, S., Matteucci, C., Matoskova, B., Nagase, T., Nomura, N., Di Fiore, P.P., and Draetta, G.F. (1998). UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J. 17, 3241–3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M.S., Madsen, P., Christensen, E.I., Nykjær, A., Gliemann, J., Kasper, D., Pohlmann, R., and Petersen, C.M. (2001). The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 20, 2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A., Fernandez, M.M., Steen, H., Blagoev, B., Nielsen, M.M., Roche, S., Mann, M., and Lodish, H.F. (2000). Identification of a novel immunoreceptor tyrosine-based activation motif-containing molecule, STAM2, by mass spectrometry and its involvement in growth factor and cytokine receptor signaling pathways. J. Biol. Chem. 275, 38633–38639. [DOI] [PubMed] [Google Scholar]
- Piper, R.C., Cooper, A.A., Yang, H., and Stevens, T.H. (1995). VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131: 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S., Sigismund, S., Faretta, M., Guidi, M., Capua, M.R., Bossi, G., Chen, H., De Camilli, P., and Di Fiore, P.P. (2002). A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416, 451–455. [DOI] [PubMed] [Google Scholar]
- Puertollano, R., Aguilar, R.C., Gorshkova, I., Crouch, R.J., and Bonifacino, J.S. (2001). Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science 292, 1712–1716. [DOI] [PubMed] [Google Scholar]
- Raiborg, C., Bache, K.G., Mehlum, A., Stang, E., and Stenmark, H. (2001). Hrs recruits clathrin to early endosomes. EMBO J. 20, 5008–5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg, C., Bache, K.G., Gillooly, D.J., Helene Madshus, I., Stang, E., and Stenmark, H. (2002). Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nat. Cell Biol. 4, 394–398. [DOI] [PubMed] [Google Scholar]
- Raymond, C.K., Howald-Stevenson, I., Vater, C.A., and Stevens, T.H. (1992). Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba, T., et al. (2002). Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415, 937–941. [DOI] [PubMed] [Google Scholar]
- Shih, S.C., Katzmann, D.J., Schnell, J.D., Sutanto, M., Emr, S.D., and Hicke, L. (2002). Epsins and Vps27p/Hrs contain ubiquitin-binding domains that function in receptor endocytosis. Nat. Cell Biol. 4, 389–393. [DOI] [PubMed] [Google Scholar]
- Shih, S.C., Sloper-Mould, K.E., and Hicke, L. (2000). Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 19, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper-Mould, K.E., Jemc, J.C., Pickart, C.M., and Hicke, L. (2001). Distinct functional surface regions on ubiquitin. J. Biol. Chem. 276, 30483–30489. [DOI] [PubMed] [Google Scholar]
- Takata, H., Kato, M., Denda, K., and Kitamura, N. (2000). A Hrs binding protein having a Src homology 3 domain is involved in intracellular degradation of growth factors and their receptors. Genes Cells 5, 57–69. [DOI] [PubMed] [Google Scholar]
- Takatsu, H., Katoh, Y., Shiba, Y., and Nakayama, K. (2001). Golgi-localizing, γ-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276, 28541–28545. [DOI] [PubMed] [Google Scholar]
- Takeshita, T., et al. (1996). Cloning of a novel signal-transducing adaptor molecule containing an SH3 domain and ITAM. Biochem. Biophys. Res. Commun. 225, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Young, P., Deveraux, Q., Beal, R.E., Pickart, C.M., and Rechsteiner, M. (1998). Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J. Biol. Chem. 273, 5461–5467. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Doray, B., Poussu, A., Lehto, V.-P., and Kornfeld, S. (2001). Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science 292, 1716–1718. [DOI] [PubMed] [Google Scholar]