Abstract
Rab7, a member of the Rab family small G proteins, has been shown to regulate intracellular vesicle traffic to late endosome/lysosome and lysosome biogenesis, but the exact roles of Rab7 are still undetermined. Accumulating evidence suggests that each Rab protein has multiple target proteins that function in the exocytic/endocytic pathway. We have isolated a new Rab7 target protein, Rabring7 (Rab7-interacting RING finger protein), using a CytoTrap system. It contains an H2 type RING finger motif at the C termini. Rabring7 shows no homology with RILP, which has been reported as another Rab7 target protein. GST pull-down and coimmunoprecipitation assays demonstrate that Rabring7 specifically binds the GTP-bound form of Rab7 at the N-terminal portion. Rabring7 is found mainly in the cytosol and is recruited efficiently to late endosomes/lysosomes by the GTP-bound form of Rab7 in BHK cells. Overexpression of Rabring7 not only affects epidermal growth factor degradation but also causes the perinuclear aggregation of lysosomes, in which the accumulation of the acidotropic probe LysoTracker is remarkably enhanced. These results suggest that Rabring7 plays crucial roles as a Rab7 target protein in vesicle traffic to late endosome/lysosome and lysosome biogenesis.
INTRODUCTION
The Rab family small G proteins appear to be a key regulator of intracellular vesicle traffic, including exocytosis and endocytosis (Novick and Zerial, 1997; Olkkonen and Stenmark, 1997; Martinez and Goud, 1998; Segev 2001; Pfeffer, 2001; Takai et al., 2001; Zerial and McBride, 2001). In mammalian cells, more than 60 members have been identified and each Rab protein localizes at the surface of a distinct intracellular compartment. Evidence is accumulating that Rab proteins regulate one (or more) specific steps of vesicle traffic by the recruitment of the tethering, docking and fusion factors and the actin- and microtubule-based motor proteins to facilitate vesicle traffic (Pfeffer, 1999, 2001; Segev, 2001; Takai et al., 2001; Zerial and McBride, 2001; Hammer and Wu, 2002). Rab proteins cycle between the GDP-bound inactive and GTP-bound active forms and between the cytosol and the membrane. This cyclical activation, inactivation, and translocation are regulated by at least three regulators, GDP dissociation inhibitor (GDI), GDP/GTP exchange protein (GEP), and GTPase-activating protein (GAP), and impose temporal and spatial regulation of vesicle traffic by Rab proteins. The GTP-bound form interacts with one or more downstream target proteins and the interaction between each Rab protein and its specific target proteins is required for each step of vesicle traffic. Therefore, it is important to identify the regulators and target proteins of Rab proteins to elucidate the mechanisms of vesicle traffic.
Twelve Rab proteins have been identified in the endocytic pathway of mammalian cells (Somsel Rodman and Wandlinrer-Ness, 2000). Some of them involved in the early steps of endocytosis have been studied extensively and the mechanisms of these steps of vesicle traffic are well understood. Rab5 is involved in the first step of internalization and subsequent fusion of vesicles with early endosomes. Rab5 also regulates the homotypic fusion of early endosomes. Rabaptin-5, EEA1, hVPS34/p150, and Rabenosyn-5 have been identified as Rab5 target proteins (Stenmark et al., 1995; Christoforidis et al., 1999a, 1999b; Callaghan et al., 1999). The characterization of these target proteins has revealed that Rab5 can regulate several steps through these multiple target proteins and that these target proteins act cooperatively to coordinate the various functions of Rab5 in early endocytic traffic.
In contrast, relatively little is known about Rab proteins involved in late endocytic traffic. Rab7 localizes to the late endocytic structures, late endosome/lysosome. A dominant negative mutant of Rab7 has been shown to strongly inhibit vesicle traffic from early to late endosomes, indicating that Rab7 is required for this step (Feng et al., 1995; Vitelli et al., 1997; Press et al., 1998). Previous data obtained with a constitutive active mutant of Rab7 suggest that Rab7 also regulates vesicle traffic from late endosome to lysosome (Meresse et al., 1995). Moreover, Rab7 has also been implicated in the lysosome biogenesis, by regulation of the heterotypic fusion between late endosomes and lysosomes and the homotypic fusion of lysosomes (Bucci et al., 2000). Thus, Rab7 is involved in several steps of late endocytic traffic, but the exact roles and the mode of actions of Rab7 in the late endocytic traffic are still unknown. Recently, a Rab7 target protein, RILP (Rab-interacting lysosomal protein) has been identified and characterized (Bucci et al., 2001; Cantalupo et al., 2001; Jordens et al., 2001). However, there seems to be multiple target proteins of Rab7 to execute their functions in several steps of the late endocytic pathway.
We identified a new Rab7 target protein, named Rabring7, by performing a CytoTrap screening. Rabring7 controls late endocytic traffic and lysosome biogenesis in a downstream of Rab7.
MATERIALS AND METHODS
Material and Chemicals
Anti-GFP and anti-6 × His monoclonal antibodies were purchased from Clontech (Palo Alto, CA). An anti-Xpress mAb was obtained from Invitrogen (Carlsbad, CA). The anti-HA mAb came from Roche (Basel, Switzerland) and anti-Sos mAb from Transduction Laboratories (Lexington, KY). An anti-Rabring7 antibody was generated in rabbits using glutathione S-transferase (GST)-tagged N-terminal fragment (1–145 amino acids [aa]) of Rabring7 (Rabring7N) as an antigen. 125I-labeled human epidermal cell growth factor (EGF) (>750 Ci/mmol, 100 μCi/ml) and [α-32P]dCTP (3000 Ci/mmol, 10 mCi/ml) were obtained from Amersham Bioscience (Piscataway, NJ) and Perkin Elmer, Boston, MA, respectively.
Construction of Plasmids
The cDNA encoding full open reading frames of the murine Rab proteins (Rab4, Rab7, and Rab8) were amplified from a murine immature B cell line (WEHI231) cDNA library by PCR using the following pairs of oligonucleotides with restriction enzyme sites (underlined) or stop codon (bold) designed on the basis of the mouse sequence in the databases: 5′-cgggatcccgatgtccgagacttacgattt-3′ (Rab4 forward) and 5′-tccccgcggctaacagccacactcct-3′ (Rab4 reverse; GenBank accession no. D86563); 5′-gtcgaccaccatgacctctaggaagaaagt-3′ (Rab7 forward) and 5′-gcggccgctcaacaactgcagctttctg-3′ (Rab7 reverse; GenBank accession no. NM_009005); 5′-gtcgaccaccatggcgaagacctacgatta-3′ (Rab8 forward) and 5′-gcggccgctcacaggagactgcaccgga-3′ (Rab8 reverse; GenBank accession no. XM_134267). The PCR products were purified from agarose gel and directly inserted into the pGEM-T Easy vector (Promega, Madison, WI). A site direct mutagenesis was carried out by a PCR-based method. A deletion mutant was also generated by a PCR-based method. The cDNA of Rab7 mutants used in this study have already been described elsewhere (Vitelli et al., 1997). The pSos/Rab7 plasmids were made by subcloning the Rab7wt, Q67L, T22N, N125I, wtΔC, Q67LΔC, T22NΔC, and N125IΔC cDNA as a SalI-NotI fragment into pSos (Stratagene, La Jolla, CA) from the pGEM/Rab7 plasmids, respectively. The pEGFP/Rab7 plasmids were made by subcloning the Rab7wt, Q67L, T22N, and N125I cDNA as a SalI-SacII fragment into pEGFP-C3 (Clontech) from the pSos/Rab7 plasmids, respectively. All expression plasmids were sequenced using an Applied Biosystems (Foster, CA) automated DNA sequencer 377.
The Rabring7 cDNA was amplified from the WEHI231 cDNA library by PCR using the following pairs of primers with restriction enzyme sites (underlined) or stop codon (bold) designed on the basis of the mouse sequence in the databases: 5′-ccggaattcaggcgaaacatggcggagg-3′ (Rabring7 forward) and 5′-ccgctcgagtcagaaagtccatcggtcatg-3′ (Rabring7 reverse; GenBank accession no. MN_026406). The truncated mutants, Rabring7N (1–145 aa) and Rabring7C (146–305 aa), were generated by PCR using the following pairs of primers with restriction enzyme sites (underlined) or stop codons (bold): Rabring7 forward and 5′-ccgctcgagtcatattccttcgatagctgg-3′ (Rabring7N reverse); 5′-ccggaattcatacaacagatctttgcagga-3′ (Rabring7C forward) and Rabring7 reverse. After digestion with EcoRI and XhoI, the PCR products were subcloned into pcDNA4/HisMaxC (Invitrogen) to obtain the pcDNA/Rabring7 plasmids. The Rabring7, Rabring7N, and Rabring7C were also inserted into pCIneoHA (Promega) to produce the pCIneoHA/Rabring7 plasmids.
Strain, Media, and Yeast Transformation
The yeast strain cdc25Hα has the following genotype: MATα ura3, lys2, leu2, trp1, his200, ade101, cdc25-2, GAL+ (Aronheim et al., 1997; Aronheim, 2001). A point mutation of the cdc25 gene in cdc25Hα prevents host growth at a restrictive temperature (37°C), but not at a permissive temperature (24°C). This strain was grown in YPAD medium or Burkholder's minimum media (BMM) with appropriate supplements at 24°C and was transformed by the standard lithium acetate method (Gietz et al., 1992). For selection of transformants, glucose base BMM lacking leucine and uracil (BMM/glucose–leu, ura) was used. For induction of protein expression from pMry (Stratagene), galactose base BMM lacking leucine and uracil (BMM/galactose–leu, ura) was used.
CytoTrap Screening
The yeast strain cdc25Hα was used in a CytoTrap system (Stratagene). To construct a WEHI231 cDNA library, oligo-dT primed cDNA fragments derived from WEHI231 cells were cloned into pMry. The screening was performed according to the manufacturer's recommendations with slight modification. Briefly, the yeast containing pSos/Rab7Q67LΔC was transformed with a WEHI231 cDNA library. The transformants were grown on a BMM/galactose–leu, ura plate at 24°C for 3 d and then grown at 37°C. The colonies grown at 37°C were picked and tested for growth on BMM/glucose–leu, ura and BMM/galactose–leu, ura plates at 37°C. The library plasmids were isolated from the clones that exhibited galactose-dependent growth at 37°C and were retransformed into the cdc25Hα cells either with pSos/Rab7Q67LΔC or with pSos/Collagenase IV as a negative control. Only plasmids that suppressed the cdc25Hα phenotype in the presence of pSos/Rab7Q67LΔC were sequenced. DNA sequencing was performed on an Applied Biosystems automated DNA sequencer 377.
Cell Culture and Transfection
BHK, COS1, MDCK, and HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal calf serum (FCS), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). WEHI231 cells were maintained in RPMI-1640 medium containing 10% FCS, 5.5 × 10 –5 M 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin. For experiments, the cells were seeded at a density of 106 cells/dish in 10-cm culture dishes and cultured for 1 d. Subsequently, the cells were transfected by Fu-GENE6 (Roche) according to the manufacturer's recommendations.
Northern Blot Hybridization
Twenty micrograms of total RNA prepared by the acid guanidine phenol chloroform method from various cell lines was separated on a 1% agarose gel containing formaldehyde, transferred to Hybond-N+(Amersham Bioscience) in 20× SSC, and immobilized by UV-cross linking. The blot was hybridized in a solution containing 5× SSC, 5× Denhardt's solution, 0.1% SDS, 50% deionized formamide, and 100 μg/ml salmon sperm DNA at 42°C, with the DNA probe corresponding to a part of coding Rabring7 (51–139 aa). The probe was prepared by PCR and labeled by random priming with [α-32P]dCTP. After hybridization, the blot was washed with 2× SSC and 0.1% SDS at room temperature and with 0.1× SSC and 0.1% SDS at 45°C. The multitissue mouse Northern blots (Clontech, cat. no.7763-1 and 7762-1) were also hybridized under the same conditions as described above. The signal was detected by a BAS2000 imaging analyzer (Fuji Film, Tokyo, Japan).
GST Pull-Down Assay
For the GST pull-down assay, GST-fusion proteins were expressed in the BLR bacteria strain after being induced by isopropyl β-d-1-thiogalactopyranoside using a pGEX vector (Amersham Bioscience) and were then purified on glutathione beads (Amersham Bioscience). Rab proteins were expressed as N-terminal EGFP-tagged Rab proteins (EGFP-Rabs) by transfection of pEGFP/Rabs in COS1 cells. The COS1 cells expressing EGFP-Rabs were lysed in 1 ml of a lysis buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 0.1% NP-40, 5 mM EDTA, 10 mM MgCl2, and 1 mM DTT) for 10 min at 4°C. The cell lysates were centrifuged and the supernatants were used as the cell extracts. The cell extracts were incubated with GST or GST-fusion proteins immobilized on glutathione beads. The beads were washed extensively with the same buffer. Samples were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking, the membrane was treated with the anti-GFP antibody and immunoreactive proteins were detected according to the enhanced chemiluminescence protocol (Amersham Bioscience) using the horseradish peroxidase-linked secondary antibody (Bio-Rad, Hercules, CA).
Coimmunoprecipitation Assay
The COS1 cells were transiently transfected with the indicated combinations of expression plasmids. Then the cells were scraped from the dishes in PBS and washed three times with the same buffer. The cells were lysed in 1 ml of a lysis buffer for 10 min at 4°C. The cell lysates were centrifuged and the supernatants were used as the cell extracts. The cell extracts were incubated with the anti-HA antibody or the control IgG for 2 h at 4°C, followed by treatment with protein A sepharose FF beads (Amersham Bioscience). The beads were washed extensively with lysis buffer. Samples were subjected to SDS-PAGE and were transferred to PVDF membranes. After blocking, the membranes were treated with the anti-HA antibody and the immunoreactive proteins were detected according to the enhanced chemiluminescence protocol (Amersham Bioscience) using the horseradish peroxidase-linked secondary antibody (BioRad). After stripping, the PVDF membrane was reprobed by the anti-GFP antibody and the immunoreactive proteins were detected as described above.
Fluorescence Microscopy
BHK cells cultured on cover glasses were transfected with the various plasmids. After 48 h, the cells were fixed with 3.7% formaldehyde in PBS for 15 min at room temperature. The cells were washed three times with PBS and were treated with 0.1% Triton X-100 in PBS for 5 min at room temperature. After blocking, the cells were incubated with the first antibodies for 1 h at room temperature and were washed three times with PBS, followed by incubation with the Alexa488- or TexasRed–conjugated secondary antibody for 30 min at room temperature. The cells were washed three times with PBS and were mounted on slide glasses. For labeling of lysosome, living cells were incubated with 100 nM LysoTracker Red DND-99 (Molecular Probes, Eugene, OR) for 30 min at 37°C. In the transferrin uptake assay, living cells were incubated with 50 μg/ml TexasRed-transferrin (Molecular Probes) for 30 min at 37°C (Meresse et al., 1995; Yoshimori et al., 2000).
Fluorescence was visualized through a Bio-Rad Radiance 2000 confocal laser scanning microscope. Z axial sections were collected at a 0.85-μm step and the projected images were represented.
EGF Degradation Assay
The EGF degradation assay was performed as previously described (Cantalupo et al., 2001). Briefly, HeLa cells cultured in six-well plates were transfected with various expression plasmids. After 24 h, the cells were incubated overnight with serum-starved medium. The cells were incubated further with 125I-labeled EGF (1 μCi/ml) on ice for 1 h. Unbound ligand was washed off and the cells were allowed to internalize and to degrade the EGF at 37°C. After 2 h, the extracellular media were collected and treated with trichloroacetic acid (TCA). The extracellular TCA-soluble 125I-labeled EGF was quantitated as a degraded EGF fraction by gamma counter.
RESULTS
Isolation of cDNA Clones Encoding Rabring7
For identification of Rab7 target proteins, we performed a CytoTrap screening from a murine immature B cell (WEHI231) cDNA library. A CytoTrap system is studied in a temperature-sensitive yeast strain, cdc25Hα, where the yeast Ras GEP, cdc25p, is inactive at the restrictive temperature (37°C). Human Ras GEP, Sos-tagged GTPase-deficient mutant of Rab7, Rab7Q67L, was used as a bait. The final three amino acids of Rab7 were deleted in this mutant to prevent the C-terminal posttranslational prenylation, because such modification caused a high background in this system (our unpublished results). Expression of Sos-Rab7Q67LΔC was confirmed by Western blot with the anti-Sos antibody (our unpublished results). The yeast cells expressing Sos-Rab7Q67LΔC were subsequently transformed with a WEHI231 cDNA library. The prey was expressed as a fusion protein with a myristylation sequence that anchors the fusion protein onto the plasma membrane. If the bait protein physically interacted with the prey protein, Sos was recruited to the membrane, thereby activating the yeast RAS-signaling pathway and allowing cdc25Hα to grow at 37°C.
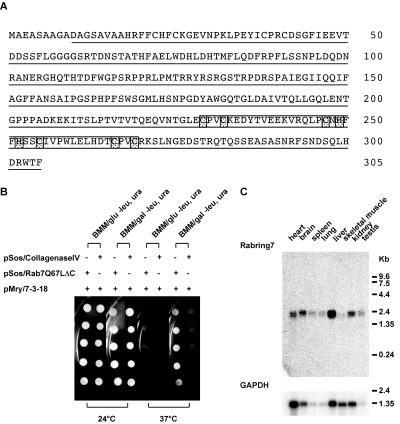
Approximately 2.0 × 106 transformants were screened and 228 positive clones were identified. Among these positive clones, 45 clones showed galactose-dependent growth at 37°C. Finally, this screen resulted in the isolation of 34 clones that specifically interacted with Rab7Q67LΔC but not with Collagenase IV. The library plasmids rescued from the clones were sequencing and 4 clones including clone 7-3-18 were identified as zinc finger protein 364 (GenBank accession no. MN_026406), a 305-aa open reading frame of unknown function. As shown in Figure 1A, clone 7-3-18 encoded a portion of the protein, lacking N-terminal 9 aa. Indeed, clone 7-3-18 interacted with Rab7Q67LΔC but not with Collagenase IV in the CytoTrap system (Figure 1B). This clone also interacted with the wild-type Rab7 lacking C-terminal 3 aa, Rab7wtΔC, but not with two mutants lacking C-terminal 3 aa, the dominant negative mutant locked in the GDP-bound form, Rab7T22NΔC, or the nucleotide-empty mutant, Rab7N125IΔC (our unpublished results). These results suggest that zinc finger protein 364 is a Rab7 target protein. Amino acid sequence analysis using PROSITE and PFAM revealed that the zinc finger protein 364 contains a C-terminal H2 type RING finger motif (229–270 aa, Figure 1A). Thus, we refer to zinc finger protein 364 as Rabring7, Rab7-interacting RING finger protein. There is no significant sequence similarity between Rabring7 and RILP that was reported as another Rab7 target protein (Bucci et al., 2001; Cantalupo et al., 2001; Jordens et al., 2001). We could not obtain any clones that encoded RILP in this screening.
Figure 1.
Identification of a novel Rab7 target, Rabring7. (A) Predicted amino acid sequence of Rabring7. The amino acid sequence deduced from the nucleotide sequence of the full-length Rabring7 cDNA is shown in single letter code. The underline shows the clone 7-3-18 encoded region. The box indicates the RING finger motif and the shaded boxes indicate conserved amino acids in the RING finger motif. (B) Specific interaction between Sos-Rab7Q67LΔC and clone 7-3-18 in the CytoTrap system. Yeast strain cdc25Hα were transformed with the indicated plasmids and the transformants were assayed for growth as described in MATERIALS AND METHODS. (C) Northern blot analysis. Mouse multiple tissue blots (Clontech) were hybridized with Rabring7 cDNA probe (top) or GAPDH cDNA probe (bottom).
The expression pattern of the Rabring7 transcript was studied by hybridization of cDNA to Northern blot (Figure 1C). The Rabring7 gene was expressed ubiquitously and indicated a single transcript, ∼2.3 kb, in all the mouse tissues examined. Rabring7 expression was also observed in various cell lines including WEHI231, MDCK, and BHK cells (our unpublished results).
Rabring7 Specifically Interacts with the GTP-bound Form of Rab7
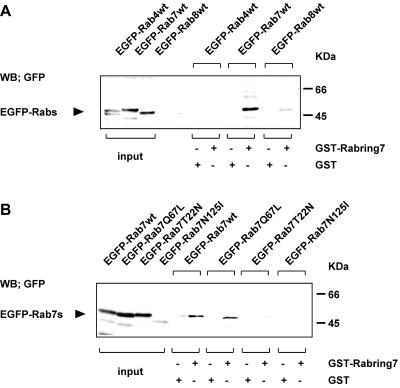
To confirm the interaction between Rab7 and Rabring7 observed in the CytoTrap system, we performed the GST pull-down assay. The cell extracts from the COS1 cells expressing EGFP-tagged Rab7wt, Rab4wt, or Rab8wt were incubated with GST-Rabring7 or GST alone immobilized on glutathione-beads. EGFP-tagged proteins bound to the beads were detected by Western blot with the anti-GFP antibody. GST-Rabring7 interacted with EGFP-Rab7wt, but not with EGFP-Rab4wt (Figure 2A). Very weak interaction was observed with EGFP-Rab8wt. GST alone did not interact with any EGFP-Rabs.
Figure 2.
GST pull-down assay. (A) Rabring7 was expressed as a GST-fusion protein and its binding to Rab4wt, Rab7wt, or Rab8wt was examined in a GST pull-down assay. The cell lysates from the EGFP-tagged Rab4wt-, Rab7wt-, or Rab8wt-expressing COS1 cells were incubated with GST-Rabring7 or GST alone immobilized on glutathione-beads. EGFP-Rabs bound to the beads were detected by Western blot with the anti-GFP antibody. The expression levels of EGFP-Rabs in the cell extracts were also monitored by Western blot with the anti-GFP antibody. (B) Rabring7 was expressed as a GST fusion protein and its binding to Rab7wt, Rab7Q67L, Rab7T22N, or Rab7N125I were examined by the same method described above.
Next, we examined the guanine nucleotide specificity of interaction of Rab7 with Rabring7. The cell extracts from the COS1 cells expressing EGFP-tagged Rab7wt or Rab7 mutants (Q67L, T22N, or N125I) were incubated with GST-Rabring7 or GST alone and the proteins bound to GST-fusion proteins were analyzed by the same method described above. As shown in Figure 2B, GST-Rabring7 interacted with EGFP-Rab7wt and EGFP-Rab7Q67L. In contrast, there was little interaction of GST-Rabring7 with the two mutants, EGFP-Rab7T22N and EGFP-Rab7N125I.
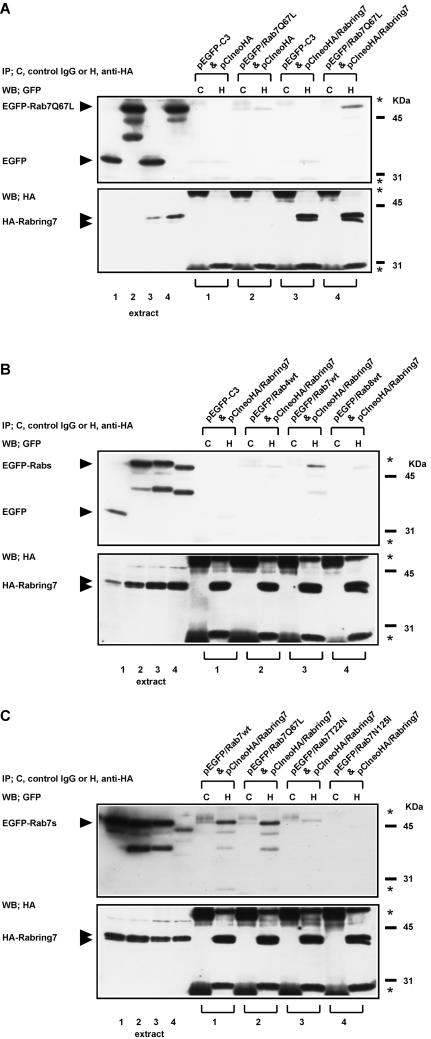
The interaction of Rab7 with Rabring7 was also analyzed by coimmunoprecipitation assay. EGFP-tagged Rab7Q67L was transiently expressed with or without HA-tagged Rabring7 (HA-Rabring7) in COS1 cells. Anti-HA antibody specifically recognized two bands in the cells transfected with the HA-Rabring7 expression plasmid, but not in the cells transfected with the control plasmid (Figure 3A, bottom panel). The sizes of both bands (39 and 41 kDa) were similar to that estimated from the amino acid sequence. The reason why the two bands appeared in the transfected cell extracts is unclear. The cell extracts were immunoprecipitated with the anti-HA antibody or the control mouse IgG, and coprecipitated EGFP-tagged Rab7Q67L was detected as the band at 51 kDa by Western blot with the anti-GFP antibody (Figure 3A, top panel). EGFP-tagged Rab7Q67L was coimmunoprecipitated with HA-Rabring7 in the cells coexpressing EGFP-tagged Rab7Q67L and HA-Rabring7. This result indicated that Rabring7 also interacted with the GTP-bound form of Rab7 in COS1 cells. The Rab and guanine nucleotide specificities were also observed in the coimmunoprecipitation assays (Figure 3, B and C). These indicate that Rabring7 binds the GTP-bound form of Rab7 more preferentially than the GDP-bound form, which supports the idea that the Rabring7 is a Rab7 target protein.
Figure 3.
Coimmunoprecipitation assay. (A) COS1 cells were transiently transfected with the indicated combinations of expression plasmids (1–4). The cell extracts (1–4) were prepared and immunoprecipitated with the anti-HA antibody or the control mouse IgG. Coprecipitated EGFP-Rab7Q67L was detected by Western blot analysis with the anti-GFP antibody. The expression levels of HA-Rabring7 and EGFP-Rab7Q67L in the cell extracts were also determined by Western blot with the anti-HA antibody (bottom) or the anti-GFP antibody (top), respectively. The positions of HA-Rabring7 and EGFP-Rab7Q67L are indicated. (B) COS1 cells were transiently transfected with expression plasmids encoding HA-tagged Rabring7 and the EGFP alone (1), EGFP-Rab4wt (2), EGFP-Rab7wt (3), or EGFP-Rab8wt (4). The cell extracts (1–4) were prepared and a coimmunoprecipitation assay was performed as described above. (C) COS1 cells were transiently transfected with expression plasmids encoding HA-tagged Rabring7 and EGFP-Rab7wt (1), EGFP-Rab7Q67L (2), EGFP-Rab7T22N (3), and EGFP-Rab7N125I (4). The cell extracts (1–4) were prepared and coimmunoprecipitation assay was performed as described above. The asterisks indicate the light and heavy chains of each antibody.
The GTP-bound Form of Rab7 Recruits Rabring7 to Late Endosome/lysosome
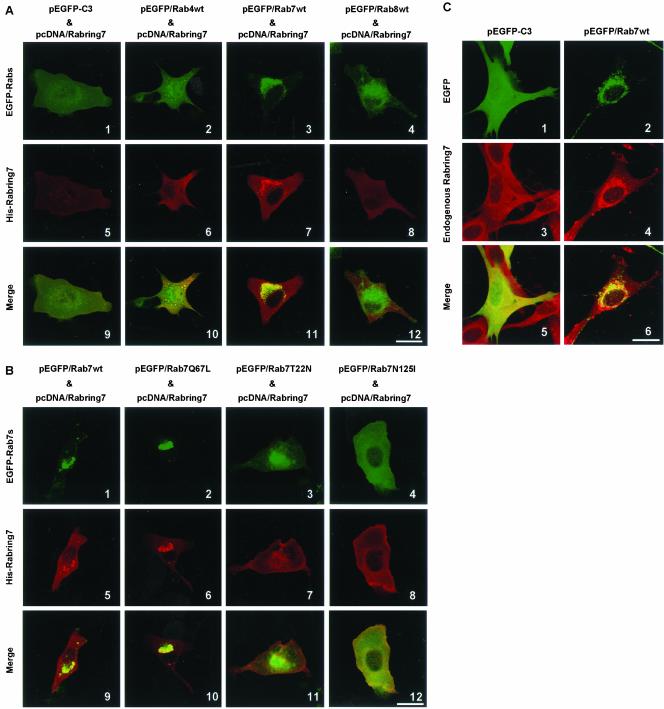
We studied subcellular localization of Rabring7 further. When BHK cells were transiently transfected with N-terminal 6× His- and Xpress-tagged full-length Rabring7 (His-Rabring7) expression plasmid, His-Rabring7 was diffusely observed in the cytosol (Figure 4A-5). EGFP-Rab7wt was mostly concentrated in the perinuclear region corresponding to late endosome/lysosome, whereas EGFP-Rab7T22N and N125I were distributed throughout the cytoplasm (our unpublished results, Bucci et al., 2000). Coexpression of EGFP-Rab7wt with His-Rabring7 caused changes in the distribution of His-Rabring7 and His-Rabring7 was accumulated in the perinuclear region where Rab7wt and Rabring7 were colocalized (Figure 4A-7). EGFP-Rab4wt and EGFP-Rab8wt did not affect the subcellular distribution of Rabring7 (Figure 4A-6 and -8). This effect of Rab7 on Rabring7 localization correlated well with the binding property of Rab7 to Rabring7 and was limited to wild-type and GTPase-deficient mutant (Figure 4B). We also confirmed that endogenous Rabring7 was distributed in the cytosol in BHK cells (Figure 4C-3). Exogenous expression of EGFP-Rab7wt also recruited endogenous Rabring7 to the perinuclear region (Figure 4C-4). These results indicate that Rabring7 is mainly cytosolic and that the GTP-bound form of Rab7 recruits Rabring7 from the cytosol onto late endosomes/lysosomes.
Figure 4.
Subcellular distribution of Rabring7. (A) BHK cells were transfected with pcDNA/Rabring7 and pEGFP-C3 or pEGFP/Rab4wt, pEGFP/Rab7wt, pEGFP/Rab8wt. After 48 h, the cells were fixed and examined by GFP fluorescence for EGFP-Rabs and by immunofluorescence labeling for His-Rabring7. Top: fluorescence images of EGFP; middle: fluorescence images of His-Rabring7; bottom: merged images. Bar, 30 μm (B) BHK cells were transfected with pcDNA/Rabring7 and pEGFP/Rab7wt or pEGFP/Rab7Q67L, pEGFP/Rab7T22N, and pEGFP/Rab7N125I. After 48 h, the cells were fixed and examined by GFP fluorescence for EGFP-Rab7s and by immunofluorescence labeling for His-Rabring7. Top: fluorescence images of EGFP; middle: fluorescence images of His-Rabring7; bottom: merged images. Bar, 30 μm (C) BHK cells were transfected with pEGFP-C3 or pEGFP/Rab7wt. After 48 h, the cells were fixed and examined by GFP fluorescence for EGFP and EGFP-Rab7 and by immunofluorescence labeling for endogenous Rabring7. Top: fluorescence images of EGFP; middle: fluorescence images of endogenous Rabring7; bottom: merged images. Bar, 30 μm.
Mapping of the Rab7-Binding Region in Rabring7
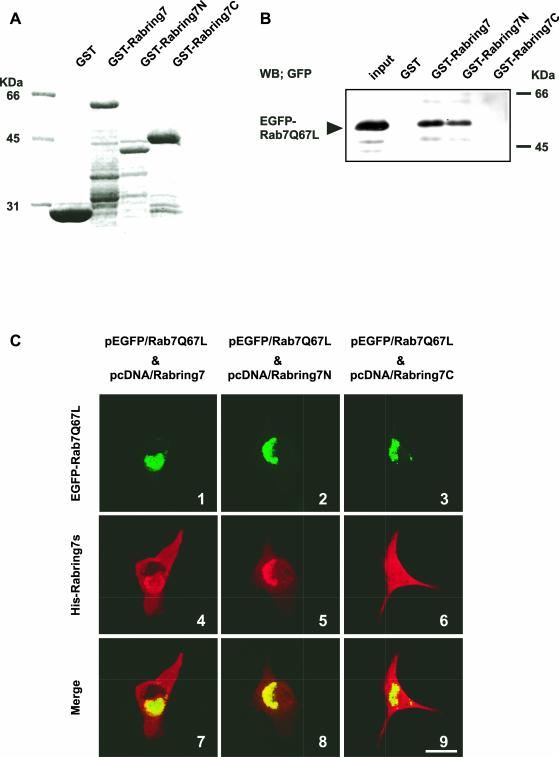
To determine the Rab7-binding region of Rabring7, GST-tagged full-length Rabring7 (GST-Rabring7) and two truncated mutants of Rabring7, GST-Rabring7N (1–145 aa) and GST-Rabring7C (146–305 aa), were used in a GST pull-down assay (Figure 5A). GST-Rabring7 and GST-Rabring7N bound Rab7Q67L, but GST-Rabring7C did not (Figure 5B). GST-7-3-18 also bound Rab7Q67L (our unpublished results). These results indicated that the N-terminal region of Rabring7 (10–145 aa) was needed for interaction with Rab7. In agreement with the in vitro binding data, Rab7Q67L also recruited Rabring7N in the perinuclear region but did not recruit Rabring7C in BHK cells (Figure 5C). However, full-length Rabring7 and both truncated mutants of Rabring7 did not affect the distribution of exogenously expressed Rab7Q67L.
Figure 5.
Mapping of the Rab7-binding region in Rabring7. (A) Expression levels of GST-fusion proteins used in the pull-down assay were checked by SDS-PAGE. A Coomassie brilliant blue stained image is represented. (B) The COS1 cells were transfected with pEGFP/Rab7Q67L. The cell extracts were prepared and incubated with GST alone, GST-Rabring7, Rabring7N, or Rabring7C immobilized on glutathione-beads. EGFP-Rab7Q67L bound to GST-fusion proteins was detected by Western blot with the anti-GFP antibody. The expression levels of EGFP-Rab7Q67L in the cell extracts were also monitored by Western blot with the anti-GFP antibody. (C) BHK cells were transfected with pEGFP/Rab7Q67L and pcDNA/Rabring7, pcDNA/Rabring7N, or pcDNA/Rabring7C. After 48 h, the cells were fixed and examined by GFP fluorescence for EGFP-Rab7Q67L and by immunofluorescence labeling for His-Rabring7s. Top: fluorescence images of EGFP; middle: fluorescence images of His-Rabring7s; bottom: merged images. Bar, 30 μm.
Effect of Rabring7 on Subcellular Distribution and Acidity of Lysosome
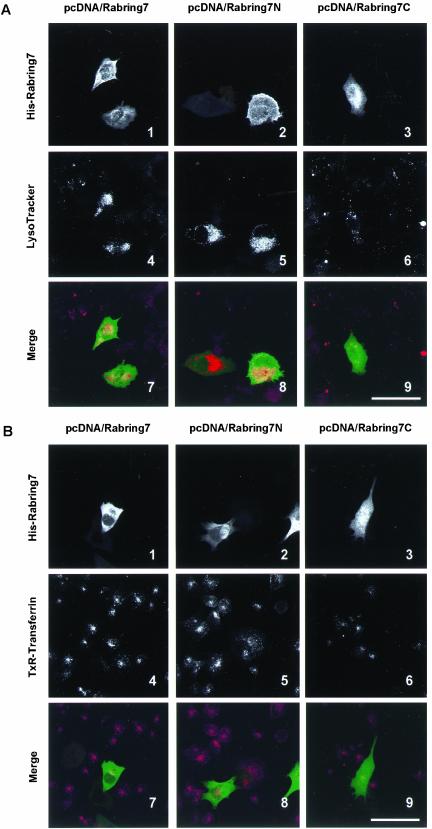
Next, we examined the effect of Rabring7 on lysosomal distribution in BHK cells. In the untransfected cells, Lyso-Tracker-accumulating vesicles were dispersed throughout the cytoplasm (Figure 6A). In contrast, LysoTracker-accumulating vesicles were aggregated in the perinuclear region of the Rabring7- and Rabring7N-expressing cells (Figure 6, A-4 and -5). In addition, these vesicles showed enhanced acidity, as shown by the very strong fluorescent signal relative to the untransfected cells. These effects were not observed in the Rabring7C-expressing cells (Figure 6A-6). On the other hand, Rabring7 did not change the distribution of internalized transferrin in BHK cells (Figure 6B), suggesting that Rabring7 did not affect the distribution of early/recycling endosomes and the early endocytic traffic.
Figure 6.
Effect of Rabring7 on subcellular distribution and acidity of lysosome. BHK cells were transfected with pcDNA/Rabring7, pcDNA/Rabring7N, or pcDNA/Rabring7C. After 48 h, living cells were incubated with 100 nM LysoTracker Red DND-99 (A) or with 50 μg/ml Alexa488-transferrin (B) for 30 min at 37°C. Then, the cells were fixed and indirect immunofluorescent staining was carried out. Top: fluorescence images of His-Rabring7; middle: fluorescence images of LysoTracker Red (A) or TexasRed-transferrin (B); bottom: merged images. Bar, 60 μm.
Effect of Nocodazole on Subcellular Distribution of Rab7 and Rabring7
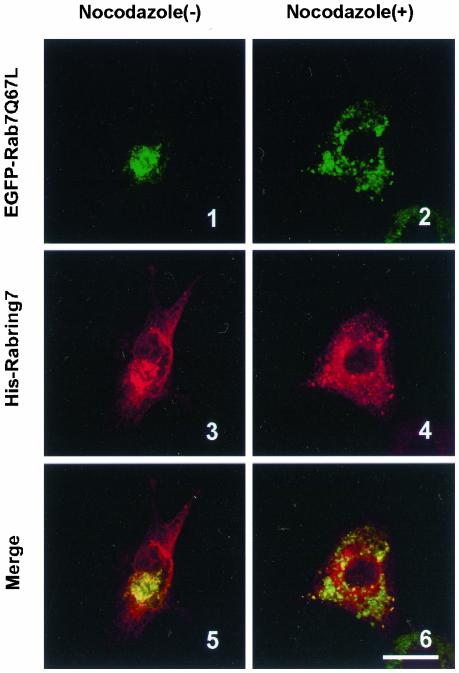
Lysosomal distribution and the late endocytic traffic have been shown to depend on microtubules (Matteoni and Kreis, 1987; Gruenberg et al. 1989; Caplan et al., 2001; Jordens et al., 2001). Therefore, we examined the effect of nocodazole, a microtubule-disrupting agent, on distribution of Rab7 and Rabring7 in BHK cells. When EGFP-Rab7Q67L was coexpressed with His-Rabring7 in the absence of nocodazole, both proteins were accumulated and colocalized in the perinuclear region (Figure 7-1, -3, and -5). Treatment with nocodazole caused the dispersion of EGFP-Rab7Q67L and His-Rabring7 to the cell periphery, but both proteins were colocalized on small clusters throughout the cytoplasm (Figure 7–2, -4, and -6). These results suggest that recruitment of Rabring7 by the GTP-bound form of Rab7 precedes lysosomal clustering through microtubules to the perninulear region.
Figure 7.
Effect of nocodazole on subcellular distribution of Rab7 and Rabring7. BHK cells were transfected with pEGFP/Rab7Q67L and pcDNA/Rabring7. After 36 h, the cells were treated with or without 0.5 μM nocodazole for 12 h at 37°C. Then, the cells were fixed and examined by GFP fluorescence for EGFP-Rab7Q67L and by immunofluorescence labeling for His-Rabring7. Top: fluorescence images of EGFP-Rab7Q67L; middle: fluorescence images of His-Rabring7; bottom: merged images. Bar, 30 μm.
Effect of Rabring7 on EGF Degradation
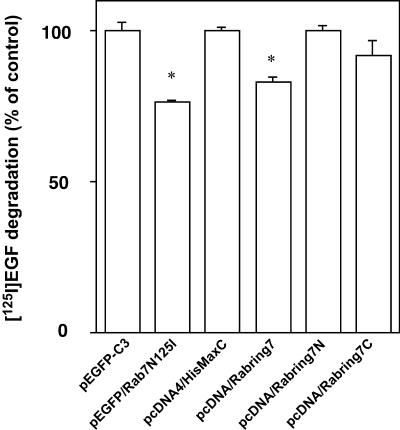
Furthermore, because Rab7 affects late endocytic traffic, we investigated whether Rabring7 plays a role in the degradative pathway. Rab7N125I inhibited EGF degradation, as previously reported (Figure 8; Feng et al., 1995; Bucci et al., 2000). Overexpression of Rabring7 significantly inhibited EGF degradation (Figure 8), although it did not affect the uptake nor recycling of EGF (our unpublished results). Because the untransfected cells, which accounted for ∼50% of total cells in this experiment, showed normal EGF degradation, its inhibitory effect seems to be modest. Rabring7N and Rabring7C did not affect EGF degradation (Figure 8). Although the effect of Rabring7N on EGF degradation was inconsistent with that on lysosomal clustering and acidification, it may have been due to the impaired transfection efficiency of pcDNA/Rabring7N compared with that of pcDNA/Rabring7 under the experimental conditions (our unpublished results).
Figure 8.
Effect of Rabring7 on EGF degradation. HeLa cells were transfected with the indicated expression plasmids. After 24 h, the serum was starved overnight and the cells were incubated with EGF for 1 h on ice. EGF degradation was measured as described in MATERIALS AND METHODS. The results are presented as a percentage of the cells transfected with the empty plasmid. The data represent three to six independent experiments with SEM. Statistical significance, *p < 0.05; Student's t test.
DISCUSSION
Here we have identified a novel protein that interacts with the GTP-bound form of Rab7 and named it Rabring7. We concluded that Rabring7 is a Rab7 target protein on the basis of the following observations: 1) it binds the GTP-bound form of Rab7 more preferentially than the GDP-bound form in the in vitro and in vivo assay systems, 2) it is recruited by the GTP-bound form of Rab7 onto late endosome/lysosome in BHK cells, and 3) Rabring7 as well as Rab7 regulates EGF degradation in HeLa cells.
Most of the Rab target proteins reported to date are recruited to particular membrane compartments by the GTP-bound form of Rab proteins, except that Rabpilin-3A, a Rab3A target protein, is associated with synaptic vesicles in a manner independent of Rab3A (Shirataki et al., 1994; McKiernan et al., 1996). A current model of the mode of actions of Rab proteins is that Rab proteins recruit the tethering, docking, and fusion factors and the actin- and microtubule-based motor proteins, most of which are their target proteins themselves or are proteins that interact with their target proteins, resulting in the assembly of a protein complex that facilitates vesicle traffic (Pfeffer, 1999, 2001; Segev, 2001; Takai et al., 2001; Zerial and McBride, 2001; Hammer and Wu, 2002). Our data are consistent with this model.
Rabring7 contains an H2 type RING finger motif at the C termini that is thought to mediate protein/protein interactions. Many RING finger proteins have been identified. Among them, yeast Vps11p, Vps18p, and Vps41p are core components of the 38S complex, called the HOPS complex (Wickner, 2002). It has been reported that the yeast counterpart of Rab7, Ypt7p, interacts with the HOPS complex (Price et al., 2000; Seals et al., 2000; Ungermann et al., 2000). The HOPS complex is also called the class C Vps complex and consists of Vps11p, Vps16p, Vps18p, Vps33p, Vps39p, and Vps41p. The functional roles of the HOPS complex are to facilitate the docking and fusion of late endosome-vacuoles (lysosome-like structure in yeast) and vacuole-vacuoles.
Recently, mammalian counterparts of HOPS complex members have been identified (Caplan et al., 2001; Huizing et al., 2001; Kim et al., 2001; McVey Ward et al., 2001). By analogy with the yeast HOPS complex, the mammalian HOPS complex is believed to regulate the heterotypic fusion between late endosomes and lysosomes and/or the homotypic fusion of lysosomes for the biogenesis and functions of lysosomes. We have shown here that exogenous expression of Rabring7 causes the perinuclear aggregation of lysosomes detected by the accumulation of the acidotropic probe LysoTracker, in the perinuclear region. Similar changes have been reported in the cells expressing the wild-type Rab7 or a constitutive active mutant of Rab7, Rab7Q67L (Bucci et al., 2001). Only a small portion of exogenously expressed Rabring7, which is recruited onto late endosomes/lysosomes by endogenous Rab7, may be sufficient to cause the lysosomal clustering, although most of exogenously expressed Rabring7 is cytosolic. Exogenous expression of mammalian Vps39 has been reported to cause lysosomal clustering in the perinuclear region, just like Rabring7 (Caplan et al., 2001). These observations suggest a possible link between Rabring7 and the HOPS complex. Thus, Rab7 may modulate the actions of the HOPS complex through Rabring7 in the biogenesis and function of lysosome.
Previously, RILP has been reported as another target protein of Rab7 (Bucci et al., 2001; Cantalupo et al., 2001; Jordens et al., 2001). No homology has been found between RILP and Rabring7, but both target proteins have similar properties: both are recruited to late endosome/lysosome by the GTP-bound form of Rab7, exogenous expression of these proteins causes perinuclear aggregation of late endosomes/lysosomes, and they affect EGF degradation. Endogenous RILP is mainly cytosolic and a part of RILP localizes to the late endosome/lysosome. We have shown here that most of endogenous Rabring7 is also cytosolic, although we could not determine whether a part of endogenous Rabring7 localizes to the late endosome/lysosome. RILP has been reported to recruit a minus-end directed dynein-dynactin motor to the late endosome/lysosome and consequently to stimulate the transport of these compartments toward the perinuclear region along microtubules (Jordens et al., 2001).
We have shown here that treatment of a microtubule-disrupting agent induces the dispersion of Rab7/Rabring7-containing late endosome/lysosome. It is likely that a part of endogenous Rabring7 may localize to the late endosome/lysosome and work together with RILP in microtubule-dependent lysosomal clustering in a downstream of Rab7. The unique feature of Rabring7 is that exogenous expression of Rabring7 shows increased acidity in the lysosome, as shown by the very strong fluorescent signal obtained with LysoTracker. It seems that RILP does not affect lysosomal acidity (Jordens et al., 2001). Previously, it has been reported that a dominant negative mutant of Rab7, Rab7T22N, decreases lysosomal acidity (Bucci et al., 2000). Based on these observations, Rabring7 might play a specific role in the Rab7-mediated lysosomal acidification. The low lysosomal pH is maintained by vacuolar proton ATPase (Marquez-Sterling et al., 1991). Rabring7 may affect the delivery and accumulation of vacuolar proton ATPase within lysosome.
In conclusion, Rabring7 is a novel Rab7 target protein that regulates vesicle traffic to late endosome/lysosome, lysosome biogenesis, and lysosomal acidification. Further studies are necessary to clarify its exact functions in late endocytic traffic.
Acknowledgments
We thank Dr. N. Nishimura in our laboratory for stimulating discussions. We also thank Dr. S. Irie (RIKEN) for giving us advice on the generation of pMry WEHI-231 cDNA library. This work was supported by grants from the Grants-in-aid for Scientific Research (2001, 2002) and for Cancer Research (2002) from the Ministry of Education, Science, Culture, Sports, Science, and Technology of Japan; by grants from the Yamanouchi Foundation Research on Metabolic Disease (2000, 2001, 2002); the Naito Foundation (2000); and the NOVARTIS Foundation (Japan) for the Promotion of Science (2000).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0495. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-08-0495.
References
- Aronheim, A., Zandi, E., Hennemann, H., Elledge, S.J., and Karin, M. (1997). Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17, 3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim, A. (2001). Ras signaling pathway for analysis of proteinprotein interactions. Methods Enzymol. 332, 260–270. [DOI] [PubMed] [Google Scholar]
- Bucci, C., Thomsen, P., Nicoziani, P., McCarthy, J., and van Deurs, B. (2000). Rab 7, a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci, C., De Gregorio, L., and Bruni, C.B. (2001). Expression analysis and chromosomal assignment of PRA1 and RILP genes. Biochem. Biophys. Res. Commun. 286, 815–819. [DOI] [PubMed] [Google Scholar]
- Callaghan, J., Nixon, S., Bucci, C., Toh, B.H., and Stenmark, H. (1999). Direct interaction of EEA1 with Rab5b. Eur. J. Biochem. 265, 361–366. [DOI] [PubMed] [Google Scholar]
- Cantalupo, G., Alifano, P., Roberti, V., Bruni, C.B., and Bucci, C. (2001). Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, S., Hartnell, L.M., Aguilar, R.C., Naslavsky, N., and Bonifacino, J.S. (2001). Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 154, 109–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis, S., McBride, H.M., Burgoyne, R.D., and Zerial, M. (1999a). The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621–625. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., Miaczynska, M., Ashman, K., Wilm, M., Zhao, L., Yip, S.S., Waterfield, M.D., Backer, J.M., and Zerial, M. (1999b). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1, 249–252. [DOI] [PubMed] [Google Scholar]
- Feng, Y., Press, B., and Wandinger-Ness, A. (1995). Rab 7, an important regulator of late endocytic membrane traffic. J. Cell Biol. 131, 1435–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg, J., Griffiths, G., and Howell, K.E. (1989). Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vivo. J. Cell Biol. 108, 1301–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, J.A., III, and Wu, X.S. (2002). Rab grab motors: defining the connections between Rab GTPases and motor proteins. Curr. Opin. Cell Biol. 14, 69–75. [DOI] [PubMed] [Google Scholar]
- Huizing, M., Didier, A., Walenta, J., Anikster, Y., Gahl, W.A., and Kramer, H. (2001). Molecular cloning and characterization of human VPS18, VPS 11, VPS16, and VPS33. Gene 264, 241–247. [DOI] [PubMed] [Google Scholar]
- Jordens, I., Fernandez-Borja, M., Marsman, M., Dusseljee, S., Janssen,. L, Calafat, J., Janssen, H., Wubbolts, R., and Neefjes, J. (2001). The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685. [DOI] [PubMed] [Google Scholar]
- Kim, B.Y., Kramer, H., Yamamoto, A., Kominami, E., Kohsaka, S., and Akazawa, C. (2001). Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J. Biol. Chem. 276, 29393–29402. [DOI] [PubMed] [Google Scholar]
- Marquez-Sterling, N., Herman, I.M., Pesacreta, T., Arai, H., Terres, G., and Forgac, M. (1991). Immunolocalization of the vacuolar-type (H+)-ATPase from clathrin-coated vesicles. Eur. J. Cell Biol. 56, 19–33. [PubMed] [Google Scholar]
- Martinez, O., and Goud, B. (1998). Rab proteins. Biochem. Biophys. Acta 1404, 101–112. [DOI] [PubMed] [Google Scholar]
- Matteoni, R., and Kreis, T.E. (1987). Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105, 1253–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKiernan, C.J., Stabila, P.F., and Macara, I.G. (1996). Role of the Rab3A-binding domain in targeting of rabphilin-3A to vesicle membranes of PC12 cells. Mol. Cell. Biol. 16, 4985–4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey Ward, D., Radisky, D., Scullion, M.A., Tuttle, M.S., Vaughn, M., and Kaplan, J. (2001). hVPS41 is expressed in multiple isoforms and can associate with vesicles through a RING-H2 finger motif. Exp. Cell Res. 267, 126–134. [DOI] [PubMed] [Google Scholar]
- Meresse, S., Gorvel, J.P., and Chavrier, P. (1995). The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci. 108, 3349–3358. [DOI] [PubMed] [Google Scholar]
- Novick, P., and Zerial, M. (1997). The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol. 9, 496–504. [DOI] [PubMed] [Google Scholar]
- Olkkonen, V.M., and Stenmark, H. (1997). Role of Rab GTPases in membrane traffic. Int. Rev. Cytol. 176, 1–85. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. (1999). Transport-vesicle targeting: tethers before SNAREs. Nat. Cell Biol. 1, E17–E22. [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R. (2001). Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 11, 487–491. [DOI] [PubMed] [Google Scholar]
- Press, B., Feng, Y., Hoflack, B., and Wandinger-Ness, A. (1998). Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 140, 1075–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, A., Seals, D., Wickner, W., and Ungermann, C. (2000). The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J. Cell Biol. 148, 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seals, D.F., Eitzen, G., Margolis, N., Wickner, W.T., and Price, A. (2000). A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA 97, 9402–9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev, N. (2001). Ypt and Rab GTPases: insight into functions through novel interactions. Curr. Opin. Cell Biol. 13, 500–511. [DOI] [PubMed] [Google Scholar]
- Shirataki, H., Yamamoto, T., Hagi, S., Miura, H., Oishi, H., Jin-no, Y., Senbonmatsu, T., and Takai, Y. (1994). Rabphilin-3A is associated with synaptic vesicles through a vesicle protein in a manner independent of Rab3A. J. Biol. Chem. 269, 32717–32720. [PubMed] [Google Scholar]
- Somsel Rodman, J., and Wandlinrer-Ness, A. (2000). Rab GTPases coordinate endocytosis. J. Cell Sci. 113, 183–192. [DOI] [PubMed] [Google Scholar]
- Stenmark, H., Vitale, G., Ullrich, O., and Zerial, M. (1995). Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell 83, 423–432. [DOI] [PubMed] [Google Scholar]
- Takai, Y., Sasaki, T., and Matozaki, M. (2001). Small GTP-binding proteins. Physiol. Rev. 81, 153–208. [DOI] [PubMed] [Google Scholar]
- Ungermann, C., Price, A., and Wickner, W. (2000). A new role for a SNARE protein as a regulator of the Ypt7/Rab-dependent stage of docking. Proc. Natl. Acad. Sci. USA 97, 8889–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli, R., Santillo, M., Lattero, D., Chiariello, M., Bifulco, M., Bruni, C.B., and Bucci, C. (1997). Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 272, 4391–4397. [DOI] [PubMed] [Google Scholar]
- Wickner, W. (2002). Yeast vacuoles and membrane fusion pathways. EMBO J. 21, 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori, T. et al. (2000). The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell 11, 747–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107–117. [DOI] [PubMed] [Google Scholar]