Abstract
The human cytomegalovirus gene product US11 causes rapid degradation of class I major histocompatibility complex (MHCI) heavy chains by inducing their dislocation from the endoplasmic reticulum (ER) and subsequent degradation by the proteasome. This set of reactions resembles the endogenous cellular quality control pathway that removes misfolded or unassembled proteins from the ER. We show that the transmembrane domain (TMD) of US11 is essential for MHCI heavy chain dislocation, but dispensable for MHCI binding. A Gln residue at position 192 in the US11 TMD is crucial for the ubiquitination and degradation of MHCI heavy chains. Cells that express US11 TMD mutants allow formation of MHCI-β2m complexes, but their rate of egress from the ER is significantly impaired. Further mutagenesis data are consistent with the presence of an alpha-helical structure in the US11 TMD essential for MHCI heavy chain dislocation. The failure of US11 TMD mutants to catalyze dislocation is a unique instance in which a polar residue in the TMD of a type I membrane protein is required for that protein's function. Targeting of MHCI heavy chains for dislocation by US11 thus requires the formation of interhelical hydrogen bonds within the ER membrane.
INTRODUCTION
Many viruses target components of the MHCI antigen presentation pathway to prevent recognition of infected cells by cytotoxic T lymphocytes (Tortorella et al., 2000). The human cytomegalovirus (HCMV) encodes two ER-resident type I membrane glycoproteins, US2 and US11, both of which specifically target MHCI heavy chains for dislocation from the ER membrane to the cytosol, where they are processed by an N-glycanase and degraded by the proteasome (Wiertz et al., 1996a, 1996b). Dislocation is rapid, occurring soon after insertion and glycosylation of the MHCI heavy chain in the ER. Many similarities exist between the sequence of events catalyzed by US2 and US11 and the means by which cells dispose of misfolded or unassembled proteins that accumulate in the ER, suggesting that the viral proteins have coopted the endogenous cellular pathway to bring about the specific degradation of MHCI heavy chains.
Disposal of MHCI heavy chains and cellular proteins involves the coordinated action of many protein complexes that recognize the substrate within the ER lumen and subsequently move it across the ER membrane into the cytosol. It is unclear how proteins that fail to fold properly or that do not assemble into their correct oligomeric state are recognized and selectively targeted for removal from the ER, but several mechanisms have been proposed (Cabral et al., 2001). After recognition, substrates for dislocation must be brought into contact with the as yet unidentified protein machinery that initiates their removal from the ER (Tsai et al., 2002).
Genetic studies in yeast and biochemical studies in mammalian cells have suggested that the Sec61 channel, involved in protein import into the ER, may also be an exit route for substrates (Tsai et al., 2002). Ubiquitin conjugation of cytosolic proteins is not only important for their degradation by the proteasome, but is also essential for ER-to-cytosol dislocation. When ubiquitin conjugation is blocked, using either genetic ablation in yeast, semi-intact mammalian cells depleted of ubiquitin or mammalian cells expressing a temperature-sensitive E1 ubiquitin activating enzyme, dislocation substrates remain within the ER (Kikkert et al., 2001; Shamu et al., 2001; Jarosch et al., 2002). The Cdc48(p97)/Npl4/Ufd1 complex acts in an ATP-dependent manner at the cytosolic face of the ER to promote removal of ubiquitinated dislocation substrates from the ER membrane (Ye et al., 2001). Proteolysis is then carried out by the 26S proteasome (Wiertz et al., 1996a). For glycoproteins, the N-linked glycan is removed before proteolysis by a cytosolic N-glycanase (PNG1), which has a preference for unfolded proteins (Hirsch et al., 2003).
US2- and US11-mediated dislocation of MHCI heavy chains resembles dislocation of misfolded cellular proteins. However, MHCI heavy chains in US2- or US11-expressing cells have a half-life of only minutes, whereas misfolded cellular proteins have a half-life of between 30 min and several hours (Wiertz et al., 1996a, 1996b; Hughes et al., 1997; Huppa and Ploegh, 1997). US2 and US11 must somehow cause MHCI heavy chains to bypass normal quality control, which grants cellular proteins a chance to assume their proper conformation. How US2 and US11 accomplish this task remains unknown, but analysis of both MHCI heavy chain mutants and the US2 protein have revealed some of the requirements for dislocation. The cytosolic portion of the MHCI heavy chain is required for dislocation by both US2 and US11, but Lys residues in this region are dispensable, suggesting that ubiquitination of the cytosolic region of the MHCI heavy chain is not required (Shamu et al., 1999; Story et al., 1999). The structure of a US2 fragment bound to a complex of MHCI heavy chain, β2m, and peptide suggests that an interaction between their lumenal domains is how US2 selectively targets ER-resident MHCI heavy chains for dislocation (Gewurz et al., 2001). However, mere interaction of US2 with MHCI heavy chains is not sufficient: US2 mutants that lack the cytosolic tail but remain membrane anchored continue to interact strongly with MHCI heavy chains, yet do not catalyze dislocation (Furman et al., 2002). US11, in all likelihood, causes dislocation in a manner distinct from US2 (Furman et al., 2002).
Extraction of the MHCI heavy chain membrane anchor represents a critical step in dislocation, but events that occur within the lipid bilayer have yet to be examined. Here, we explore interactions within the lipid bilayer by examining the role of the US11 transmembrane domain (TMD). MHCI heavy chain dislocation is blocked by mutation of the single Gln residue within the US11 TMD, suggesting that interhelical hydrogen bonds formed by the US11 TMD are essential for US11's function. Our results suggest that US11 uses interactions within the ER lipid bilayer to manipulate the cellular quality control pathway to bring about MHCI heavy chain degradation.
MATERIALS AND METHODS
Cell Lines, Antibodies, and Chemicals
U373-MG astrocytoma cells transfected with US11 and US11–180 have been described (Jones et al., 1995; Rehm et al., 2001). All astrocytoma cell lines were cultured in DMEM as described (Tortorella et al., 1998), and the retroviral packaging cell line 293GPG was maintained as described (Ory et al., 1996). Astrocytoma cell lines that stably express the US11 variants generated in this study were initially selected and subsequently maintained in DMEM containing 0.5 mg/ml Geneticin (Life Technologies, Rockville, MD). Antibodies used in this study have been described (Parham et al., 1979; van de Rijn et al., 1983; Stam et al., 1986; Hochstenbach et al., 1992; Tortorella et al., 1998; Rehm et al., 2001). 12CA5 (anti-HA) was coupled to CNBr-activated Sepharose 4 Fast Flow (Amersham-Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's specifications. Reagents used for immunofluorescence analysis were: biotinylated 12CA5 (Roche, Indianapolis, IN), Alexa-Fluor 488–conjugated goat anti-mouse (Molecular Probes, Eugene, OR) and Cy3-conjugated streptavidin (Rockland, Gilbertsville, PA). Horseradish peroxidase (HRP)-conjugated goat anti-mouse and goat anti-rabbit antibodies (Jackson ImmunoResearch, West Grove, PA) as well as HRP-conjugated streptavidin (Roche) were used according to the manufacturer's specifications. Digitonin was purchased from Merck (Whitehouse Station, NJ) and was purified as described (Gorlich and Rapoport, 1993).
Plasmid Constructs and Retroviral Infections
A pcDNA3.1+ (Invitrogen, Carlsbad, CA) construct containing the US11 gene from the AD169 strain of human cytomegalovirus (Rehm et al., 2001) was used as a template for the introduction of all missense mutations which were generated using the Quick Change method (Stratagene, La Jolla, CA). The 11-C-11 construct is a chimeric molecule in which the TMD of US11 (amino acids 179–199) was replaced with that of the human CD4 protein (residues 395–418). In the constructs specified, the signal sequence of US11 (amino acids 1–17) was replaced with the signal sequence of the H-2Kb molecule. HA epitope-tagged versions of US11 were generated by inserting US11 sequence (amino acids 18–215) downstream of a sequence containing (from 5′ to 3′) the H-2Kb signal sequence and the HA epitope. All US11-related constructs were verified by sequencing and were subcloned into a modified pLNCX-based (Clontech, Palo Alto, CA) retroviral expression vector to be described elsewhere. 293GPG retroviral packaging cells were transfected with the retroviral plasmids using FuGene6 transfection reagent (Roche) according to the manufacturer's specifications. Cell supernatants containing retrovirus were used to infect U373 astrocytoma cells.
Metabolic Labeling, Pulse-chase Analysis, and Immunoprecipitation
Metabolic labeling, pulse-chase analysis, detergent solubilization, and immunoprecipitation were performed as described (Tortorella et al., 1998; Rehm et al., 2001). Immune complexes were recovered from digitonin lysates (1% digitonin, 25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2 with 1 mM PMSF, 1 μM leupeptin, and 1.5 μg/ml aprotinin) using Protein A-agarose (RepliGen, Needham, MA) and were washed in 0.2% digitonin in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA. Where indicated, immune complexes were treated with endoglycosidase H (EndoH, New England Biolabs, Beverly, MA).
Gel Electrophoresis, Immunoblotting, and Immunofluorescence
Immune complexes were analyzed using SDS-PAGE and fluorography (Ploegh, 1995). Quantitation of radiolabeled MHCI heavy chains was performed using a STORM PhosphorImager and Imagequant software (Molecular Dynamics, Sunnyvale, CA). In immunoblotting experiments, proteins were transferred electrophoretically on to PVDF membranes (NEN, Boston, MA), and were probed with the specified antibodies. The analysis of ubiquitinated MHCI heavy chains was performed essentially as described (Shamu et al., 1999) except 10 μM biotinylated ubiquitin (Ub-bio; Mitsui and Sharp, 1999) was substituted for iodinated ubiquitin and immune complexes were recovered with Protein A-agarose and were analyzed using SDS-PAGE and immunoblotting. For immunofluorescence analysis, cells were seeded onto glass coverslips 18 h before fixation with 4% paraformaldehyde in PBS for 10 min at room temperature. Immunohistochemistry and epifluorescence microscopy were performed as described (Tirabassi and Ploegh, 2002).
RESULTS
Features of the US11 TMD Are Essential for Causing Dislocation of MHCI Heavy Chains
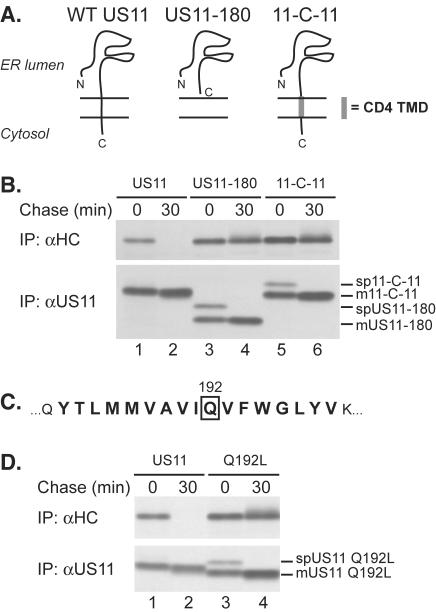
To investigate how the US11 protein causes dislocation of MHCI molecules, we examined the minimal sequence requirements necessary for this function. A US11 molecule that lacks the cytosolic portion of the protein still catalyzes the dislocation of MHCI heavy chains (Furman et al., 2002). We produced a C-terminally truncated US11 molecule from which both the cytosolic and transmembrane domains of US11 (US11–180) were lacking (Rehm et al., 2001). We explored the importance of the identity of the TMD of US11 by replacing it with that of the human CD4 protein, an unrelated type I membrane protein used in other studies of TMD function (Cocquerel et al., 1998), resulting in a chimera designated 11-C-11. We then assessed the stability of MHCI heavy chains in cells expressing wild-type US11, US11–180, or 11-C-11. Lysates were prepared under fully denaturing conditions to ensure recovery of all MHCI heavy chains by immunoprecipitation with the αHC serum (Shamu et al., 1999).
MHCI heavy chains are completely degraded within 30 min of synthesis in cells that express wild-type US11, but are stable in cells expressing US11–180 or 11-C-11 (Figure 1B). Expression levels of US11–180 and 11-C-11 were comparable to that of US11. At the onset of the chase, we observe both the signal peptide-containing and mature US11–180 and 11-C-11 proteins (Figure 1B, bottom panel, lanes 3–6), as reported for the US11–180 molecule (Rehm et al., 2001). Wild-type US11 binds to complexes of MHCI heavy chains and β2m, recognized by the conformation-specific antibody W6/32 (Story et al., 1999). Can US11–180 and 11-C-11 proteins bind to such MHCI complexes? Immunoprecipitation using W6/32 showed coprecipitation of US11–180 and 11-C-11 with MHCI complexes in nondenaturing lysis buffers (NP-40 or digitonin, our unpublished results). Therefore, although US11 mutants lacking a TMD or containing a heterologous TMD retain the ability to bind to MHCI heavy chains, specific features of the US11 TMD itself are required for dislocation.
Figure 1.
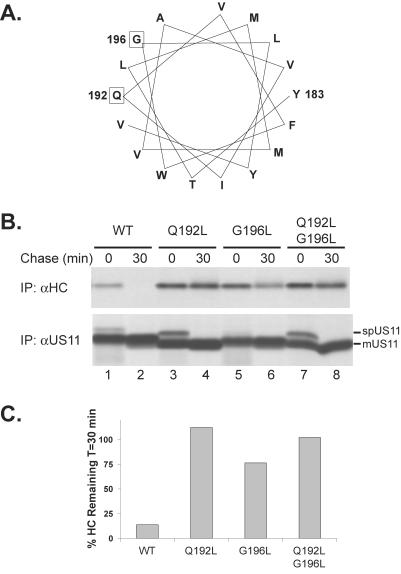
Gln192 within the US11 TMD is required for MHCI heavy chain dislocation. (A) Schematic representation of US11, US11–180, and 11-C-11. (B) U373 cells expressing wild-type US11 (lanes 1 and 2), US11–180 (lanes 3 and 4), or 11-C-11 (lanes 5 and 6) were pulse labeled for 10 min and chased for the indicated times. Immunoprecipitations were performed with antisera against either MHCI heavy chains (αHC, top panel) or US11 (αUS11, bottom panel). The positions of the signal peptide-containing (spUS11-x) and the mature forms of US11–180 and 11-C-11 (mUS11-x) are indicated. (C) The amino acid sequence of the predicted US11 TMD is indicated in bold and Gln192 is boxed. (D) U373 cells expressing US11 (lanes 1 and 2) or US11 Q192L (lanes 3 and 4) were analyzed as in B, and the signal peptide-containing (spUS11 Q192L) and the mature form of US11 Q192L (mUS11 Q192L) are indicated. The US11 Q192L molecule consistently migrated faster than wild-type US11 in SDS-PAGE, attributable to the introduction of the Leu residue.
The US11 sequence contains a Gln residue at position 192, predicted to lie in the center of the US11 TMD (Figure 1C). The TMDs of bitopic proteins usually contain few, if any, polar or charged residues (Landolt-Marticorena et al., 1993). Hydrogen bonding between polar amino acids can mediate strong interactions between alpha-helices within the lipid bilayer (Gratkowski et al., 2001; Zhou et al., 2001). Polar or charged amino acid side chains in the single TMDs of type I and type II membrane proteins likely form favorable contacts with residues of other proteins within the cellular lipid bilayer and facilitate assembly of protein complexes (Popot and Engelman, 2000). Replacement of Gln192 with Leu, a hydrophobic residue with a size roughly similar to that of Gln yielded US11 Q192L. MHCI heavy chains were rapidly degraded in cells expressing wild-type US11, yet they were stable in cells expressing US11 Q192L (Figure 1D, top panel). Analysis of the US11 Q192L protein shows an additional, more slowly migrating form of US11 Q192L present at the onset of the chase, which corresponds to the US11 Q192L molecule that has retained its signal peptide post-translationally (spUS11 Q192L), but is processed during the chase into the mature form (mUS11 Q192L; Figure 1D, lower panel). This behavior resembles that of US11–180 (Rehm et al., 2001) and 11-C-11 (Figure 1B, bottom panel). At present, we do not understand the reason for the delayed signal sequence cleavage phenotype observed for US11 TMD mutants, but this phenomenon will be addressed in detail elsewhere. Delayed signal peptide cleavage of US11 Q192L did not contribute to its inability to cause MHCI heavy chain dislocation. Replacement of the signal sequence of the US11 Q192L molecule with that of H-2Kb resulted in cotranslational signal sequence cleavage (Rehm et al., 2001), but failed to rescue US11 Q192L's activity (our unpublished results).
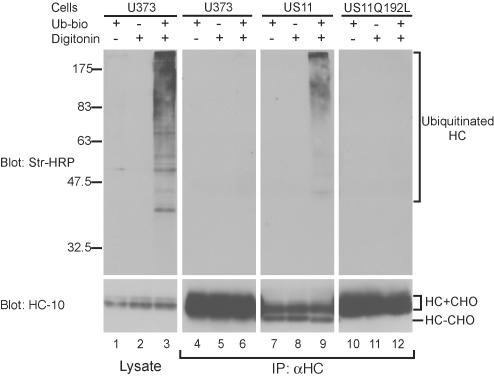
During US11-mediated dislocation, ubiquitin conjugation is required to move the MHCI heavy chain from the ER membrane into the cytosol, and MHCI heavy chains are themselves ubiquitinated before degradation by the proteasome (Shamu et al., 1999, 2001; Kikkert et al., 2001). A small fraction of these ubiquitinated MHCI heavy chains is associated with the membrane, indicating that they may be exposed to the cytosol, yet still inserted in the membrane. Poly-ubiquitination may occur on the lumenal domain of the MHCI heavy chain upon exposure to the cytosol and may serve to prevent MHCI heavy chains from moving back into the ER lumen (Shamu et al., 2001). We therefore examined whether MHCI heavy chains in cells expressing US11 Q192L were ubiquitinated by using a permeabilized cell system (Shamu et al., 1999).
Only when cells were permeabilized with a low concentration of digitonin in the presence of biotinylated ubiquitin (Ubbio) and an ATP-regenerating system did Ub-bio conjugation to cellular proteins occur (Figure 2, left panel). Control U373 cells, US11-expressing cells and US11 Q192L-expressing cells were treated with proteasome inhibitor and permeabilized in the presence of Ub-bio, and immunoprecipitations for MHCI heavy chains from denaturing lysates were performed and analyzed by immunoblotting for Ub-bio and MHCI heavy chains (Figure 2, top and bottom panels, respectively). In US11-expressing cells, the characteristic deglycosylated MHCI heavy chain species was recovered, indicating dislocation to the cytosol (Figure 2, bottom panel, lanes 7–9; Wiertz et al., 1996a). A fraction of the recovered heavy chains was poly-ubiquitinated (Figure 2, top panel, lane 9). No deglycosylated or poly-ubiquitinated species were recovered from control U373 or US11 Q192L cells (Figure 2, top panel, lanes 6 and 12). MHCI heavy chains in US11 Q192L cells are apparently not exposed to the cytosol, where ubiquitination and deglycosylation occur. Gln192 in the US11 TMD is thus essential for dislocation at a stage that precedes exposure of MHCI heavy chains to the cytosol.
Figure 2.
Gln192 is required for MHCI heavy chain ubiquitination. U373 control cells (lanes 1–3 and 4–6), US11-expressing cells (lanes 7–9), or US11 Q192L-expressing cells (lanes 10–12) were treated with the proteasome inhibitor carboxybenzyl-leucyl-leucyl-leucine vinyl sulfone (ZL3VS; Bogyo et al., 1997) at a concentration of 50 μM for 1 h. Cells were then incubated in a buffer containing an ATP-regenerating system and were permeabilized with 0.025% digitonin in the presence or absence of Ub-bio, as indicated. After lysis, a fraction of the lysate was removed and analyzed directly by SDS-PAGE and immunoblot (left panels), and the remainder was immunoprecipitated with αHC serum (right panels). Samples were transferred to a PVDF membrane and analyzed by probing with HRP-conjugated streptavidin (top panels), followed by reprobing with the mAb HC-10 (bottom panels; Stam et al., 1986), which recognizes unfolded MHCI heavy chains. HC+CHO and HC–CHO: heavy chains with and without their N-linked glycan, respectively. Ubiquitinated heavy chains and the positions of molecular weight standards are indicated.
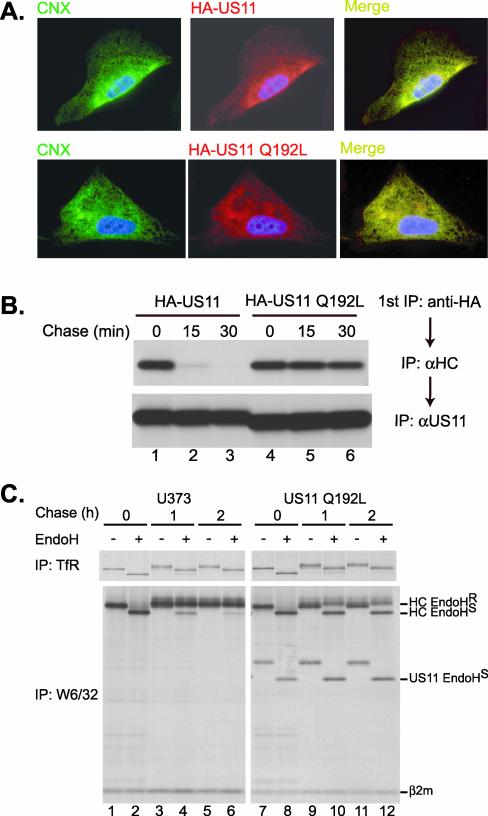
US11 Q192L Is an ER-resident Protein and Causes Retention of MHCI Complexes in the ER
US11 resides in the ER where it acts on MHCI heavy chains (Wiertz et al., 1996a). Polar residues within TMDs of single spanning proteins can affect localization and cause retention of proteins within the ER (Bonifacino et al., 1991). Does replacement of Gln192 alter the subcellular localization of US11? Because the αUS11 serum used for immunoprecipitations was not suitable for immunohistochemistry, we analyzed cells expressing HA epitope-tagged versions of US11 and US11 Q192L (termed HA-11 and HA-11 Q192L, respectively). These tagged versions are as effective at MHCI heavy chain dislocation as their untagged counterparts (our unpublished results). We observed complete colocalization of HA-11 and HA-11 Q192L with the ER-resident protein calnexin (Figure 3A). The mechanism by which US11 is retained in the ER is unclear, because US11 lacks a known ER retention motif, but the lumenal domain of US11 appears to be sufficient to confer ER localization to the protein (B.L., unpublished results).
Figure 3.
(A and B) Mutation of Gln192 does not affect subcellular localization or MHCI heavy chain binding. (A) HA-US11 cells (top panels) or HA-US11 Q192L cells (bottom panels) were stained with the AF8 mAb (Hochstenbach et al., 1992) recognizing the ER chaperone calnexin (left panels, green), biotinylated anti-HA (middle panels, red), and DAPI (blue, all panels). A merge of the left and middle panels (right panels) shows colocalization of calnexin and US11. (B) HA-US11 cells (lanes 1–3) or HA-US11 Q192L cells (lanes 4–6) were labeled for 10 min and chased for 0, 15, or 30 min. A first immunoprecipitation was performed from lysates (1% [wt/vol] digitonin in 25 mM Tris-HCE, pH 7.4, 150 mM NaCl, 5 mM MgCl2) with 12CA5-coupled Sepharose (anti-HA). Bound material was eluted from the beads and denatured by boiling in 1% [wt/vol] SDS with 5 mM DTT. After dilution with NP-40 lysis mix, a second immunoprecipitation was carried out with αHC (top panel) and αUS11 (bottom panel). (C) MHCI-β2m complexes are stable in cells expressing US11 Q192L, but are retained in the ER. Control U373 cells (lanes 1–6) and US11 Q192L cells (lanes 7–12) were pulse-labeled for 15 min and were chased for 0, 1, or 2 h. Immunoprecipitations were performed with antitransferrin receptor (van de Rijn et al., 1983; top panels) or W6/32 (Parham et al., 1979; bottom panels). Immune complexes were treated with EndoH where indicated. The positions of the EndoH-resistant (EndoHR) and EndoH-sensitive (EndoHS) MHCI heavy chains, US11 Q192L, and β2m are indicated.
The US11 Q192L protein is present at the correct site to mediate dislocation, but is there a role for Gln192 in binding to MHCI heavy chains? We examined the kinetics of association of MHCI heavy chains with HA-11 and HA-11 Q192L in a pulse-chase experiment. MHCI heavy chains were recovered in a complex with HA-11 after the onset of the chase, and the binding of MHCI heavy chains to HA-11 was rapidly lost, with kinetics that resembled that of MHCI heavy chain dislocation (Figure 3B, lanes 1–3 and our unpublished results). At the onset of the chase, we recovered similar levels of MHCI heavy chains in HA-11 Q192L cells as seen for HA-11. However, the complex of MHCI heavy chains and HA-11 Q192L was stable and persisted throughout the chase (Figure 3B, lanes 4–6). Therefore, Gln192 is not required for the interaction of US11 with its substrate, and in fact, mutation of Gln192 to Leu results in a persistent interaction with MHCI heavy chains.
We next examined the effect, if any, of US11 Q192L expression on MHCI complex assembly and stability. Early after insertion into the ER and chaperone-assisted folding, MHCI heavy chains associate with β2m to form a heterodimeric MHCI complex recognized by the mAb W6/32 (Parham et al., 1979). In US11-expressing cells, this complex can be recovered only transiently before dislocation and degradation, consistent with its disassembly during dislocation (Wiertz et al., 1996a; Tortorella et al., 1998). Disassembly most likely occurs before exposure of the MHCI complex to the cytosol (Tortorella et al., 1998). In control U373 cells, MHCI complexes were stable throughout a 2-h chase period and >90% of the complexes acquired EndoH resistance (Figure 3C, bottom left panel), indicating passage through the Golgi. In cells expressing US11 Q192L, MHCI complexes retained W6/32 reactivity and were stable during the 2-h chase (Figure 3C, lower right panel). Therefore, consistent with its role in MHCI heavy-chain dislocation and ubiquitination, the Gln residue at position 192 is also essential for destabilizing the MHCI heavy chain-β2m complex.
Although MHCI complexes were stable, their trafficking through the secretory pathway was dramatically slowed in US11 Q192L cells, as only ∼50% of the MHCI complexes acquired EndoH resistance, compared with full resistance seen in U373 cells (Figure 3, compare lanes 5 and 6 with 11 and 12). The expression of US11 Q192L did not affect trafficking of the transferrin receptor, which acquired resistance to EndoH at equivalent rates in control cells and US11 Q192L cells (Figure 3C, top panels). Also, the US11 Q192L protein itself was coimmunoprecipitated with MHCI complexes at all time points tested and remained fully EndoH sensitive, consistent with its ER localization. We also observed retention of MHCI complexes in the ER in cells expressing US11-180 and 11-C-11 as well as coimmunoprecipitation of the US11 TMD mutants with MHCI complexes (our unpublished results). Therefore, the ER lumenal domain of US11 must be responsible for both MHCI heavy chain binding and retention in the ER.
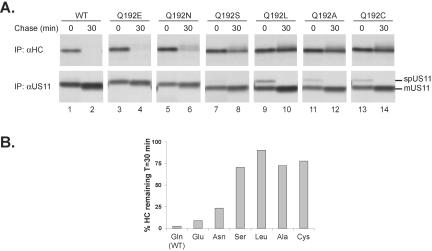
Additional Polar Residues Can Substitute for Gln192 in Mediating MHCI Heavy-chain Degradation
The Gln side chain contains a carboxamide capable of acting as both a hydrogen bond donor and acceptor. Mutant forms of US11 in which Gln192 was substituted with Asn, Glu, Cys, Ala, and Ser were stably expressed in U373 cells and their ability to cause MHCI heavy chain dislocation was examined. When we mutated Gln192 to Leu, Ser, Cys, and Ala, the activity of US11 was largely eliminated, because little or no MHCI heavy chain degradation occurred in cells expressing these mutants (Figure 4). Changing Gln192 to Asn had a modest effect on US11 activity. Mutation of Gln192 to Glu did not significantly alter the rate of MHCI heavy chain degradation. Therefore, a residue containing either a carboxamide or carboxylate, both of which can promote strong interhelical interactions (Zhou et al., 2001), is required at position 192 to mediate rapid MHCI heavy chain dislocation.
Figure 4.
Other polar residues can substitute for Gln at position 192. (A) Pulse-chase immunoprecipitation analysis of cell lines expressing wild-type US11 (WT, lanes 1 and 2), or with Gln192 mutated to Glu (lanes 3 and 4), Asn (lanes 5 and 6), Ser (lanes 7 and 8), Leu (lanes 9 and 10), Ala (lanes 11 and 12), or Cys (lanes 13 and 14) was performed as in Figure 1. The αHC and αUS11 immunoprecipitations were run on separate SDS-PAGE gels, and the data shown are from nonconsecutive lanes of the individual gels. The positions of the signal peptide-containing form (spUS11) and the mature form (mUS11) are indicated. Differences in the mobilities of the US11 mutants were consistently observed in SDS-PAGE gels, attributable to the introduction of the various residues at position 192. (B) Quantitation of the MHCI heavy chain amounts recovered in the experiment shown in A was performed using PhosphorImager analysis for each individual cell line. The values shown represent the amount of MHCI heavy chain recovered at 30 min as a percentage of that recovered at the 0-min time point.
The US11 TMD Contains a Structural Feature That Is Essential for MHCI Heavy chain Dislocation
Although polar amino acids can mediate strong interactions between TM helices in model systems, additional structural elements of TM helices also play an important role (Popot and Engelman, 2000). Based on a helical wheel projection of the US11 TMD, Gly196 is predicted to be on the same face of the helix as Gln192 (Figure 5A). Gly residues, frequently found in membrane-spanning helices, allow for close packing and favorable Van der Waals contacts between helices that have a complementary interface (Russ and Engelman, 2000). We thus examined the role of Gly196 in MHCI heavy chain dislocation. In cells expressing a US11 mutant with Leu substituted for Gly196 (US11 G196L), only minimal MHCI heavy chain degradation was seen (Figure 5, B and C). A double mutant (US11 Q192L/G196L) behaved indistinguishably from US11 Q192L. Expression of the different US11 mutants was equivalent to that of wild-type US11, ruling out low US11 G196L expression as the cause for impaired MHCI heavy chain degradation (Figure 5B, bottom panel). Thus, there are requirements for specific amino acids at multiple points within the US11 TMD.
Figure 5.
The US11 TMD contains a structure that is required for MHCI dislocation. (A) Helical wheel projection of the US11 TMD. Gln192 and Gly196 are boxed. (B) Pulse-chase immunoprecipitation analysis of cell lines expressing wild-type US11 (WT, lanes 1 and 2), US11 Q192L (lanes 3 and 4), and mutants G196L (lanes 5 and 6) and Q192L/G196L (lanes 7 and 8) was performed as in Figure 1. The positions of the signal peptide-containing form (spUS11) and the mature form (mUS11) are indicated. (C) Quantitation of MHCI heavy chain amounts from the experiment shown in B was performed using PhosphorImager analysis as in Figure 4B.
DISCUSSION
The study of US11- and US2-mediated dislocation of MHCI heavy chains has thus far focused on either the interactions between the lumenal domain of MHCI heavy chains and the viral proteins or on their cytosolic portions. The lumenal domains of both US11 and US2 mediate a recognition event that most likely allows them to target MHCI heavy chains selectively for dislocation (Gewurz et al., 2001). The lumenal domains of US2 and US11 may have other, as yet undefined, roles.
However, events that occur within the ER lipid bilayer have not been examined in any detail. During dislocation, the MHCI heavy chain, a type I membrane protein, is removed from the membrane and can be retrieved from the cytosol when the proteasome is inhibited (Wiertz et al., 1996a, 1996b). Other membrane proteins known to be dislocated due to their misfolding or improper assembly have also been identified as soluble species in the cytosol (Hughes et al., 1997; Huppa and Ploegh, 1997; Johnston et al., 1998). Therefore, removal of the stably integrated TMDs from the lipid bilayer likely represents a crucial, though energetically unfavorable, step in the disposal of membrane proteins. How the cell accomplishes this is unclear, but may involve partitioning of the TMD from the lipid environment into an aqueous, proteinaceous channel. This would represent the reverse of what happens during TMD insertion into the lipid bilayer, where the Sec61 channel opens laterally to allow hydrophobic sequences to insert (Heinrich et al., 2000). The energetic cost of such an event, regardless of the precise mechanism, would be high because of the removal of a hydrophobic sequence from the lipid phase into an aqueous phase. Accordingly, dislocation is ATP-dependent (Wiertz et al., 1996b).
Despite the importance of events that must occur within the ER lipid bilayer, the roles of the US2 and US11 TMDs in dislocation had not been examined. Suprisingly, when the US11 TMD was replaced with that of the CD4 protein, a typical, nonpolar membrane anchor, dislocation was abrogated (Figure 1B). Thus, the identity of the US11 TMD is essential.
TMDs of bitopic membrane proteins function as more than simple membrane anchors, by promoting protein assembly and folding and regulating subcellular localization (Cocquerel et al., 2000; Call et al., 2002). There are many documented cases where TMDs form homo- or hetero-oligomers, essential for the function of the respective proteins (MacKenzie et al., 1997; Cocquerel et al., 2000; Constantinescu et al., 2001; Call et al., 2002). In the striking case of the T cell receptor complex, lateral associations formed by the TMDs of the individual subunits contribute to the formation of a macromolecular signaling complex (Call et al., 2002). Many of the known instances of TMD interactions involve Gly motifs or charge pair interactions (Cosson et al., 1991; Russ and Engelman, 2000), but interhelical hydrogen bonding by polar residues can also provide a major force for TMD association, as judged from the behavior of model TMDs in vitro (Gratkowski et al., 2001; Zhou et al., 2001). Although the functional relevance of such interactions has been shown for a small number of proteins only, there are two notable examples. The TMD of the bovine papillomavirus E5 protein contains a Gln residue that contributes to the formation of an E5 homodimer. The E5 dimer then forms a complex with the platelet-derived growth factor β receptor, inducing ligand-independent autophosphorylation and cellular transformation (Klein et al., 1998). A Gln residue in the TMD of the invariant chain (Ii) contributes to the assembly of Ii trimers, the formation of which is required to mediate Ii-class II MHC association in the ER (Ashman and Miller, 1999). In both of these instances, Gln residues in the TMDs are required for homo-oligomerization, a prerequisite for subsequent assembly into a hetero-oligomeric complex.
Here, we show an absolute requirement for a helical structure encoded by the US11 TMD. Without a polar residue capable of forming hydrogen bonds at position 192 of the US11 TMD, MHCI heavy chains, complexed with US11, are not exposed to the cytosol. Instead, they persist as folded complexes that are retained in the ER. The importance of the Gly residue at position 196, on the same face of the US11 TM helix as Gln192, suggests that the US11 TMD forms multiple contacts, each of them required for MHCI heavy chain dislocation. Gln192 in the US11 TMD may act in a manner similar to the Gln residues in the E5 and Ii TMDs by promoting the association of US11 with other ER membrane proteins involved in dislocation. However, the binding of US11 to its one known ligand, the MHCI heavy chain, is not dependent on Gln192. We are currently investigating the nature of the interactions mediated by the US11 TMD and potential cellular proteins that associate with US11 in a Gln192-dependent manner.
What sort of interaction might the US11 TMD form within the ER membrane? Because of the rapid kinetics of US11-induced degradation, there may be a direct interaction between US11 and a component of the dislocation machinery. Candidates include the Sec61 complex or its associated components. Such an interaction would place US11 in an ideal location for catalyzing dislocation through the Sec61 channel. The US11 TMD could influence the lateral gating of a proteinaceous channel such as Sec61, facilitating extraction of the MHCI heavy chain membrane anchor. Of note, when ubiquitin conjugation is blocked, MHCI heavy chains are not dislocated, but form complexes with US11 that remain in the ER (Kikkert et al., 2001). We observe similar stable complexes that form between US11 Q192L and MHCI heavy chains. This similarity indicates that the US11 TMD positions the MHCI heavy chain for ubiquitin conjugation, possibly by forming interactions with the ER-associated components of the ubiquitin conjugation machinery.
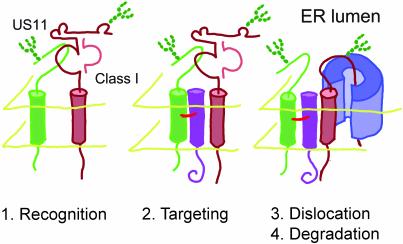
US11 is not itself a substrate for the ER-to-cytosol dislocation pathway, suggesting that US11 directs dislocation in trans via interactions with cellular components. We observe US11-MHCI heavy chain complexes that form after MHCI synthesis, but are rapidly lost with kinetics that resemble those of dislocation to the cytosol (Figure 3B). Such a US11-MHCI heavy chain complex may be a dislocation intermediate that forms within the ER before exposure to the cytosol. We hypothesize that the interactions mediated by the US11 TMD are required for bringing the MHCI heavy chain into contact with the machinery that physically removes the MHCI heavy chain from the ER (Figure 6). When the interactions formed by the US11 TMD are disrupted, MHCI heavy chains can no longer access this machinery. Consistent with this hypothesis, we observe stable association of MHCI heavy chains with US11 Q192L, whereas MHCI heavy chains associate only transiently with wild-type US11 (Figure 3B).
Figure 6.
Model for the US11 TMD mode of action. (1) After insertion into the ER, the lumenal domain of US11 binds to the MHCI heavy chain substrate, possibly in a complex with β2m (recognition). (2) Through interactions with a transmembrane helix of an as yet unidentified ER protein mediated by the US11 TMD, US11 targets the MHCI heavy chain to the dislocation machinery. The MHCI heavy chain is likely unfolded before movement across the ER membrane. (3) The MHCI heavy chain is translocated across the ER membrane, possibly through the Sec61 channel. This step requires ubiquitination and may involve the action of the p97/Npl4/Ufd1 complex. (4) Once released into the cytosol, the substrate is deglycosylated and degraded by the 26S proteasome.
We propose that US11 and its functional analog, US2, act as molecular links between MHCI heavy chains and the proteins that remove misfolded cellular proteins from the ER. The normal recognition and targeting machinery that acts on misfolded cellular substrates may thus be bypassed. Although both US2 and US11 bind to their MHCI substrate through their lumenal domains and trigger a similar series of biochemical events, the elements of the MHCI heavy chain that are recognized by US2 are different from US11. US11 preferentially recognizes incompletely folded MHCI heavy chains (J. Loureiro and B.L., unpublished observations) in contrast to US2, which recognizes a folded MHCI-β2m complex (Gewurz et al., 2001). Unlike US11, US2 is itself a substrate of the ER-to-cytosol dislocation pathway and US2 may accompany the MHCI heavy chain on its way out of the ER (Wiertz et al., 1996b). The differences in the details of US11- and US2-mediated dislocation indicate that these viral proteins may use distinct mechanisms to cause MHCI heavy chain degradation. The definition of US11 and US2 mutants that are defective in dislocation will aid in the identification of the cellular proteins involved in this remarkable reaction.
Acknowledgments
The authors thank members of the Ploegh lab for discussions and M. Lorenzo for Ub-bio. This work was supported by grants from the National Institutes of Health. B.N.L. is a Howard Hughes Medical Institute predoctoral fellow.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–03–0192. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-03-0192.
Abbreviations used: β2m, β2-microglobulin; EndoH, endoglycosidase H; ER, endoplasmic reticulum; MHCI, class I major histocompatibility complex; TMD, transmembrane domain; Ubbio, biotinylated ubiquitin.
References
- Ashman, J.B., and Miller, J. (1999). A role for the transmembrane domain in the trimerization of the MHC class II-associated invariant chain. J. Immunol. 163, 2704–2712. [PubMed] [Google Scholar]
- Bogyo, M., McMaster, J.S., Gaczynska, M., Tortorella, D., Goldberg, A.L., and Ploegh, H. (1997). Covalent modification of the active site threonine of proteasomal beta subunits and the Escherichia coli homolog HslV by a new class of inhibitors. Proc. Natl. Acad. Sci. USA 94, 6629–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., Cosson, P., Shah, N., and Klausner, R.D. (1991). Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 10, 2783–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral, C.M., Liu, Y., and Sifers, R.N. (2001). Dissecting glycoprotein quality control in the secretory pathway. Trends Biochem. Sci. 26, 619–624. [DOI] [PubMed] [Google Scholar]
- Call, M.E., Pyrdol, J., Wiedmann, M., and Wucherpfennig, K.W. (2002). The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 111, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerel, L., Meunier, J.C., Pillez, A., Wychowski, C., and Dubuisson, J. (1998). A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72, 2183–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerel, L., Wychowski, C., Minner, F., Penin, F., and Dubuisson, J. (2000). Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74, 3623–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu, S.N., Keren, T., Socolovsky, M., Nam, H., Henis, Y.I., and Lodish, H.F. (2001). Ligand-independent oligomerization of cell-surface erythropoietin receptor is mediated by the transmembrane domain. Proc. Natl. Acad. Sci. USA 98, 4379–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson, P., Lankford, S.P., Bonifacino, J.S., and Klausner, R.D. (1991). Membrane protein association by potential intramembrane charge pairs. Nature 351, 414–416. [DOI] [PubMed] [Google Scholar]
- Furman, M.H., Ploegh, H.L., and Tortorella, D. (2002). Membrane-specific, host-derived factors are required for US2- and US11-mediated degradation of major histocompatibility complex class I molecules. J. Biol. Chem. 277, 3258–3267. [DOI] [PubMed] [Google Scholar]
- Gewurz, B.E., Gaudet, R., Tortorella, D., Wang, E.W., Ploegh, H.L., and Wiley, D.C. (2001). Antigen presentation subverted: Structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl. Acad. Sci. USA 98, 6794–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich, D., and Rapoport, T.A. (1993). Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75, 615–630. [DOI] [PubMed] [Google Scholar]
- Gratkowski, H., Lear, J.D., and DeGrado, W.F. (2001). Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. USA 98, 880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich, S.U., Mothes, W., Brunner, J., and Rapoport, T.A. (2000). The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 102, 233–244. [DOI] [PubMed] [Google Scholar]
- Hirsch, C., Blom, D., and Ploegh, H.L. (2003). A role for N-glycanase in the cytosolic turnover of glycoproteins. EMBO J. 22, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstenbach, F., David, V., Watkins, S., and Brenner, M.B. (1992). Endoplasmic reticulum resident protein of 90 kilodaltons associates with the T- and B-cell antigen receptors and major histocompatibility complex antigens during their assembly. Proc. Natl. Acad. Sci. USA 89, 4734–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, E.A., Hammond, C., and Cresswell, P. (1997). Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl. Acad. Sci. USA 94, 1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppa, J.B., and Ploegh, H.L. (1997). The alpha chain of the T cell antigen receptor is degraded in the cytosol. Immunity 7, 113–122. [DOI] [PubMed] [Google Scholar]
- Jarosch, E., Taxis, C., Volkwein, C., Bordallo, J., Finley, D., Wolf, D.H., and Sommer, T. (2002). Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139. [DOI] [PubMed] [Google Scholar]
- Johnston, J.A., Ward, C.L., and Kopito, R.R. (1998). Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.R., Hanson, L.K., Sun, L., Slater, J.S., Stenberg, R.M., and Campbell, A.E. (1995). Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J. Virol. 69, 4830–4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert, M., Hassink, G., Barel, M., Hirsch, C., van der Wal, F.J., and Wiertz, E. (2001). Ubiquitination is essential for human cytomegalovirus US11-mediated dislocation of MHC class I molecules from the endoplasmic reticulum to the cytosol. Biochem. J. 358, 369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, O., Polack, G.W., Surti, T., Kegler-Ebo, D., Smith, S.O., and DiMaio, D. (1998). Role of glutamine 17 of the bovine papillomavirus E5 protein in platelet-derived growth factor beta receptor activation and cell transformation. J. Virol. 72, 8921–8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt-Marticorena, C., Williams, K.A., Deber, C.M., and Reithmeier, R.A. (1993). Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 229, 602–608. [DOI] [PubMed] [Google Scholar]
- MacKenzie, K.R., Prestegard, J.H., and Engelman, D.M. (1997). A transmembrane helix dimer: structure and implications. Science 276, 131–133. [DOI] [PubMed] [Google Scholar]
- Mitsui, A., and Sharp, P.A. (1999). Ubiquitination of RNA polymerase II large subunit signaled by phosphorylation of carboxyl-terminal domain. Proc. Natl. Acad. Sci. USA 96, 6054–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory, D.S., Neugeboren, B.A., and Mulligan, R.C. (1996). A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 93, 11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham, P., Barnstable, C.J., and Bodmer, W.F. (1979). Use of a monoclonal antibody (W6/32) in structural studies of HLA-A, B, C, antigens. J. Immunol. 123, 342–349. [PubMed] [Google Scholar]
- Ploegh, H.L. (1995). One-dimensional isoelectric focusing of proteins in slab gels. In: Current Protocols in Protein Science, Vol. 1, ed. J.E. Coligan, B.M. Dunn, H.L. Ploegh, D.W. Speicher, and P.T. Wingfield. New York: John Wiley & Sons, 10.12.11–10.12.18. [DOI] [PubMed] [Google Scholar]
- Popot, J.L., and Engelman, D.M. (2000). Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69, 881–922. [DOI] [PubMed] [Google Scholar]
- Rehm, A., Stern, P., Ploegh, H.L., and Tortorella, D. (2001). Signal peptide cleavage of a type I membrane protein, HCMV US11, is dependent on its membrane anchor. EMBO J. 20, 1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ, W.P., and Engelman, D.M. (2000). The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296, 911–919. [DOI] [PubMed] [Google Scholar]
- Shamu, C.E., Flierman, D., Ploegh, H.L., Rapoport, T.A., and Chau, V. (2001). Polyubiquitination is required for US11-dependent movement of MHC class I heavy chain from endoplasmic reticulum into cytosol. Mol. Biol. Cell 12, 2546–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu, C.E., Story, C.M., Rapoport, T.A., and Ploegh, H.L. (1999). The pathway of US11-dependent degradation of MHC class I heavy chains involves a ubiquitin-conjugated intermediate. J. Cell Biol. 147, 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam, N.J., Spits, H., and Ploegh, H.L. (1986). Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137, 2299–2306. [PubMed] [Google Scholar]
- Story, C.M., Furman, M.H., and Ploegh, H.L. (1999). The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc. Natl. Acad. Sci. USA 96, 8516–8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirabassi, R.S., and Ploegh, H.L. (2002). The human cytomegalovirus US8 glycoprotein binds to major histocompatibility complex class I products. J. Virol. 76, 6832–6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella, D., Gewurz, B.E., Furman, M.H., Schust, D.J., and Ploegh, H.L. (2000). Viral subversion of the immune system. Annu. Rev. Immunol. 18, 861–926. [DOI] [PubMed] [Google Scholar]
- Tortorella, D., Story, C.M., Huppa, J.B., Wiertz, E.J., Jones, T.R., Bacik, I., Bennink, J.R., Yewdell, J.W., and Ploegh, H.L. (1998). Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J. Cell Biol. 142, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, B., Ye, Y., and Rapoport, T.A. (2002). Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell. Biol. 3, 246–255. [DOI] [PubMed] [Google Scholar]
- van de Rijn, M., Geurts van Kessel, A.H., Kroezen, V., van Agthoven, A.J., Verstijnen, K., Terhorst, C., and Hilgers, J. (1983). Localization of a gene controlling the expression of the human transferrin receptor to the region q12 leads to qter of chromosome 3. Cytogenet. Cell Genet. 36, 525–531. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., Jones, T.R., Sun, L., Bogyo, M., Geuze, H.J., and Ploegh, H.L. (1996a). The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz, E.J., Tortorella, D., Bogyo, M., Yu, J., Mothes, W., Jones, T.R., Rapoport, T.A., and Ploegh, H.L. (1996b). Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Ye, Y., Meyer, H.H., and Rapoport, T.A. (2001). The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656. [DOI] [PubMed] [Google Scholar]
- Zhou, F.X., Merianos, H.J., Brunger, A.T., and Engelman, D.M. (2001). Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. USA 98, 2250–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]