Abstract
Muscle myosin heavy chain (MHC) rod domains intertwine to form alpha-helical coiled-coil dimers; these subsequently multimerize into thick filaments via electrostatic interactions. The subfragment 2/light meromyosin “hinge” region of the MHC rod, located in the C-terminal third of heavy meromyosin, may form a less stable coiled-coil than flanking regions. Partial “melting” of this region has been proposed to result in a helix to random-coil transition. A portion of the Drosophila melanogaster MHC hinge is encoded by mutually exclusive alternative exons 15a and 15b, the use of which correlates with fast (hinge A) or slow (hinge B) muscle physiological properties. To test the functional significance of alternative hinge regions, we constructed transgenic fly lines in which fast muscle isovariant hinge A was switched for slow muscle hinge B in the MHC isoforms of indirect flight and jump muscles. Substitution of the slow muscle hinge B impaired flight ability, increased sarcomere lengths by approximately 13% and resulted in minor disruption to indirect flight muscle sarcomeric structure compared with a transgenic control. With age, residual flight ability decreased rapidly and myofibrils developed peripheral defects. Computational analysis indicates that hinge B has a greater coiled-coil propensity and thus reduced flexibility compared to hinge A. Intriguingly, the MHC rod with hinge B was ~5 nm longer than myosin with hinge A, consistent with the more rigid coiled-coil conformation predicted for hinge B. Our study demonstrates that hinge B cannot functionally substitute for hinge A in fast muscle types, likely as a result of differences in the molecular structure of the rod, subtle changes in myofibril structure and decreased ability to maintain sarcomere structure in indirect flight muscle myofibrils. Thus alternative hinges are important in dictating the distinct functional properties of myosin isoforms and the muscles in which they are expressed.
Keywords: myosin, S2 hinge, indirect flight muscle, myofibril, Drosophila
Introduction
Myosin II, the molecular motor responsible for muscle contraction, is a hexameric protein consisting of two myosin heavy chains (MHCs), two essential light chains and two regulatory light chains (Figure 1a). Each MHC monomer has an actin-binding N-terminal globular head, an alpha-helical rod, and a non-helical C-terminal tailpiece. Two MHC rods intertwine to form an alpha-helical coiled-coil. Charged regions on the surface of the coiled-coils allow myosin rods to electrostatically interact and polymerize into the thick filament backbone.1 Thick filaments interdigitate with actin-containing thin filaments, and these serve as the key contractile elements of striated muscle sarcomeres. During the contractile cycle, the catalytic myosin head hydrolyzes ATP and subsequently forms crossbridges with actin. The subfragment 2 (S2) portion of the myosin rod tethers the head to the thick filament. Just prior to the release of inorganic phosphate, crossbridges swing causing thin filaments to slide past thick filaments, resulting in sarcomere contraction (for review see reference 2).
Figure 1.
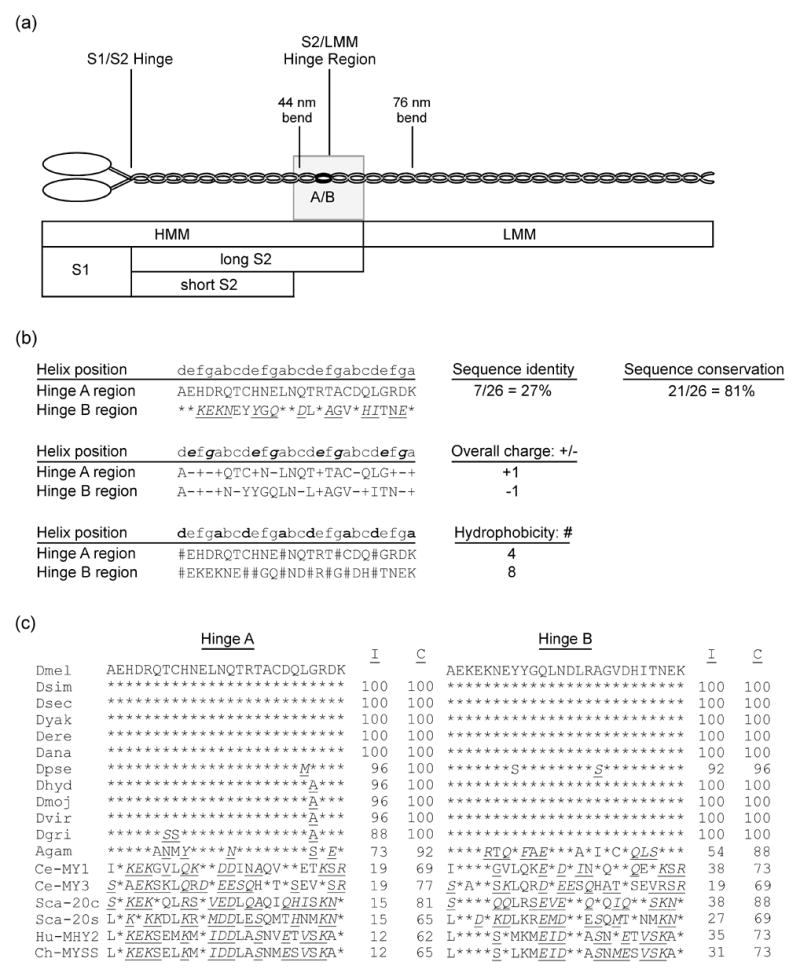
Common proteolytic sites of myosin, highlighting the S2/LMM hinge region, and analysis of its variant sequences in Drosophila and other organisms. (a) MHC molecules dimerize by intertwining of their alpha-helical coiled-coil tails. Each MHC consists of a globular head, an alpha-helical rod, and a non-helical C-terminal tailpiece. The essential and regulatory light chains are not shown. The MHC rod is ~155 nm long5,51,79 and can be proteolytically cleaved into heavy meromyosin (HMM) and light meromyosin (LMM). HMM can be further digested to generate subfragment 1 (S1), containing the head-neck region, as well as (long) subfragment 2 (S2). S2 can be further digested into short S2 and the C-terminal S2 hinge region also designated as the S2/LMM hinge (shaded rectangle). Bends in the rod are observed at 44 nm and 76 nm from the head-rod junction.5 The black oval labeled A/B indicates the approximate location and relative size of the 26-residue region encoded by alternative exons 15a and 15b in Drosophila muscle myosin. Scale is approximate. (b) Comparison of sequence, charge and hydrophobic variation of hinge A with hinge B. Lower case letters indicate amino acid residue positions in the alpha-helical heptad repeat.1 Top: * indicates identical residues; underline indicates conserved residues [italic type: conservation of strong groups (suggesting both hydropathy and side group size conservation), roman type: conservation of weak groups (suggesting either hydropathy or side group size conservation)]. Middle: bold italic indicates positions e and g, which are typically occupied by charged residues.1 (+) or (−) indicates charged residues in hinges A and B. Bottom: bold indicates positions a and d, which typically contain hydrophobic residues.1 # indicates hydrophobic residues in hinges A and B. (c) Sequence comparison of hinges A and B of D. melanogaster with those of muscle myosin from other Drosophila species, mosquito, worm, bay scallop, chicken and human. * indicates identical residues; underline indicates conserved residues (italic type: conservation of strong groups, roman type: conservation of weak groups). I, % identical residues, C, % conserved residues (percentage of residues that are identical, or show strong or weak conservation with the D. melanogaster hinge sequence below which they are listed). Abbreviations: Dmel, D. melanogaster; Dsim, D. simulans; Dsec, D. sechellia; Dyak; D. yakuba; Dere; D. erecta; Dana, D. ananassae; Dpse, D. pseudoobscura; Dhyd, D. hydei; Dmoj, D. mojavensis; Dvir, D. virulis; Dgri, D. grimshawi; Agam, Anopheles gambiae; Ce-MY1, Caenorhabditis elegans MHC D; Ce-MY3, C. elegans MHC A; Sca-20c, bay scallop, Aequipecten irradians, catch muscle; Sca-20s, A. irradians striated muscle; Hu-MHY2, human, MHC-IIa; Ch-MYSS, chicken adult skeletal muscle MHC.
Assembly of MHC dimers and thick filaments depends on the sequence periodicity of the myosin rod.1 The rod contains a heptapeptide repeat (a, b, c, d, e, f, g), with hydrophobic residues concentrated at positions a and d. Hydrophobic interactions form a “seam” between the two alpha-helices of the coiled-coil, which is stabilized by salt-bridges between charged residues at positions e and g. Positions b, c and f are mainly charged at the surface of the rod and are free to interact with surrounding molecules. The heptad repeats can be further grouped into 40 zones of non-identical 28-residue repeats. Each 28-residue repeat contains alternating regions of positive and negative charge, causing myosin rods to assemble in a staggered arrangement within the thick filament.1,3 These 40 zones are interrupted at four positions by an additional “skip” residue which breaks the heptad repeat1,3 and may serve to control the stagger of neighboring parallel myosins during thick filament assembly.4
Myosin molecules are characterized by the presence of two “hinges” that lie C-terminal to the globular head (Figure 1a). The S1/S2 hinge is located at the junction of subfragment 1 (S1) and S2 and may help to position myosin heads interacting with the thin filament. The S2/LMM hinge region of the MHC rod is located between the C-terminus of short S2 and the N-terminus of light meromyosin (LMM), spanning ~152 amino acid residues. Two of the four “skip” residues described above flank the S2/LMM hinge domain5 and may correspond to the location of bends in the rod observed in electron micrographs of negatively-stained myosin molecules.6 The S2/LMM hinge is proposed to be a flexible domain that allows myosin S1 to lift away from the thick filament and reach toward the thin filament.7–9 Proteolysis of thick filaments suggests that the C-terminal two thirds of the MHC rod form the thick filament backbone and the N-terminal third is loosely attached.1,10,11 The S2/LMM hinge may serve as a flexible linker connecting these regions, but it is important to note that the distinct bends in the myosin rod flank the S2/LMM hinge, while the “hinge” itself is not a known site of rod bending.
The S2/LMM hinge region has a reduced propensity to form a coiled-coil. Lu and Wong12 predicted that the coiled-coil formed from the rabbit skeletal MHC S2/LMM hinge is relatively unstable, since it is rich in basic and deficient in hydrophobic residues. The significantly lower stability of the S2/LMM hinge coiled-coil compared to other portions of the rod leads to preferential melting (helix to random-coil transition) when temperature, pH or ionic strength is perturbed. The helix to random-coil transition is implicated in shortening of the rod during temperature or pH alteration, with the principal melting site being the S2/LMM hinge.13,14 Recent experimental evidence supports the flexibility of this hinge region.15–17
The S2/LMM hinge region may influence mechanochemical energy transduction. By undergoing a helix-coil transition in vivo, the resulting shortening of the rod could contribute to force generation.18 Harrington and others obtained evidence of helix-coil transition in muscle fibers upon activation or when crossbridges are released from the thick filament backbone, yielding a more open and proteolytically accessible state.19–24 Antibodies to S2 or the hinge greatly reduce isometric force generation, decrease muscle stiffness, suppress the movement of actin filaments in in vitro motility assays and reduce active shortening of sarcomeres, without altering the Mg-ATPase activity of myofibrils.25,26
Differences in S2/LMM hinge sequences among myosin isoforms correlate with muscle-specific properties, implying that this domain may define some aspects of muscle function. Sequence differences in the hinge region are among the relatively few residues that vary between rat alpha- and beta-cardiac MHCs, which have unique enzymatic and mechanical properties. 27,28 Thirteen non-conserved differences are located in the putative hinge and its immediate N-terminal region (residues 1075 to 1352). In scallop, the central region of this MHC hinge is encoded by alternative exons; one version is used in fast striated muscle while the other is used in the relatively slow catch smooth muscle.29
Drosophila melanogaster MHCs also have isovariant S2/LMM hinge regions that correlate with muscle-specific properties. In Drosophila, alternative RNA splicing of five sets of alternative exons as well as inclusion or exclusion of an alternative 3′ exon yield functionally-different MHC isoforms that are expressed in a developmental and tissue-specific manner.30–33 The central 26 amino acids of the S2/LMM hinge are encoded by one set of these alternative exons, 15a and 15b. For simplicity we use the terminology “hinge A” and “hinge B” to describe the encoded regions; however, note that only 26 amino acids of the entire S2/LMM hinge regions differ (Figure 1a). The use of the alternative hinge regions correlates to muscle contraction speed.34,35 MHC isoforms in fast muscles (the synchronous jump muscle and the asynchronous indirect flight muscles) exclusively use hinge A.34,35 Only hinge B is expressed in slow muscle types, i.e., embryonic and adult body wall muscle. Transcripts containing exon 15a or 15b are both found in intermediate muscle types such as those in the legs and proboscis.34,35 Alternative versions of these hinges and/or variations in other regions of the myosin molecule may define the observed differences in contractile and structural properties of various muscle types.
To address the structural and functional roles of alternative S2/LMM hinges in Drosophila, we substituted hinge B for hinge A in MHC isoforms expressed in two fast muscle types, the indirect flight muscle (IFM) and the tergal depressor of the trochanter (TDT) or jump muscle. We found hinge B increased the length of the MHC rod, increased IFM sarcomere lengths, weakened myofibril stability, and impaired the function of the IFM and TDT compared to hinge A. Our results demonstrate that the Drosophila muscle myosin S2/LMM hinge region is important for myofibril assembly and stability and that the slow hinge cannot functionally substitute in fast muscle myosin. Thus differences in the S2/LMM hinge region of myosin are critical for dictating muscle-specific properties.
Results
Hinge sequences analysis
We compared Drosophila melanogaster S2/LMM alternative hinge regions A and B to define structural variations that could differentially affect myosin function (Figure 1b). Nineteen of 26 amino acid residues differ between the alternative hinge domains, and 9 of these have hydrophobicity or charge changes. The net charge of the hinge A specific domain is +1, while that of hinge B is −1. Hinge A has charged residues in 6 of the 8 e and g positions, whereas hinge B has 4. Hinge A has hydrophobic residues in only 3 of 8 a and d positions, while hinge B has them in 6. PCOILS structure-prediction software36–38 (http://toolkit.tuebingen.mpg.de/index.php?view=coil) estimates that hinge A (the 26 residues encoded by exon 15a plus one residue on either side) shows a 8% tendency to dimerize into a coiled-coil, whereas hinge B’s propensity is 82%. However, in the context of the surrounding rod sequence, i.e., 28 or more residues both upstream and downstream of the exon 15 encoded region, both alternative hinge regions are predicted to have greater than 95% coiled-coil propensity.
Genome sequence comparisons reveal conserved alternative hinges in ten other Drosophila species as well as in the mosquito, Anopheles gambiae (Figure 1c). D. melanogaster hinge A sequence shows 100% similarity (88–100% identity) over ~40 million years divergence time from the other Drosophila species. The D. melanogaster hinge B sequence is identical with that of nine Drosophila species, and is 96% conserved (92% identical) with D. pseudoobscura hinge B. The A. gambiae MHC sequence reveals isovariant hinges with 92% (EAA43619.2) and 88% (XM_319310) sequence conservation with D. melanogaster hinge A and hinge B, respectively. Identity values are 73% and 54%. Although not as distinct as in Drosophila, the mosquito hinges follow the same trend in coiled-coil propensity, 60% for A. gambiae hinge A and 79% for hinge B. Evolutionary conservation of the alternative S2/LMM hinges in insects suggests they serve distinct functions.
We also compared D. melanogaster hinges A and B to muscle myosin hinges from other phyla (Figure 1c). In general, identity and conservation values are higher compared to hinge B. Interestingly, both hinges derived from alternative splicing of scallop myosin RNA more closely resemble Drosophila hinge B. The only exception noted is that the hinge encoded by C. elegans myo-3 is more closely related to Drosophila hinge A. This myosin (MHC-A) nucleates thick filament assembly in C. elegans body wall muscles.39 Overall, however, it appears that the specialized adult hinge A of insects has diverged from its ancestral form.
Generation of transgenic lines and analysis of transgene expression
To determine if alternative versions of the S2/LMM hinge region of Drosophila MHC differentially affect muscle structure and function in vivo, we expressed MHC with hinge B in muscles that normally express hinge A. We designed a P element-based transgene (pWMHC2-15b) containing Mhc genomic DNA in which exons 15a and 15b and their flanking introns were replaced with cDNA containing exons 14, 15b and 16. We performed P element-mediated germline transformation by injecting 2,400 embryos with pWMHC2-15b. We obtained thirteen independently transformed fly lines and genetically mapped their transgenes to specific chromosomes. We retained five transformed lines that did not have a transgene on their second chromosomes (the site of the endogenous Mhc gene), did not display recessive lethality, and did not have more than one transgene on the same chromosome. We crossed the five pWMHC2-15b transgene inserts into an Mhc10 genetic background. The Mhc10 mutant is null for IFM and TDT MHC35, allowing exclusive expression of hinge-switch MHCs in these two muscle types. The PwMhc2 fly line, which expresses wild-type MHC isoforms from a transgenic source40, served as a control for possible aberrant phenotypes resulting from transgene expression.
We compared expression levels of MHC in the IFM of the control and hinge-switch transgenic lines to that of wild-type flies by SDS-PAGE analysis of upper thorax preparations. We normalized MHC levels to actin levels for each line and compared this to the myosin:actin ratio of wild-type flies (expressed as 100%). Three of the hinge-switch fly lines (15b-3, -47, and -108) and the control line produced wild-type or nearly wild-type levels of MHC (Figure 2a).
Figure 2.
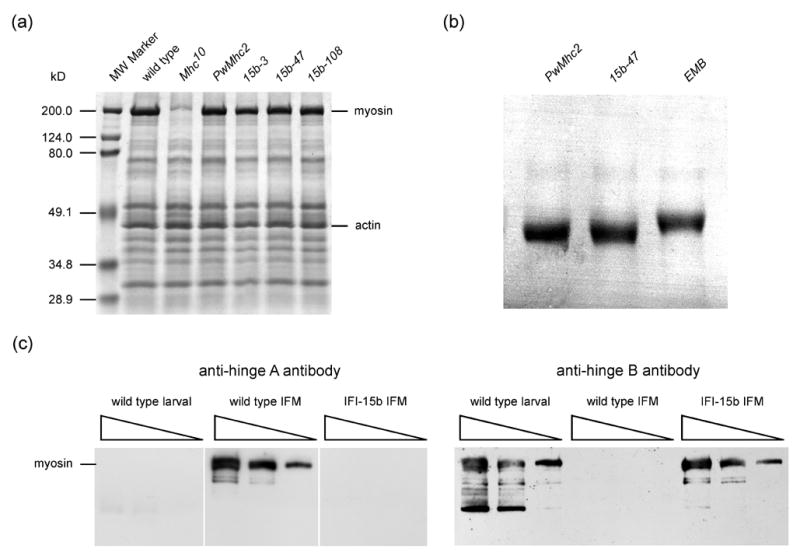
Expression of myosin in the IFI-15b hinge-switch transgenic lines. (a) 10% SDS-PAGE analysis of proteins from the upper thorax of wild-type (yw), Mhc10 (IFM/TDT MHC-null line), PwMhc2 (transgenic control) and each of the high expressing hinge-switch lines, indicated as 15b-3, 15b-47, and 15b-108, in the Mhc10 genetic background. MHC level relative to actin level was designated as 100% for wild type and found to be 99.7 ± 3.7 (n = 11) for PwMhc2, 99.9 ± 2.2 (10) for 15b-3, 107.0 ± 5.3 (6) for 15b-47, and 102.3 ± 4.7 (6) for 15b-108. Mean value ± SEM is reported. (b) IFM MHC from PwMhc2 control, 15b-47 (IFI-15b), and EMB fly lines, all in the Mhc10 genetic background. Proteins were loaded on a 5% SDS-polyacrylamide (1% bis-acrylamide) mini-gel. Samples were subjected to prolonged electrophoresis for 5 hours at a constant current of 25 mA to resolve migration differences resulting from primary sequence variation in the MHC isoforms. EMB flies express a transgenic embryonic MHC isoform32, in which the C-terminal tailpiece is 26 residues longer than the wild-type indirect flight muscle or IFI-15b isoforms. EMB MHC therefore has a lower mobility through the gel matrix. MHC from both PwMhc2 and 15b-47 IFM appear to have similar molecular masses and higher mobilities than EMB MHC. (c) Immunoblots of whole wild-type (yw) larvae, wild-type (yw) IFM and IFI-15b-expressing (15b-3) IFM in the Mhc10 genetic background, probed with either a hinge A- or hinge B-specific polyclonal antibody. Three different volumes of each sample were loaded into neighboring lanes on the gel. The hinge A-specific antibody recognized MHC from wild-type IFM and failed to recognize larval MHC or an MHC isoform in hinge-switch IFM (all samples were from the same gel and blot). Conversely, the hinge B-specific antibody recognized larval MHC and MHC expressed in hinge-switch fly IFM, but not MHC from wild-type IFM. Bands migrating faster than myosin are likely to be proteolytic products that are more readily detected on immunoblots due to their high efficiency transfer relative to full-length MHC. However, the identity of a major low molecular weight larval band is unknown.
Transgenic IFM exclusively express MHC with hinge B
We verified that transgenic MHC was produced from correctly spliced mRNA by comparing alternative exon usage in Mhc transcripts from wild-type (yw) and transgenic IFM (PwMhc2, and 15b-3, -47, and -108). To identify each alternative exon used, we synthesized partial cDNAs from portions of Mhc transcripts flanking alternative exons by a one-step RT-PCR method and performed restriction enzyme digests as indicated in Materials and Methods. For example, to verify exon 15b use in hinge-switch lines and 15a use in controls, we performed RT-PCR using primers to exons 13 and 16. We digested half the resulting cDNA with Pvu I (specific to exon 15a) and half with Sal I (specific to exon 15b). Similar analyses confirmed that IFM isoform-specific alternative exons, 3b, 7d, 9a, 11e, 15a (in controls) or 15b (in hinge-switch IFM) and 18 were used in amplified cDNAs (data not shown). In some instances, we cloned and sequenced RT-PCR products to ascertain their identities.
While most transcripts in the hinge-switch RNA contained exon 15b, we observed small amounts of two RT-PCR products containing exon 15a. DNA sequencing verified that both cDNAs derived from Mhc10 transcripts. One contained the intron between exons 14 and 15a along with the Mhc10 point mutation at the 3′ splice site of this intron.35 The other product lacked the intron but contained the translational frameshift hypothesized to occur if the intron was excised at an alternative splice site generated by the Mhc10 point mutation.35 Each of these aberrant transcripts would produce MHC truncated just before or within the exon 15 coding region. Truncated MHC protein from the endogenous Mhc10 allele does not accumulate to detectable levels in these hinge-switch lines, based on immunoblots of hinge-switch IFM probed with anti-myosin polyclonal antibodies (data not shown).
Unexpectedly, the PCR and sequence analysis detected some transcripts lacking exon 18 in the IFM of both wild-type and transgenic lines. Transcripts excluding exon 18 produce a myosin isoform with 26 additional residues at the C-terminus. To date, such transcripts have not been reported in IFM.41 To determine if MHC isoforms differing by 26 residues accumulate in IFM, we subjected IFM proteins from 15b-47 and two control lines to high resolution SDS-PAGE analysis (Figure 2b). MHC from PwMhc2 and 15b-47 IFM co-migrate, while an embryonic MHC isoform expressed in the IFM of the EMB transgenic line32 shows a lower electrophoretic mobility. This is expected, since EMB has the C-terminal tailpiece that is 26 residues longer than isoforms encoded by exon 18. We conclude that the IFM of the transgenic control and hinge-switch lines produce significant quantities of the expected smaller MHC isoform, and little, if any, of a larger MHC isoform. Previous antibody analysis also failed to detect the larger protein in wild-type IFM.42
We verified the exclusive expression of an MHC isoform with the hinge B substitution in IFM (IFI-15b: indirect flight muscle isoform of MHC with embryonic exon 15b region) by immunoblotting using affinity-purified polyclonal antibodies prepared to synthesized peptides corresponding to each hinge subdomain (Figure 2c). SDS-PAGE blots probed with hinge A antibodies demonstrated accumulation of hinge A-containing MHC in the IFM of adult wild-type flies, but not in larvae or in IFM of hinge-switch transgenics in the Mhc10 genetic background. SDS-PAGE blots probed with hinge B antibodies revealed the presence of MHC with hinge B in larvae and the IFM of hinge-switch transgenics, but not in wild-type IFM.
Hinge B expression impairs flight and jump muscle function
We performed flight tests on hinge-switch lines to determine if hinge B expression supports wild-type IFM function. Comparison of young and aged flies allowed assessment of use-related IFM dysfunction. The flight performance of each fly tested received a flight index (FI) value from 0 to 6.0, with 6 for upward flight, 4 for horizontal flight, 2 for downward flight and 0 for a lack of flight; we calculated an average FI for each line43 (Table 1).
Table 1.
Effect of hinge B expression on flight ability
| Age/Line | Flight ability (%) | Flight index (n) | |||
|---|---|---|---|---|---|
| U | H | D | N | ||
| 2 days | |||||
| wild type (yw) | 79.0 | 7.6 | 7.6 | 5.7 | 5.2 ± 1.7 (105) |
| PwMhc2 | 53.1 | 8.0 | 27.4 | 11.5 | 4.1 ± 2.3 (113) |
| 15b-3 | 0.0 | 0.0 | 11.7 | 88.3 | 0.2 ± 0.6 (103) |
| 15b-47 | 4.7 | 10.4 | 19.8 | 65.1 | 1.1 ± 1.7 (106) |
| 15b-108 | 0.0 | 0.0 | 7.4 | 92.6 | 0.1 ± 0.5 (149) |
| 7 days | |||||
| wild type (yw) | 84.7 | 8.5 | 4.2 | 2.5 | 5.5 ± 1.3 (118) |
| PwMhc2 | 46.0 | 8.0 | 37.0 | 9.0 | 3.8 ± 2.2 (100) |
| 15b-3 | 0.0 | 0.0 | 5.9 | 94.1 | 0.1 ± 0.5 (102) |
| 15b-47 | 1.7 | 0.8 | 21.5 | 76.0 | 0.3 ± 0.7 (129) |
| 15b-108 | 0.0 | 0.0 | 10.8 | 89.2 | 0.2 ± 0.6 (120) |
Percentage of flies tested that flew up (U), horizontal (H), down (D), or not at all (N). Flight index scale from 0–6 ± SEM; (n), number of flies tested.
Flight ability in hinge-switch lines was clearly impaired compared with the transgenic control or wild type (Table 1). Regardless of age, over 50% of transgenic control flies were able to fly upwards or horizontal, while 15b-3 and 15b-108 flies completely lacked this ability. A small percentage of flies (15.1%) from line 15b-47 demonstrated upward or horizontal flight ability at two days of age.
The flight ability of hinge-switch transgenic line 15b–47 decreased more rapidly over time than that of the control. At one week old, the majority of PwMhc2 transgenic control flies retained upward or horizontal flight ability, i.e, the flight index dropped only 7% from 4.1 to 3.8. However, the 15b–47 flight index dropped 73% from 1.1 to 0.3. These results suggest that hinge B expression causes a use-dependent decrease of flight muscle function.
To determine if hinge B expression can support wild-type jump muscle function, we assayed jumping ability of young adults from each of the hinge-switch lines. Compared with the PwMhc2 transgenic control, jump ability in all hinge-switch lines was significantly reduced (p < 0.001). Control flies jumped a mean distance of 5.7 ± 0.2 cm (SEM). The mean distances jumped by hinge-switch flies were 5.1 ± 0.2 cm (15b-3), 4.7 ± 0.2 cm (15b-47), and 5.2 ± 0.6 cm (15b-108). The average jump distance for hinge-switch flies was 9% to 18% less than that of the transgenic control, indicating that hinge B expression impairs the function of the jump muscle.
Neuronal response of the flight muscle is intact
We considered the possibility that the failure of muscles to function in hinge-switch transgenic organisms could arise from defects in neuromuscular connectivity. An abnormality in IFM development or function could affect its ability to secrete a neurotrophic factor, and such factors can influence innervation44 We therefore examined whether the neural circuitry between the giant fiber (which links the brain to the thorax) and the dorsologitudinal IFM (DLM) is intact in transgenic flies. By impaling the head of an adult fly with a pair of stimulating electrodes and inserting a recording electrode onto the surface of a DLM fiber, we could stimulate the giant fiber pathway at different voltages and record muscle response.45–47 We determined both short and long latency responses of these fibers. The short latency response occurs at high stimulating voltages and arises from stimulation of a brain neuron followed by conduction of the signal down the giant fiber into the thorax.46 The giant fiber transmits the signal via an electrical synapse with an interneuron. Acetylcholine released from the interneuron stimulates the motor neuron that innervates the DLM. The motor neuron releases glutamate to activate muscle contraction.48 The long latency response occurs at lower stimulating voltages and is thought to stimulate the giant fiber through a more circuitous pathway of neurons in the brain.46,48
Our results show that the DLMs in 15b-3 flies can be stimulated through the giant fiber pathway, despite the fact that this line showed extremely poor flight ability (Table 1) and the most severe structural defects (see below). Both short (1.78 ± 0.08 ms) and long (3.98 ± 0.07 ms) latencies were observed. Compared to wild-type Canton S flies (1.56 ± 0.03 ms and 3.78 ± 0.06 ms, for short and long latency, respectively), the short latency difference is statistically significant (p < 0.05). It is unclear if a 0.2 ms difference in short latency could disable IFM function. However, the critical test of connectivity is the percentage of stimulations that evoke responses. This value was 100% (n = 43), clearly indicating functional innervation.
Expression of MHC with hinge B causes minor IFM structural defects
To ascertain if expression of IFI-15b produced ultrastructural abnormalities, we examined the IFMs of 2-day old PwMhc2-15b adults by transmission electron microscopy and compared their structure to those of PwMhc2 transgenic control flies (Figure 3). IFM ultrastructure in the control fly line (Figure 3c, d) appeared very similar to wild type (yw; Figure 3a, b), with the exception of more variability in myofibril areas and sarcomere lengths, as previously reported.49
Figure 3.
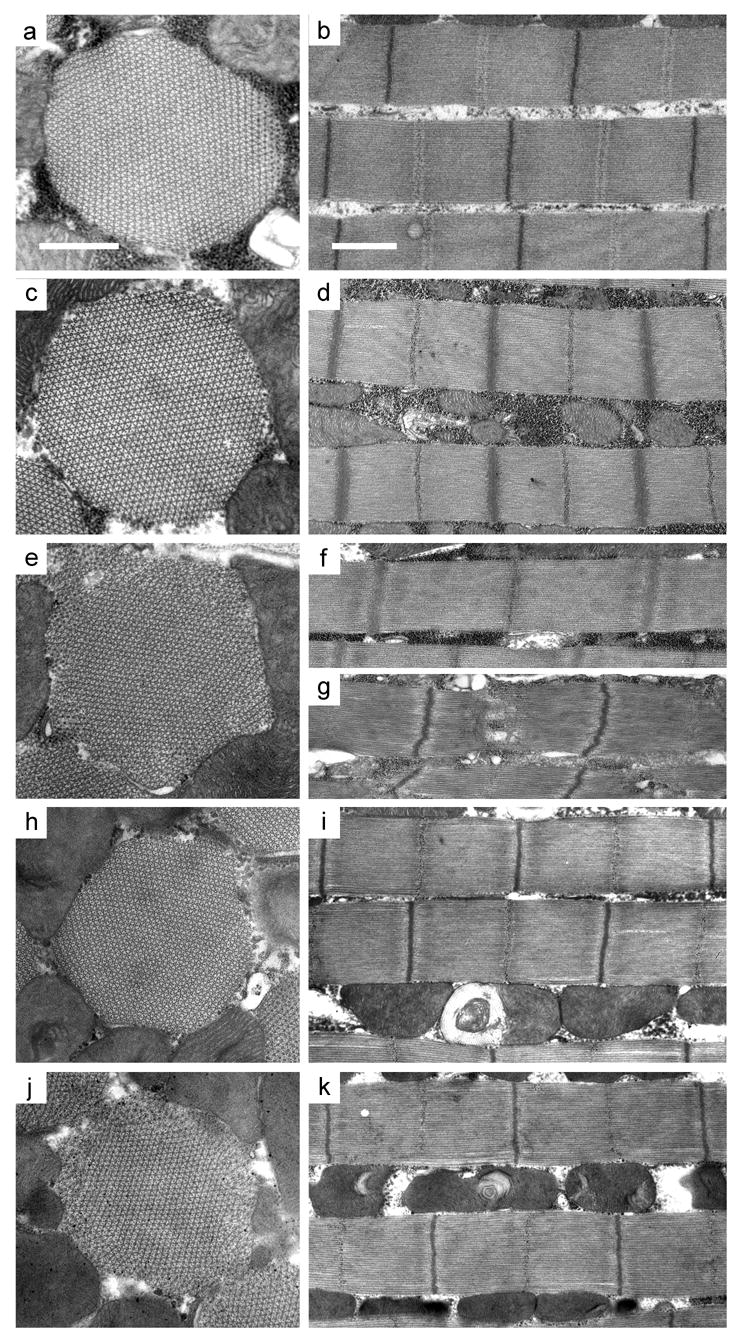
Electron micrographs of myofibrils from 2-day old wild-type (yw), PwMhc2 control and PwMhc2-15b hinge-switch IFM. Transverse (left panels) and longitudinal (right panels) sections are depicted. (a) IFM of yw (wild-type) has regular hexagonal thick and thin filament arrays and (b) intact sarcomeres with granule-containing M-lines and electron-dense Z-lines. (c, d) PwMhc2 myofibrils have a nearly wild-type structure; (c) 90–95% of myofibrils have intact double hexagonal packing of thick and thin filaments throughout and (d) normal M-line and Z-disk structure. There is some variation in sarcomere length in both yw and PwMhc2 lines, but the mean sarcomere lengths do not differ statistically (2.94 ± 0.02 μm for yw, 2.87 ± 0.02 μm for PwMhc2, mean ± SEM). Myofibrils in 15b-3 (e, f), −47 (h, i), and −108 (j, k) have varying degrees of minor disruption at 2 days post-eclosion, including peripheral filament packing defects. Some hinge-switch sarcomeres (f) are obviously longer than wild type (b) or control (d). Occasional severe disruption, particularly in the M-line region, is observed in the hinge-switch lines (g, line 15b-3). Transverse images, bar = 0.5 μm. Longitudinal images, bar = 1.0 μm.
Expression of MHC containing hinge B resulted in myofibrils with generally wild-type structure (Figure 3e–k). However, there were a variety of minor defects not observed in the control line (Figure 3c, d) including myofibrils with disrupted M-lines (Figure 3g) and rough edges (Figure 3e, j). The frequency and severity of defects varied among the three hinge-switch lines with 15b-3 flies showing the most severe and frequent defects. Most 15b-3 myofibrils had disrupted peripheries with filaments often appearing to be sloughing off from myofibril edges (Figure 3e). Sarcomeric structure was largely intact, although Z-disks often had jagged regions and M-lines were sometimes difficult to distinguish (Figure 3g). Defects in approximately 7% of myofibrils were so severe that myofibril boundaries were difficult to define (data not shown). Although myofibril edges were also rough in IFM of 15b-47 flies (Figure 3h), thick and thin filament packing at myofibril peripheries was disrupted the least compared to the other hinge-switch lines. The IFM from 15b-108 flies showed defects similar to but less severe than those in 15b-3 since myofibril boundaries were more defined (Figure 3j).
We compared the dimensions of myofibrils from 2-day old hinge-switch flies to those of the transgenic control to quantitatively assess morphological changes caused by hinge B expression in IFM. Myofibril dimensions of all hinge-switch lines were altered in a similar manner (Table 2). Total myofibril area within muscle fibers decreased by 6% (15b-3) to 13% (15b-47) relative to the control. On average, sarcomere length (SL) in the hinge-switch lines increased by ~13% over the control and showed a broader distribution. SL in the transgenic control ranged from 2.5–3.4 μm, while SL of hinge-switch line IFM ranged from 2.2–4.5 μm. The increased average SLs may be due to increases in thick filament lengths since similar magnitude increases in A-band lengths were observed in all hinge-switch lines. While we did not discern differences in thick filament length variation within half-sarcomeres or for sarcomeres within a myofibril, analysis of the coefficient of variation (standard deviation/mean × 100) indicates a trend toward increased variation in A-band lengths between sarcomeres in different myofibrils in two of the three hinge-switch lines [15b-3 (16%), 15b-47 (7%), 15b-108 (18%)] as compared to the transgenic control (7%). Myofibril areas were 7% (15b-3) to 26% (15b-47) smaller than for the control. In contrast, myofibril thick filament density (number of thick filaments/μm2) increased by 9% (15b-108) to 28% (15b-47) compared to the control. Interestingly, increases in thick filament density were roughly proportional to decreases in myofibril areas, so that the number of thick filaments per myofibril was similar for myofibrils with areas from 1–2 μm2. Hinge-switch myofibrils that were freshly prepared in isotonic solution and observed by light microscopy had smaller diameters and longer sarcomere lengths than those from the transgenic control50, verifying that altered myofibril morphology in myofibrils from hinge-switch fly lines did not arise from fixation artifacts.
Table 2.
Effects of hinge B expression on myofibril dimensions of 2-day old adults
| Line | Total myofibril area as a % of fiber area (n) | Sarcomere length μm (n) | A-band length μm (n) | Myofibril area μm2 (n) | TF/μm2 (n) | TF/myofibril (n) |
|---|---|---|---|---|---|---|
| PwMhc2 | 55.4 ± 0.9 (28) | 2.87 ± 0.02 (132) | 2.60 ± 0.02 (116) | 1.77 ± 0.03 (353) | 648.9 ± 22.2 (18) | 1178.4 ± 52.8 (18) |
| 15b-3 | 52.1 ± 0.6 (27) a | 3.22 ± 0.03 (130) a | 2.91 ± 0.03 (130) a | 1.64 ± 0.02 (376) a | 738.5 ± 32.9 (18) a | 1369.2 ± 50.4 (18) a |
| 15b-47 | 48.2 ± 0.6 (27) a | 3.23 ± 0.02 (148) a | 3.00 ± 0.02 (111) a | 1.31 ± 0.01 (438) a | 832.1 ± 22.8 (18) a | 1063.9 ± 45.2 (18) |
| 15b-108 | 52.0 ± 0.8 (27) a | 3.24 ± 0.02 (166) a | 2.95 ± 0.02 (166) a | 1.44 ± 0.01 (442) a | 705.1 ± 36.0 (18) | 1087.6 ± 26.5 (18) |
Mean values ± SEM; TF/μm2, number of thick filaments per μm2 within myofibrils; TF/myofibril, number of thick filaments per myofibril, calculated by multiplying myofibril areas by TF/μm2; (n), number of fields counted for Total myofibril area and TF/μm2 values, number of sarcomeres measured for Sarcomere length and A-band length values, and number of myofibrils for Myofibril area values.
p < 0.05 compared to corresponding PwMhc2 value.
To determine if defects in IFM myofibrils result from abnormalities in myofibril assembly, we examined ultrastructure of control and hinge-switch IFM myofibrils in late-stage pupae and in adults less than 2 hours old (data not shown). Myofibrils had variable diameters in both PwMhc2 control and hinge-switch transgenics, as was previously noted for 2-day old adults from all of the lines. Transverse sections of IFM myofibrils from pupal samples revealed myofibrils with smooth edges and wild-type hexagonal packing in the control and hinge-switch lines. In newly eclosed flies, myofibrils had similar but less severe and less frequent defects than in older adults from the same line. Because myofibrils in hinge B-expressing IFM lacked apparent defects prior to their use, i.e., in pupae, and because defects increased with age, we conclude that most defects present in 2-day old hinge-switch IFM are use-dependent.
To determine if differences in dimensions between hinge-switch and control myofibrils were present at or near the time of eclosion, we measured sarcomere and A-band lengths in adults less than 2 hours old (Table 3). Compared to the control line, sarcomere A-band lengths were increased in the IFM of all three hinge-switch lines indicating that hinge B alters myofibrillogenesis.
Table 3.
Effects of hinge B expression on myofibril dimensions of newly eclosed adults
| Line | Sarcomere length μm (n) | A-band length μm (n) | Myofibril area μm2 (n) |
|---|---|---|---|
| PwMhc2 | 3.01 ± 0.02 (106) | 2.65 ± 0.02 (106) | 0.72 ± 0.02 (233) |
| 15b-3 | 3.04 ± 0.04 (105) | 2.75 ± 0.04 (105) a | 1.04 ± 0.01 (344) |
| 15b-47 | 3.20 ± 0.06 (83) a | 2.89 ± 0.06 (83) a | 0.77 ± 0.01 (582) |
| 15b-108 | 3.28 ± 0.02 (255) a | 2.98 ± 0.02 (255) a | 0.81 ± 0.01 (421) |
Mean values ± SEM; (n), sample number
p < 0.05, compared to corresponding PwMhc2 value
Expression of MHC with hinge B influences myosin rod length
To ascertain the structural effects of hinge B expression on single molecule dimensions, we purified myosin hexamers from IFMs of wild-type (yw), PwMhc2 control, and 15b-47 Drosophila. We observed these molecules by electron microscopy after metal shadowing (Figure 4). Individual myosin molecules appeared well preserved and consisted of two globular N-terminal heads connected to a single tail. In many cases both the head to tail junction and the extreme C-terminus of the rod were well resolved and easily distinguishable from the granular background (Figure 4a). We measured molecules that met such criteria and determined the average contour lengths and standard errors of the mean of myosin tails from each line. yw (Figure 4b) and PwMhc2 control (Figure 4c) myosin rods had similar mean lengths of 149.28 ± 0.26 nm (n = 204) and 149.21 ± 0.24 nm (n = 202) respectively. Interestingly, replacement of hinge A by hinge B led to an increase in myosin tail length. The mean tail length for molecules isolated from 15b-47 IFMs (Figure 4d) was 154.05 ± 0.29 nm (n = 201). Histograms representing the data clearly illustrate the ~5 nm difference in mean length (Figure 4, lower panel). This length difference, imparted solely by the different hinges, is highly significant (p < 0.005).
Figure 4.
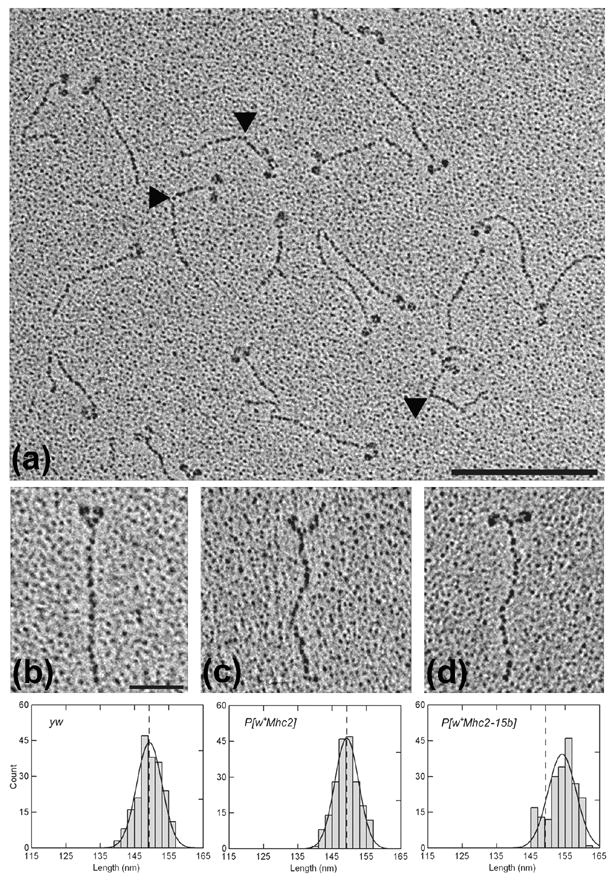
Electron micrographs of rotary shadowed myosin molecules from wild-type (yw), PwMhc2, and PwMhc2-15b hinge-switch Drosophila IFM. (a) A representative population of wild-type IFM myosin. Bar = 200 nm. Note the presence of molecules containing two clearly resolved N-terminal globular heads connected to a single C-terminal tail. A triangular, electron dense head to tail junction can be seen in several molecules. Furthermore, the extreme C-terminus of the rod can frequently be distinguished from the surrounding granular background. A sharp bend often found in the rod (denoted by black triangles) appears to represent the skip residue at the N-terminus of the S2/LMM hinge region.6 It is located 44 nm from the head to tail junction in both hinge A- and hinge B-containing molecules [44.02 ± 0.74 nm for yw (n = 54) and 43.98 ± 0.76 nm for 15b-47 (n = 55); mean ± SEM]. (b–d) Enlarged micrographs of single myosin molecules from yw (b), PwMhc2 (c), and PwMhc2-15b-47 (d) IFM that are suitable for rod contour length determinations. Bar = 50 nm. Over 200 myosin tails from each line were individually measured from the head to tail junction to the extreme C-terminus and averaged together to ascertain the mean contour rod lengths. As shown by the frequency distribution of rod lengths (lower panel), the mean contour lengths of yw and PwMhc2 tails (dotted lines) are the same and significantly shorter than the mean length of PwMhc2-15b tails.
Discussion
The strict use of alternative MHC S2/LMM hinges in different muscle types of Drosophila melanogaster suggests that the variable central domains of these hinges impart specialized properties to the muscles in which they are expressed. Hinge A is well conserved in other insects, including various Drosophila species and other dipterans such as a mosquito, but has a lower degree of conservation with muscle myosin IIs of other organisms (Figure 1c). It is unusual in that it is positively charged with an extremely low coiled-coil propensity (8%, compared to 82% for hinge B). Hinge A thus may be uniquely adapted for the demands of insect flight and jump muscles. We experimentally addressed this hypothesis by creating transgenic lines that have their fast muscle MHC hinge substituted by the isovariant hinge that normally is expressed in slow body wall muscle MHC. The hinge-switch increased the length of the MHC rod, increased sarcomere length (SL), caused a broader SL distribution, decreased myofibril stability, and impaired fast muscle function.
Different structural properties of the regions encoded by exons 15a and 15b are likely to cause the observed transgenic phenotypes. Nine of the 26 amino acid residues in the alternative domains differ by either charge or hydrophobicity (Figure 1b). The increased positive charge and decreased hydrophobicity of hinge A may weaken coiled-coil stability in this and surrounding regions of the rod while hinge B may impart local structural stability. This could increase the length of dimers containing hinge B relative to those with hinge A. The ~5 nm longer tail length observed with IFI-15b myosins (Figure 4) supports this notion.
We did not observe an obvious difference in flexibility between the hinge A- and hinge B-containing Drosophila myosin isoforms. The platinum shadowing technique might not resolve such a difference, since it does not resolve the 76 nm bend induced by a skip residue,5 In contrast, the shadowing method in this study did resolve the skip residue-induced bend at 44 nm in both types of molecules. It may be that a more flexible hinge does not induce a distinct kink, but rather a subtle variation in bending throughout the entire S2/LMM hinge region. This subtle variation would be unlikely to be obvious in our preparations, given the observed gentle bending of myosin rods.5,51
It is tempting to speculate that the hinge B-containing rods lead to increased thick filament and sarcomere lengths when this hinge-switch MHC is expressed in IFM (Figure 3, Tables 2 and 3). However, this possibility must be tempered by several considerations. First, the mean SL in each of the PwMhc2-15b lines is comparable to SLs reported for some wild-type strains.52,53 While this suggests that the PwMhc2 control SL may be unexpectedly low, we found that the SL length for yw (the wild-type strain used in the current study) is 2.94 ± 0.02 μm, verifying that PwMhc2-15b SL is indeed longer than an appropriate wild-type control line. The similarity of SLs between PwMhc2 and yw IFMs was further verified by light microscopy of myofibrils in isotonic solution.50 Another consideration is whether the S2/LMM hinge domain could serve as part of a length-determining element of the thick filament. If the hinge domain is loosely attached to the thick filament backbone so as to allow the myosin head to be presented to the thin filament, it may neither be integral in thick filament assembly nor play a direct role in length determination. However, a recent electron tomography study of insect flight muscle in rigor found that only 16% of S2 acts to tether bound S1 to the thick filament54, implying that the hinge would be part of the thick filament backbone. Even if the hinge is a constituent of the thick filament backbone during the assembly process, it is not certain that the different rod lengths observed in vitro correspond to differences in rod lengths that occur during myofibrillogenesis in vivo. Given these caveats, it is intriguing to note that in wild-type flies, muscles possessing hinge B can have longer thick filaments and sarcomeres than hinge-A-containing IFM or TDT muscle.55 This raises the possibility that the S2/LMM hinge could directly or indirectly serve as a length-determining element of the thick filament, perhaps by its ability to affect the level of polymerization of myosin molecules into thick filaments and/or by its affinity for projectin, which appears to tether the thick filament to the Z-band in IFM, but binds at A-bands in synchronous muscle.56 Alternative hinges could have different affinities for projectin and cause its differential localization. This, in turn, might result in projectin differentially regulating thick filament length, Thus, the increased lengths we have observed in IFM could arise from an S2/LMM hinge-dependent mechanism that is used in vivo to regulate thick filament and sarcomere lengths.
Age-dependent degeneration occurs in hinge-switch transgenic IFMs, particularly at the periphery of myofibrils (Figure 3), implying that the structural integrity of IFM myofibrils is compromised by mis-expression of the embryonic MHC hinge. Although these structural defects are clearly present, they are subtle compared to those seen in some other myosin mutants and isoform switch lines.32, 49, 57–59 Thus while hinge A is important for maintaining IFM myofibril integrity, it is not as critical as other regions of the myosin molecule.
Removal of hinge A from IFM myosin could delete the binding site of a protein required for normal assembly and/or stability of myofibrils. It is possible that hinge A normally binds to the adult-specific C-terminus34 of neighboring myosin molecules, and this interaction would be disrupted in the hinge-switch line. Further, IFM specific proteins that have strong potentials for interacting with the S2/LMM hinge region could turn over as a result of the lack of a myosin-binding site. Two possibilities in this regard are flightin and stretchin-klp. Flightin is a novel 20 kD IFM-specific protein that binds to the MHC rod in the LMM region.60–63 It is important for myofibril assembly and maintenance, since its elimination results in abnormally long sarcomeres and severe degeneration during aging.53 Absence of flight ability in young adult flightin nulls arises from reduced IFM myofibril stiffness.64 Interestingly, mutations in the Drosophila MHC rod, located C-terminal to the hinge region, can alter accumulation of flightin and its post-translationally modified variants.57 However, no changes in flightin levels or in the production of its isovariants occurs in hinge-switch flies (J.O. Vigoreaux, personal communication). Thus flightin accumulation and modification are not dependent upon hinge A. However, transient interactions may still occur.
Stretchin-klp’s are A-band proteins with multiple immunoglobulin domains.65 An IFM-specific isoform fails to accumulate when the embryonic myosin rod is expressed in place of the IFM rod.65 Since the only areas unique to IFM rods are hinge A and a short C-terminus, this suggests that one or both of these myosin domains is required for stability of the IFM-specific stretchin-klp isoform. Preliminary western blot analysis of hinge-switch IFM with a stretchin-klp antibody showed accumulation of both the larger IFM-specific and the smaller stretchin-klp isoforms, similar to what is observed in wild type (S. Patel, personal communication). This suggests that interaction of stretchin-klp with hinge A is not essential for the stability of the larger IFM-specific isoform.
Use-dependent loss of myofibril integrity may be the result of the inability of hinge-switch myofibrils to resist mechanical strain. This could explain the increased myofibril disruption and the decreased flight ability as hinge-switch flies age (Figure 3, Table 1). If the hinge is part of the thick filament backbone, hinge B’s tendency to remain in a coiled-coil configuration could increase the stiffness of IFM thick filaments, which may cause additional strain on other sarcomeric components during IFM use. Alternatively, if the hinge lifts off the surface of the thick filament, then hinge B’s relative stability might decrease the flexibility of the crossbridge, which could be detrimental to crossbridge kinetics. Crossbridge function might also be impaired if hinge B is not able to transition from a coiled-coil to a random coil during the contractile cycle, as hinge A might. When crossbridge kinetics are not matched to the oscillation frequency of the flight system, resultant abnormal stresses could lead to myofibril deterioration.66 Studies of fiber mechanical properties suggest that the hinge switch does not affect myofibril and fiber passive stiffness50, but that some alterations in crossbridge kinetics occur (Miller et al., in preparation). Such alterations could lead to reduction in flight and jump muscle ability (Table 1), which is the most dramatic effect of the hinge swap.
Overall, our results clearly demonstrate that the two small alternative versions of the Drosophila S2/LMM hinge region of myosin are not interchangeable. Differences in molecule, thick filament and sarcomere length resulting from the hinge switch suggest that the hinge is important for regulating these properties. Electrophysiological analysis implies that the impaired function of hinge switch muscles does not arise from defective neuromuscular activity. The relatively subtle structural changes that occur from the hinge switch apparently lead to myofibrillar defects that result in the muscle dysfunction that we have documented. Studies focusing on filament assembly, fiber mechanical properties and high-resolution sarcomere structure should provide further insight into the critical role that alternative hinges play in determining muscle fiber-type-specific structure and function.
Materials and Methods
DNA and protein sequence analyses
Drosophila Mhc genes were aligned using Multi-LAGAN67 and the database developed by the Eisen laboratory at the University of California, Berkeley (http://rana.lbl.gov/drosophila/alignments_eisenlab.html). Alignments were kindly provided by Dr. Venky Iyer. These were analyzed using the Jalview Java Alignment Editor.68 D. melanogaster hinge A and B sequences submitted to a BLAST search (www.ncbi.nlm.nih.gov/BLAST/) against the translated protein database (tblastn) produced hits to D. hydei (Genbank accession number X77570) and A. gambiae (Genbank accession numbers XM_319312 and XM_319309). Other muscle myosin hinge sequences were downloaded from SWISSPROT. Protein sequences were aligned and subjected to pairwise sequence comparisons via CLUSTAL W69 (version1.81), using default parameters and the BLOSSUM matrix selection (program accessed through the Biology Workbench: http://workbench.sdsc.edu).
The propensity of protein sequences to form coiled coils was determined using the default settings of the PCOILS program36–38 (http://toolkit.tuebingen.mpg.de/index.php?view=coil). For analysis of alternative hinge domains, the 26 amino acid residues of the alternative hinge plus one residue on each side (minimum window allowed) was used.
Construction of the pWMHC-15b transgene
Plasmid pWMHC2-15b was created by modification of PWMHC2 using standard cloning techniques40, 70 PWMHC2 contains the entire Drosophila melanogaster myosin heavy chain gene (Mhc) with 0.45 kb of sequence upstream of the transcriptional start site and 1.6 kb of sequence downstream of terminal exon 19. The plasmid vector is pCaSpeR 4 which contains P element termini for germline insertion.71 PWMHC2 was digested with Eag I to create a 19.1 kb 5′ fragment and a 12.6 kb 3′ Mhc fragment. The 3′ Mhc fragment, spanning from within exon 11e through 1.6 kb downstream of exon 19, was subcloned into the Eag I site of pKS Mod (a modified pBluescript KS vector (Stratagene, La Jolla, CA) which had been ligated after digestion with EcoR V and Sma I, resulting in the loss of both enzyme recognition sites). A 2.5 kb EcoR V/Stu I subfragment (containing exons 14, 15a, 15b, a portion of 16 and intervening introns), was subsequently replaced with the corresponding 1.6 kb EcoR V/Stu I fragment from an embryonic Mhc cDNA clone32 in which exons 14, 15b and 16 are fused (exon 15a and 3 introns are absent). The 3′ Mhc fragment with the exon 14-15b-16 fusion was excised from pKS Mod by Eag I digestion and ligated to the original 19.1 kb 5′ fragment to generate pWMHC2-15b. The plasmid was purified using QIAfilter Plasmid Maxi Kit (Qiagen Inc, Valencia, CA) according to the manufacturer's instructions. All manipulated regions of the plasmid were sequenced to insure that no unexpected alterations had been introduced.
Germline transformation and genetic crosses
Transgenic lines were produced by the method of Rubin and Spadling72, using techniques explained in detail by Cripps and Bernstein.73 In brief, embryos (generation G0) were collected from yw (yellow body, white-eyed) flies and injected with pWMHC2-15b (0.6 μg/μl) and 2–3 helper plasmid (0.08 μg/μl). The G1 generation was screened for orange eye color, indicating the presence of the P[w+Mhc2-15b] transgene carrying the miniwhite (w+) marker.
Transformants were crossed with w1118; CyO/Bl1; TM2/TM6B flies to map the chromosomal location of each insert using standard genetic techniques. Descriptions of gene and chromosome symbols can be found in Lindsley and Zimm74. Five independent hinge-swapped lines (P[w+Mhc2-15b] -3, -47, -108, -120, -150) were crossed into the Mhc10 (flight and jump muscle myosin-null) genetic background.35 P[w+Mhc2-15b] flies that were homozygous for the transgene and the Mhc10 allele were used for all further studies, along with a transgenic control, P[w+Mhc2], also in the Mhc10 background.40 For brevity, the transgenic control line in the Mhc10 background is designated as PwMhc2 in the text, figures and tables. Similarly, the hinge-switch lines in the Mhc10 background are designated 15b-3, 15b-47, etc.
RNA Isolation from Flight Muscle
Indirect flight muscle (IFM) was dissected as in Swank et al.75 Thoraces from 80 young adult flies were bisected and placed into a dissection dish on ice containing York Modified Glycerol without Triton X-100 (YMG-Tx: 20 mM potassium phosphate, pH 7.0, 2 mM MgCl2, 1 mM EGTA, 8 mM DTT, 50% v/v glycerol) with a protease inhibitor cocktail (Complete Mini, Roche Applied Science, Indianapolis, IN). The dorsologitudinal (DLM) set of IFM was removed from each thorax into the solution, and remaining tissue was discarded. The DLMs were collectively transferred to a microcentrifuge tube, pelleted by centrifugation at 8,000 × g for 5 min at 4°C and washed with YM (York Modified Glycerol without glycerol or Triton X-100). Following a second centrifugation at 8,000 × g for 5 min at 4°C, the YM was removed from the muscle pellet.
Total RNA (larger than 200 base pairs) was isolated using RNeasy Mini Kit (Qiagen Inc) with the following modifications/options to the protocol supplied by the manufacturer for 20–30 mg of animal tissue. Six μl of β-mercaptoethanol were mixed with 600 μl of Buffer RLT (supplied with the kit), and added to the DLM pellet from above. The tissue was disrupted in a microcentrifuge tube with a pestle at room temperature prior to its homogenization with a 20 gauge needle and 1 cc syringe. Contaminating DNA was digested on the silica-gel-based spin column using the Qiagen RNase-Free DNase Set. RNA was eluted from the column in a single step by addition of 40 μl of RNase-free water. The DLM from 80 flies typically yielded 22 μg of total RNA.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
One-step RT-PCR was performed on 1.2-1.5 μg of RNA using C. therm Polymerase OneStep RT-PCR System (Roche) according to the manufacturer's instructions. Primer pairs and sequences are as follows with (+) or (−) indicating forward or reverse primers, respectively:
Exon 2 (+) 5′-ATGCCGAAGCCAGTCGCAAAT-3′ and
Exon 6 (−)5′-GGAATTCTGGT-AGTAGATGTGG-3′;
Exon 5 (+)5′-GGCTGGTGCTGATATTGAGA-3′ and
Exon 10 (−)5′-TGTTGGTCAGCTTCTCCGACA-3′;
Exon 10 (+)5′-GTTCCCCAAGGCCTCCGA-TCA-3′ and
Exon 13 (−)5′-GCGGTCTTCTCAGCCAAAAGC-3′;
Exon 13 (+)5′-CTGC-ATGCCGCTGAAGTGAAG-3′ and
Exon 16 (−)5′-GGGTGACAGACGCTGCTT-GGT-3′;
Exon 17 (+)5′-GAACAGCTGGGTATCTCCGAGCG-3′ and
Exon 19 (−)5′-GGTCGAATCTTGGTGGGAAGGCC-3′;
Exon 18 (+) 5′-GGCCGCAAG-AGCGCGCTGC-3′ with Exon 19 (−) above.
For the exon 2/exon 6, primer pairs the following program was run on a DNA Engine thermocycler (MJ Research Inc., Waltham, MA): 60°C, 30 min; 95°C, 2 min.; 11 rounds of (95°C, 30 sec; 50°C, 30 sec; 72°C, 2 min); 26 rounds of (95°C, 30 sec; 50°C, 30 sec; 72°C, 2 min plus an additional 5 sec of extension time per cycle); 72°C, 7 min. This program was adjusted for use with the remaining primer pairs by substituting different annealing temperatures in place of the 50°C annealing temperature above. A 65°C annealing temperature was used with the exon 5/exon 10 and exon 10/exon 13 primer pairs; a 60°C annealing temperature was used for the exon 13/exon 16 primer pair, and a 62°C annealing temperature for the exon 17/exon 19 and exon 18/exon 19 primer pairs.
RT-PCR Product Analysis
RT-PCR products were electrophoresed on agarose gels, purified using the QIAEX II Gel Extraction System (Qiagen) and digested with restriction endonucleases (New England Bio Labs, Beverly, MA) appropriate to the IFM-specific alternative exon of interest. The purified fragment containing Exon 3b was digested with Bgl II; exon 7d with Eco RI; exon 9a with Pst I; exon 11e with Bam HI; exon 15a with Pvu I or for product from hinge-switch IFM, exon 15b with Sal I.
The presence or absence of exon 18 was determined by cloning and sequencing of the purified product generated from the exon 17/exon 19 primer pair and the 90 bp product amplified with an exon 18-specific primer, since this region lacks convenient restriction sites. Gel-purified RT-PCR products were ligated into pGEM-T Easy vector (Promega Corp., Madison, WI) and cloned using DH5α E. coli (Invitrogen, Carlsbad, CA). Plasmid DNA was isolated using the UltraClean plasmid miniprep kit (Mo Bio Laboratories Inc., Carlsbad, CA) according to the manufacturer's instructions. Sequencing was performed at the San Diego State University Microchemical Core Facility (San Diego, CA) using universal primers. Additionally, undigested DNA from the exon 13/16 Sal I reaction was cloned and sequenced in this manner.
Quantification of transgene expression levels
Transgenic protein expression levels were estimated by SDS-PAGE.76 For each experimental sample, the upper thorax from a 2-day old female fly was homogenized in 60 μl of sample buffer and boiled for 5 min prior to loading. For each line, 10 or 12 μl of six experimental samples were run on a Tris-glycine minigel containing 10% polyacrylamide with a 4% polyacrylamide stacking gel (BioRad, Richmond, CA). An additional four to six wells were loaded with 8–15 μl of a wild-type standard created by combining the upper thoraces of six yw female flies and homogenizing in 360 μl of sample buffer. Gels were stained with Coomassie blue, scanned on an Expression 636 flatbed scanner (Epson America, Inc., Torrance, CA), and analyzed using the gel plotting macros of the NIH Image software package (http://rsb.info.nih.gov/nih-image/). For each gel, the average myosin to actin ratio for each experimental line was determined and expressed relative to the average myosin to actin ratio for the wild-type samples.
Hinge-specific antibodies and Western blotting
Polyclonal hinge-specific antibodies were produced in rabbits from synthetic peptides consisting of hinge A or hinge B sequence (produced by Sigma Genesys, The Woodlands, TX). Antibodies were affinity purified from antisera using the appropriate hinge peptide conjugated to CNBr-activated Sepharose (Amersham Biosciences, Piscataway, NJ) according to the manufacturer’s protocol.
Western blotting was performed as described in Sambrook et al.70 Proteins from samples containing either larvae or upper thoraces from adult flies were separated by 7.5 % SDS-PAGE mini gels (Bio-Rad), according to Laemmli76, and blotted to nitrocellulose using a Mini Trans-blot Eletrophoretic Transfer Cell (Bio-Rad). Blots were probed with affinity-purified anti-hinge antibodies followed by horseradish-peroxidase conjugated secondary antibody using the SuperSignal West Pico Complete Rabbit IgG Detection Kit (Pierce, Rockford, IL) and signals were detected on Kodak X-Omat Blue-XB-1 film (Eastmond Kodak, Rochester, NY).
Locomotory studies
Flight testing was performed as in Drummond et al.77 at room temperature. Newly eclosed flies were aged for 2 or 7 days at 25°C. Several hours prior to testing, flies were placed at room temperature to acclimate. Individual flies were released into the center of a flight test chamber, a Plexiglas box (43 cm H × 27.5 cm W × 43 cm L), with a bright light source positioned at the top. Each fly was assigned a flight index value43 based on its ability to fly up toward the light (6.0), horizontal (4.0), downward (2.0), or not at all (0.0). The average flight index was calculated by dividing the sum of the individual flight index values by the number of individuals tested for each line.
Jump testing was performed as in Swank et al.58 In preparation for jump testing, newly eclosed flies with their wings removed were placed into glass vials, and aged 2 days at 25°C. Several hours prior to testing, the flies were placed at room temperature to acclimate. Individual flies were encouraged with a paint brush to jump from the edge of a 9.5 cm tall inverted plastic vial onto a piece of paper with concentric rings printed 0.5 cm apart. The three farthest jump distances out of ten trials per fly were averaged for 50 flies per line.
Electrophysiological Analysis
Evoked action potentials were intracellularly recorded in IFM fibers following brain stimulation of the fly giant fiber pathway as in Zordan et al.78 About 10 flies were analyzed for each genotype. Measurements were made from 3–5 fibers per fly with 2 responses per fiber, yielding about 50–100 evoked responses. The ratio of evoked responses to stimulatory input was calculated as a measure of synaptic transmission and muscle responsiveness. Latencies were calculated from the end of the square wave stimulus to the start of the evoked response at the muscle membrane. Student’s t-test was used for statistical evaluation.
In brief, flies anaesthetized in ice were placed in a slab on the bottom of a Plexiglas recording chamber and then blocked and covered with dental wax (Bellevue), except for the dorsal thorax. Flies were covered with physiological saline and a constant physiological oxygen afflux to IFM fibers was maintained. Experiments were conducted at room temperature (20–22° C). After a 15 min recovery from anesthesia, an intracellular glass recording microelectrode was inserted in IFM fibers through a hole made in the scutellum cuticola. One tungsten stimulating electrode was inserted in each fly eye. Square pulse stimuli (0.2 ms duration, 4–15 V intensity) were delivered to the fly brain from a Grass S88 stimulator connected to a stimulus isolation unit (Grass SIU5). An Ag/AgCl electrode in the bath was used as a ground electrode. Signals from the recording electrode were amplified using an intracellular current-voltage clamp amplifier (NPI Turbo Tec, Germany) fed to an A/D interface (Digidata 1200, Axon Instruments). Amplified signals were digitized and recorded on a computer using Pclamp v.6 software (Axon Instruments). Data were analyzed using the same software, as well as Mini v 5 (Synaptosoft). Usually, stimuli above 7–9 V elicited short-latency IFM evoked action potentials, while stimuli below 7–9 V evoked long-latency IFM action potentials.
IFM ultrastructural studies and image analysis
Thoraces from late pupal and 2-day old female flies were isolated and prepared for transmission electron microscopy according to Cripps et al.52 Fixatives and Embed812 resin were purchased from Electron Microscopy Sciences (Fort Washington, PA); other reagents were purchased from Sigma (St. Louis, MO).
Late pupal and 2-day old adult samples were examined on a Philips 410 transmission electron microscope operating at 80 kV. Images were recorded on film and later digitized using an Epson 1640SU PHOTO flatbed scanner. Young adult and some pupal samples were examined with a FEI Tecnai 12 transmission electron microscope operating at 120 kV. Digital images were taken with a TVIPS (Tietz) TemCam-F214 high-resolution digital camera. Microscope magnifications were calibrated using a diffraction grating replica and latex calibration standard (Ted Pella, Redding, CA).
Myofibril measurements were collected using NIH ImageJ software (http://rsb.info.nih.gov/ij/) and imported into Microsoft Excel for statistical analyses. Myofibril areas and percentage of myofibril area within each muscle fiber were determined by analyzing 9 transverse images collected from 3 different DLMs (3 images per fiber) from 3 flies. The dimensions of each field were approximately 8.3 × 7.4 μm. Each field contained whole myofibrils toward its center and peripheral myofibrils falling partially outside of the defined boundary (termed “partial myofibrils”). Additionally, mitochondria, glycogen granules, membranous structures (e.g. sarcoplasmic reticulum), and occasional organelle-free space were distributed throughout each field. The areas of individual whole or partial myofibrils were determined by outlining each myofibril with the freehand tool in ImageJ and measuring the area outlined. To estimate the myofibril area within each muscle fiber, the sum of the areas of all partial and whole myofibrils was divided by the area of the field. The density of thick filaments within myofibrils (thick filaments/μm2) was determined by analyzing 6–7 transverse myofibril images from 2 of the DLMs (3 images per fiber) selected from the same fibers as above. The number of thick filaments within a single 0.5 μm2 area of each myofibril was counted. Sarcomere lengths were determined by measuring the distance from the mid-point of one Z-disk to that of the next Z-disk from images of longitudinal myofibrils collected from DLM (and a dorsoventral IFM in some cases) from 3 or more flies per line. Dorsoventral IFM measurements did not deviate from DLM measurements. Thick filament lengths were determined from the length of the A-band of each sarcomere. When possible, a straight line tracing one thick filament from the edge of one I-band and across the M-line to the thick filament terminus near the opposite I-band was measured. Otherwise a straight line was drawn equidistant from the top and bottom edges of the sarcomere, from one A-I junction, across (and perpendicular to-) the M-line to the second A-I junction of the same sarcomere and measured.
Myosin Tail Length Determination
Myosin molecules were purified from approximately 120 DLMs of 12–24 hr-old yw, PwMhc2, and 15b-47 flies according to Swank et al.75 15b-47 flies were chosen as the most stringent test for this analysis since their IFM myofibrils have the best structural integrity of the three lines (Figure 3), they display the best flight ability (Table 1) and because their MHC migrates at the same mobility as PwMhc2 (Figure 2b) and wild-type (not shown) IFM isoforms. Myosin was resuspended in 0.6 M ammonium acetate in 50% (v/v) glycerol at a final concentration of 15–30 μg/ml. Individual samples were sprayed from a nebulizer onto freshly cleaved mica squares (~1 cm) and air dried at 23°C. The mica squares were placed in a Denton 501A evaporator for rotary shadowing. ~10 mg of platinum from a 20 mm (0.2 mm diameter) platinum wire, wrapped around a 10.5 cm (0.5 mm diameter) tungsten wire, were evaporated at a 6° tilt onto the rotating samples under high vacuum. A carbon support was then evaporated onto the non-rotating samples from 90°. The platinum-carbon replicas were floated onto the surface of water and picked up with 400 mesh copper grids.
The samples were examined with a FEI Tecnai 12 transmission electron microscope operating at 120 kV. Digital images were taken with a TVIPS (Tietz) TemCam-F214 high-resolution digital camera. All specimens were carefully maintained at the eucentric height. The magnification was held constant throughout all phases of microscopy to avoid lens hysteresis and the introduction of magnification errors. All grids, including standards and myosin, were inserted and exchanged, searched and photographed at a continuous magnification of 21000 X. The grids of yw, PwMhc2, and 15b-47 myosin were examined alternately and immediately after each other, to minimize any systematic difference in magnification. Furthermore, myosin was obtained from several different generations and preparations of Drosophila IFM to insure purely random sampling.
Additional steps were performed to insure accurate length determination of myosin tails. The major 8.75 nm lattice repeat from the center of numerous catalase crystals was used as a standard. NIH ImageJ software was employed to repeatedly measure the distance in pixels across 2 – 50 lattice lines corresponding to 17.5 to 437.5 nm. Once the set scale (pix/nm) reproduced consistent measured distances over variable numbers of lattice lines, the scale was then cross checked against NIST traceable NANOSPHERE polystyrene standards (Electron Microscopy Sciences) with a certified mean diameter of 199 nm by measuring ~50 polystyrene spheres and confirming a mean distance of 199 nm. This scale was used to measure the contour lengths of the myosin tails from the base of the head to tail junction through the extreme C-terminus. Individual, well-resolved myosin tails were magnified by 300 – 400% with NIH ImageJ software. The segmented line tool was used to create a selection of anchor points that outlined the rod contour. A line connecting the selection was created and individual points adjusted to insure their central locations along the entire myosin tail length. The length of the contour line was then determined for each molecule based on the set scale (pix/nm). Numerous values were averaged together to yield the mean myosin rod length from each Drosophila line. Standard errors of the mean were calculated and Student’s t-tests were performed to determine statistical significance.
Acknowledgments
We are grateful to Mr. Allen Church and Dr. Michelle Mardahl-Dumesnil for their technical expertise and to Mr. Gregory Aselis for initial work on the project. We thank Dr. Jim Vigoreaux (University of Vermont) and Dr. Sunita Patel (Brigham and Women’s Hospital) for unpublished data and Dr. Venky Iyer (University of California, Berkeley) for Drosophila Mhc gene alignments. We appreciate the advice of Dr. Roger Craig (University of Massachusetts Medical School) and Dr. Steve Barlow regarding sample preparation and electron microscopy. We thank Dr. Douglas Deutschman for consultation on statistical analysis. We appreciate the helpful discussions and manuscript review provided by Dr. Douglas Swank (Rensselaer Polytechnic Institute), Dr. Jim Vigoreaux, Dr. Mark Miller (University of Vermont), Dr. David Maughan (University of Vermont), Dr. Yudong Hao (University of Washington), Dr. Gerald Pollack (University of Washington) and Dr. Aileen Knowles. This work was supported by grant R01-AR43396 to SIB from the NIH/National Institute of Arthritis and Musculoskeletal Disease and a postdoctoral research supplement from NIH to AC.
Abbreviations used
- MHC
myosin heavy chain
- IFM
indirect flight muscle
- TDT
tergal depressor of the trochanter
- S1
myosin subfragment 1
- S2
myosin subfragment 2
- LMM
light meromyosin
- HMM
heavy meromyosin
- IFI-15b
indirect flight muscle isoform of MHC with embryonic exon 15b region
- DLM
dorsolongitudinal indirect flight muscle
- SL
sarcomere length
- RT-PCR
reverse transcriptase polymerase chain reaction
- SEM
standard error of the mean
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature. 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- 2.Takagi Y, Shuman H, Goldman YE. Coupling between phosphate release and force generation in muscle actomyosin. Philos Trans R Soc Lond B Biol Sci. 2004;359:1913–1920. doi: 10.1098/rstb.2004.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLachlan AD, Karn J. Periodic features in the amino acid sequence of nematode myosin rod. J Mol Biol. 1983;164:605–626. doi: 10.1016/0022-2836(83)90053-0. [DOI] [PubMed] [Google Scholar]
- 4.Straussman R, Squire JM, Ben-Ya’acov A, Ravid S. Skip residues and charge interactions in myosin II coiled-coils: implications for molecular packing. J Mol Biol. 2005;353:613–628. doi: 10.1016/j.jmb.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Walker M, Knight P, Trinick J. Negative staining of myosin molecules. J Mol Biol. 1985;184:535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- 6.Offer G. Skip residues correlate with bends in the myosin tail. J Mol Biol. 1990;216:213–218. doi: 10.1016/S0022-2836(05)80309-2. [DOI] [PubMed] [Google Scholar]
- 7.Huxley HE. The mechanism of muscular contraction. Science. 1969;164:1356–1365. [PubMed] [Google Scholar]
- 8.Burke M, Himmelfarb S, Harrington WF. Studies on the “hinge” region of myosin. Biochemistry. 1973;12:701–710. doi: 10.1021/bi00728a020. [DOI] [PubMed] [Google Scholar]
- 9.Tsong TY, Himmelfarb S, Harrington WF. Stability and melting kinetics of structural domains in the myosin rod. J Mol Biol. 1983;164:431–450. doi: 10.1016/0022-2836(83)90060-8. [DOI] [PubMed] [Google Scholar]
- 10.Weeds AG, Pope B. Studies on the chymotryptic digestion of myosin. Effects of divalent cations on proteolytic susceptibility. J Mol Biol. 1977;111:129–157. doi: 10.1016/s0022-2836(77)80119-8. [DOI] [PubMed] [Google Scholar]
- 11.Lowey S, Slayter HS, Weeds AG, Baker H. Substructure of the myosin molecule. I Subfragments of myosin by enzymic degradation. J Mol Biol. 1969;42:1–29. doi: 10.1016/0022-2836(69)90483-5. [DOI] [PubMed] [Google Scholar]
- 12.Lu RC, Wong A. The amino acid sequence and stability predictions of the hinge region in myosin subfragment 2. J Biol Chem. 1985;260:3456–3461. [PubMed] [Google Scholar]
- 13.Walker M, Trinick J. Electron microscope study of the effect of temperature on the length of the tail of the myosin molecule. J Mol Biol. 1986;192:661–667. doi: 10.1016/0022-2836(86)90283-4. [DOI] [PubMed] [Google Scholar]
- 14.Walzthony D, Eppenberger HM, Ueno H, Harrington WF, Wallimann T. Melting of myosin rod as revealed by electron microscopy. II Effects of temperature and pH on length and stability of myosin rod and its fragments. Eur J Cell Biol. 1986;41:38–43. [PubMed] [Google Scholar]
- 15.Gundapaneni D, Xu J, Root DD. High flexibility of the actomyosin crossbridge resides in skeletal muscle myosin subfragment-2 as demonstrated by a new single molecule assay. J Struct Biol. 2005;149:117–126. doi: 10.1016/j.jsb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 16.Scholz T, Altmann SM, Antognozzi M, Tischer C, Horber JK, Brenner B. Mechanical properties of single myosin molecules probed with the photonic force microscope. Biophys J. 2005;88:360–371. doi: 10.1529/biophysj.104.047795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Root DD, Yadavalli VK, Forbes JG, Wang K. Coiled-coil nanomechanics and uncoiling and unfolding of the superhelix and alpha-helices of myosin. Biophys J. 2006;90:2852–2866. doi: 10.1529/biophysj.105.071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrington WF. On the origin of the contractile force in skeletal muscle. Proc Natl Acad Sci USA. 1979;76:5066–5070. doi: 10.1073/pnas.76.10.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno H, Harrington WF. Conformational transition in the myosin hinge upon activation of muscle. Proc Natl Acad Sci USA. 1981;78:6101–6105. doi: 10.1073/pnas.78.10.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Applegate D, Reisler E. Crossbridge release and alpha-helix-coil transition in myosin and rod minifilaments. J Mol Biol. 1983;169:455–468. doi: 10.1016/s0022-2836(83)80061-8. [DOI] [PubMed] [Google Scholar]
- 21.Ueno H, Harrington WF. An enzyme-probe study of motile domains in the subfragment-2 region of myosin. J Mol Biol. 1984;180:667–701. doi: 10.1016/0022-2836(84)90032-9. [DOI] [PubMed] [Google Scholar]
- 22.Ueno H, Harrington WF. Temperature-dependence of local melting in the myosin subfragment-2 region of the rigor cross-bridge. J Mol Biol. 1986;190:59–68. doi: 10.1016/0022-2836(86)90075-6. [DOI] [PubMed] [Google Scholar]
- 23.Ueno H, Harrington WF. Local melting in the subfragment-2 region of myosin in activated muscle and its correlation with contractile force. J Mol Biol. 1986;190:69–82. doi: 10.1016/0022-2836(86)90076-8. [DOI] [PubMed] [Google Scholar]
- 24.Bertazzon A, Tsong TY. Effects of ions and pH on the thermal stability of thin and thick filaments of skeletal muscle: high-sensitivity differential scanning calorimetric study. Biochemistry. 1990;29:6447–6452. doi: 10.1021/bi00479a016. [DOI] [PubMed] [Google Scholar]
- 25.Margossian SS, Krueger JW, Sellers JR, Cuda G, Caulfield JB, Norton P, Slayter HS. Influence of the cardiac myosin hinge region on contractile activity. Proc Natl Acad Sci USA. 1991;88:4941–4945. doi: 10.1073/pnas.88.11.4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugi H, Kobayashi T, Gross T, Noguchi K, Karr T, Harrington WF. Contraction characteristics and ATPase activity of skeletal muscle fibers in the presence of antibody to myosin subfragment 2. Proc Natl Acad Sci USA. 1992;89:6134–6137. doi: 10.1073/pnas.89.13.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNally EM, Kraft R, Bravo-Zehnder M, Taylor DA, Leinwand LA. Full-length rat alpha and beta cardiac myosin heavy chain sequences. Comparisons suggest a molecular basis for functional differences. J Mol Biol. 1989;210:665–671. doi: 10.1016/0022-2836(89)90141-1. [DOI] [PubMed] [Google Scholar]
- 28.McNally EM, Gianola KM, Leinwand LA. Complete nucleotide sequence of full length cDNA for rat alpha cardiac myosin heavy chain. Nucleic Acids Res. 1989 Sep 25;17(18):7527–8. doi: 10.1093/nar/17.18.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nyitray L, Jancso A, Ochiai Y, Graf L, Szent-Gyorgyi AG. Scallop striated and smooth muscle myosin heavy-chain isoforms are produced by alternative RNA splicing from a single gene. Proc Natl Acad Sci USA. 1994;91:12686–12690. doi: 10.1073/pnas.91.26.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Donnell PT, Collier VL, Mogami K, Bernstein SI. Ultrastructural and molecular analyses of homozygous-viable Drosophila melanogaster muscle mutants indicate there is a complex pattern of myosin heavy-chain isoform distribution. Genes Dev. 1989;3:1233–1246. doi: 10.1101/gad.3.8.1233. [DOI] [PubMed] [Google Scholar]
- 31.Hastings GA, Emerson CP., Jr Myosin functional domains encoded by alternative exons are expressed in specific thoracic muscles of Drosophila. J Cell Biol. 1991;114:263–276. doi: 10.1083/jcb.114.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells L, Edwards KA, Bernstein SI. Myosin heavy chain isoforms regulate muscle function but not myofibril assembly. EMBO J. 1996;15:4454–4459. [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/s0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- 34.George EL, Ober MB, Emerson CP., Jr Functional domains of the Drosophila melanogaster muscle myosin heavy-chain gene are encoded by alternatively spliced exons. Mol Cell Biol. 1989;9:2957–2974. doi: 10.1128/mcb.9.7.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collier VL, Kronert WA, O’Donnell PT, Edwards KA, Bernstein SI. Alternative myosin hinge regions are utilized in a tissue-specific fashion that correlates with muscle contraction speed. Genes Dev. 1990;4:885–895. doi: 10.1101/gad.4.6.885. [DOI] [PubMed] [Google Scholar]
- 36.Parry DA. Coiled-coils in alpha-helix-containing proteins:analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled-coils in other proteins. Biosci Rep. 1982;2:1017–1024. doi: 10.1007/BF01122170. [DOI] [PubMed] [Google Scholar]
- 37.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 38.Lupas A. Prediction and analysis of coiled coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 39.Miller DM, 3rd, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- 40.Swank DM, Wells L, Kronert WA, Morrill GE, Bernstein SI. Determining structure/function relationships for sarcomeric myosin heavy chain by genetic and transgenic manipulation of Drosophila. Microsc Res Tech. 2000;50:430–442. doi: 10.1002/1097-0029(20000915)50:6<430::AID-JEMT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 41.Kazzaz JA, Rozek CE. Tissue-specific expression of the alternately processed Drosophila myosin heavy-chain messenger RNAs. Dev Biol. 1989;133:550–561. doi: 10.1016/0012-1606(89)90057-2. [DOI] [PubMed] [Google Scholar]
- 42.Crough EM, Kazzaz JA, Rozek CE. Tissue specific distribution of Drosophila sarcomeric myosin heavy-chain protein isoforms. Insect Mol Biol. 1994;3:15–26. doi: 10.1111/j.1365-2583.1994.tb00146.x. [DOI] [PubMed] [Google Scholar]
- 43.Tohtong R, Yamashita H, Graham M, Haeberle J, Simcox A, Maughan D. Impairment of muscle function caused by mutations of phosphorylation sites in myosin regulatory light chain. Nature. 1995;374:650–653. doi: 10.1038/374650a0. [DOI] [PubMed] [Google Scholar]
- 44.English AW. Cytokines, growth factors and sprouting at the neuromuscular junction. J Neurocytol. 2003;32:943–960. doi: 10.1023/B:NEUR.0000020634.59639.cf. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JB, Wyman RJ. Mutations altering synaptic connectivity between identified neurons in Drosophila. J Neurosci. 1984;4:530–538. doi: 10.1523/JNEUROSCI.04-02-00530.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Engel JE, Wu CF. Altered habituation of an identified escape circuit in Drosophila memory mutants. J Neurosci. 1996;16:3486–3499. doi: 10.1523/JNEUROSCI.16-10-03486.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin M, Nash HA. Influence of general anesthetics on a specific neural pathway in Drosophila melanogaster. Proc Natl Acad Sci USA. 1996;93:10446–10451. doi: 10.1073/pnas.93.19.10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanouye MA, Wyman RJ. Motor outputs of giant nerve fiber in Drosophila. J Neurophysiol. 1980;44:405–421. doi: 10.1152/jn.1980.44.2.405. [DOI] [PubMed] [Google Scholar]
- 49.Miller BM, Zhang S, Suggs JA, Swank DM, Littlefield KP, Knowles AF, Bernstein SI. An alternative domain near the nucleotide-binding site of Drosophila muscle myosin affects ATPase kinetics. J Mol Biol. 2005;353:14–25. doi: 10.1016/j.jmb.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 50.Hao Y, Miller MS, Swank DM, Liu H, Bernstein SI, Maughan DW, Pollack GH. Passive stiffness in Drosophila indirect flight muscle reduced by disrupting paramyosin phosphorylation, but not by embryonic myosin S2 hinge substitution. Biophys J. 2006;91:4500–4506. doi: 10.1529/biophysj.106.088492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elliott A, Offer G. Shape and flexibility of the myosin molecule. J Mol Biol. 1978;123:505–519. doi: 10.1016/0022-2836(78)90204-8. [DOI] [PubMed] [Google Scholar]
- 52.Cripps RM, Suggs JA, Bernstein SI. Assembly of thick filaments and myofibrils occurs in the absence of the myosin head. EMBO J. 1999;18:1793–1804. doi: 10.1093/emboj/18.7.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reedy MC, Bullard B, Vigoreaux JO. Flightin is essential for thick filament assembly and sarcomere stability in Drosophila flight muscles. J Cell Biol. 2000;151:1483–1500. doi: 10.1083/jcb.151.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, Wu S, Reedy MC, Winkler H, Lucaveche C, Cheng Y, Reedy MK, Taylor KA. Electron tomography of swollen rigor fibers of insect flight muscle reveals a short and variably angled S2 domain. J Mol Biol. 2006;362:844–860. doi: 10.1016/j.jmb.2006.07.084. [DOI] [PubMed] [Google Scholar]
- 55.O’Donnell PT, Bernstein SI. Molecular and ultrastructural defects in a Drosophila myosin heavy chain mutant: differential effects on muscle function produced by similar thick filament abnormalities. J Cell Biol. 1988;107:2601–2612. doi: 10.1083/jcb.107.6.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vigoreaux JO, Saide JD, Pardue ML. Structurally different Drosophila striated muscles utilize distinct variants of Z-band-associated proteins. J Muscle Res Cell Motil. 1991;12:340–354. doi: 10.1007/BF01738589. [DOI] [PubMed] [Google Scholar]
- 57.Kronert WA, O’Donnell PT, Fieck A, Lawn A, Vigoreaux JO, Sparrow JC, Bernstein SI. Defects in the Drosophila myosin rod permit sarcomere assembly but cause flight muscle degeneration. J Mol Biol. 1995;249:111–125. doi: 10.1006/jmbi.1995.0283. [DOI] [PubMed] [Google Scholar]
- 58.Swank DM, Knowles AF, Suggs JA, Sarsoza F, Lee A, Maughan DW, Bernstein SI. The myosin converter domain modulates muscle performance. Nat Cell Biol. 2002;4:312–316. doi: 10.1038/ncb776. [DOI] [PubMed] [Google Scholar]
- 59.Swank DM, Maughan DW. Rates of force generation in Drosophila fast and slow muscle types have opposite responses to phosphate. Adv Exp Med Biol. 2003;538:459–467. doi: 10.1007/978-1-4419-9029-7_42. [DOI] [PubMed] [Google Scholar]
- 60.Vigoreaux JO, Saide JD, Valgeirsdottir K, Pardue ML. Flightin, a novel myofibrillar protein of Drosophila stretch-activated muscles. J Cell Biol. 1993;121:587–598. doi: 10.1083/jcb.121.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vigoreaux JO, Perry LM. Multiple isoelectric variants of flightin in Drosophila stretch-activated muscles are generated by temporally regulated phosphorylations. J Muscle Res Cell Motil. 1994;15:607–616. doi: 10.1007/BF00121068. [DOI] [PubMed] [Google Scholar]
- 62.Vigoreaux JO. Alterations in flightin phosphorylation in Drosophila flight muscles are associated with myofibrillar defects engendered by actin and myosin heavy-chain mutant alleles. Biochem Genet. 1994;32:301–314. doi: 10.1007/BF00555832. [DOI] [PubMed] [Google Scholar]
- 63.Ayer G, Vigoreaux JO. Flightin is a myosin rod binding protein. Cell Biochem Biophys. 2003;38:41–54. doi: 10.1385/CBB:38:1:41. [DOI] [PubMed] [Google Scholar]
- 64.Henkin JA, Maughan DW, Vigoreaux JO. Mutations that affect flightin expression in Drosophila alter the viscoelastic properties of flight muscle fibers. Am J Physiol Cell Physiol. 2004;286:C65–72. doi: 10.1152/ajpcell.00257.2003. [DOI] [PubMed] [Google Scholar]
- 65.Patel SR, Saide JD. Stretchin-klp, a novel Drosophila indirect flight muscle protein, has both myosin dependent and independent isoforms. J Muscle Res Cell Motil. 2005;26:213–224. doi: 10.1007/s10974-005-9012-y. [DOI] [PubMed] [Google Scholar]
- 66.Swank DM, Kronert WA, Bernstein SI, Maughan DW. Alternative N-terminal regions of Drosophila myosin heavy chain tune muscle kinetics for optimal power output. Biophys J. 2004;87:1805–1814. doi: 10.1529/biophysj.103.032078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S NISC Comparative Sequencing Program. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003;13:721–731. doi: 10.1101/gr.926603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinformatics. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- 69.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N. Y: 1989. [Google Scholar]
- 71.Thummel CS, Pirrotta V. New pCaSpeR P element vectors. Drosophila Infor Serv. 1992;71:150. [Google Scholar]
- 72.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 73.Cripps RM, Bernstein SI. Generation of transgenic Drosophila melanogaster by P element-mediated germline transformation. In: Natick MA, editor. Gene Transfer Methods:Introducing DNA into Living Cells and Organisms. Eaton Publishing; 2000. pp. 93–125. [Google Scholar]
- 74.Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. Academic Press; San Diego: 1992. [Google Scholar]
- 75.Swank DM, Bartoo ML, Knowles AF, Iliffe C, Bernstein SI, Molloy JE, Sparrow JC. Alternative exon-encoded regions of Drosophila myosin heavy chain modulate ATPase rates and actin sliding velocity. J Biol Chem. 2001;276:15117–15124. doi: 10.1074/jbc.M008379200. [DOI] [PubMed] [Google Scholar]
- 76.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 77.Drummond DR, Hennessey ES, Sparrow JC. Characterisation of missense mutations in the Act88F gene of Drosophila melanogaster. Mol Gen Genet. 1991;226:70–80. doi: 10.1007/BF00273589. [DOI] [PubMed] [Google Scholar]
- 78.Zordan MA, Massironi M, Ducato MG, Te Kronnie G, Costa R, Reggiani C, Chagneau C, Martin JR, Megighian A. Drosophila CAKI/CMG protein, a homolog of human CASK, is essential for regulation of neurotransmitter vesicle release. J Neurophysiol. 2005;94:1074–1083. doi: 10.1152/jn.00954.2004. [DOI] [PubMed] [Google Scholar]
- 79.Stewart M, Edwards P. Length of myosin rod and its proteolytic fragments determined by electron microscopy. FEBS Lett. 1984;168:75–8. doi: 10.1016/0014-5793(84)80209-4. [DOI] [PubMed] [Google Scholar]


