Abstract
A cellular role and the mechanism of action for small GTPase Arl1 have been defined. Arl1-GTP interacts with the GRIP domains of Golgin-97 and Golgin-245, a process dependent on conserved residues of the GRIP domains that are important for Golgi targeting. The switch II region of Arl1 confers the specificity of this interaction. Arl1-GTP mediates Golgi recruitment of Golgin-97 in a switch II-dependent manner, whereas tethering Arl1-GTP onto endosomes can mediate endosomal targeting of Golgin-97. Golgin-97 and Golgin-245 are dissociated from the Golgi when Arl1 is knocked-down by its siRNA. Arl1-GTP thus functions to recruit Golgin-97 and Golgin-245 onto the Golgi via interacting with their GRIP domains.
INTRODUCTION
Members of the Ras-like small GTPase superfamily participate in many cellular processes ranging from signal transduction, cell cycle control, membrane traffic, cytoskeleton organization, transport between cytosol and nucleus, and development (Boman and Kahn, 1995; Bar-Sagi and Hall, 2000; Aridor et al., 2001; Zerial and McBride, 2001; Hetzer et al., 2002). ADP ribosylation factors (ARF1–6) and ARF-like proteins (Arl1–7) consist a unique family of proteins sharing significant amino acid identities and have been shown to participate in signaling, membrane traffic, cell motility, and mitochondria function (Boman and Kahn, 1995; Sharer et al., 2002). The ARFs are >60% identical to each other and have been classified into three classes. Class I members consisting of ARF1, 2, and 3 are mainly associated with the Golgi apparatus and endosomes and regulate vesicle budding of several transport events. Class II consists of ARF4 and 5, whose locations and functions are less well defined. ARF6 is the only member of class III and has been shown to coordinate actin cytoskeleton with membrane traffic and cell motility (Turner and Brown, 2001). Arls are ∼40–60% identical to each other and to ARFs (Schurmann et al., 1994; Boman and Kahn, 1995; Lowe et al., 1996; Ingley et al., 1999; Jacobs et al., 1999). The major differences between Arls and ARFs are that ARFs but not Arls are able to serve as cofactors in cholera toxin-catalyzed ADP-ribosylation of Gαs, to activate phospholipase D and to rescue simultaneous deletion of ARF1 and ARF2 genes in yeast. Among Arls, only Arl1 has been shown to be associated with the Golgi apparatus (particularly the trans-side of the Golgi apparatus; Lowe et al., 1996; Lu et al., 2001). Overexpression of mutant forms of Arl1 affected Golgi structure and function (Lu et al., 2001; Van Valkenburgh et al., 2001). Drosophila Arl1 plays an essential role in fly development (Tamkun et al., 1991), whereas Arl4 plays a regulatory role in the development of normal sperm count and fertility in mice (Schurmann et al., 2002). Other members of Arls are distributed in the cytosol, mitochondria, and/or nucleus (Schurmann et al., 1994; Boman and Kahn, 1995; Lowe et al., 1996; Ingley et al., 1999; Jacobs et al., 1999) and their biological and physiological functions remain to be further established.
Golgin-97 and Golgin-245/p230 are two autoantigens identified by antibodies from patients suffering from Sjogren's syndromes and they are enriched in the trans-Golgi network (TGN; Fritzler et al., 1995; Erlich et al., 1996; Griffith et al., 1997). Golgin-97, Golgin-245, and other Golgins such as GM130, Golgin-45, p115, and giantin contain extensive coiled-coil regions. Golgin-97 and Golgin-245 are targeted to the trans-face of the Golgi by their C-terminal GRIP domains, an evolutionally conserved motif of ∼50 amino acids (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999; Brown et al., 2001). Four mammalian proteins (Golgin-97, Golgin-245, GCC1p, and KIAA0336) contain GRIP domains (Luke et al., 2002). The molecular mechanism underlying Golgi targeting by the GRIP domain remains elusive, although a possible role of Rab6 has been implicated (Barr, 1999).
In this report, we provide insightful understanding of the role and mechanism of Arl1 action as well as GRIP domain-mediated Golgi targeting of Golgin-97 and Golgin-245. Our results have thus established a key cellular role for Arl1 and have defined the underlying mechanism.
MATERIALS AND METHODS
Yeast Two-hybrid Analysis
Construction of Arl1(Q71L) in pGBKT7 and its subsequent yeast two-hybrid screening of human brain cDNA library in pretransformed yeast Y187 (Clontech, Palo Alto, CA) were described before (Lu et al., 2001). Human Golgin-97 was one of the positive clones from the screening. The GRIP domains of human Golgin-97 (EST clone, Accession number AI803928), human Golgin-245/p230 (EST clone, Accession number AA626910), human GCC1p (EST clone, Accession number Z54284) and human KIAA0336 (KIAA0336, Kazusa DNA Research Institute) were retrieved by PCR and cloned into GAL4-AD fusion vector pGADT7 between EcoRI and BamHI sites. Full-length cDNA for Golgin-97 in pET28a (Novagen, Madison, WI; Griffith et al., 1997) was a gift from Dr. Marvin J. Fritzler and Dr. Edward K.L. Chan. The clone was digested by NotI (end blunted) and EcoRI, and the insert was subsequently ligated with BamHI- (end blunted) and EcoRI-digested pGADT7. The GRIP domain deleted Golgin-97 (Golgin-97Δ GRIP) was generated by standard PCR reaction (primers: 5′-GACTCGCATATGTTTGCAAAAVTGAAGAAGAAAATTGCAGAAGAGACTGC-3′ and 5′-GCACACGAATTCTTAGACTTCGAAGAGCTCATTATC-3′), digested by NdeI and EcoRI and cloned into pGADT7 using correspondent sites. These pGADT7 clones were subsequently transformed into Y187 yeast cells. Y187 yeast cells transformed with human full-length GGA1 in pGADT7 and human full-length POR1 in pACT2 were described previously (Lu et al., 2001). Each of these transformed Y187 yeast cells was individually mated with four types of AH109 yeast cells containing Arl1(T31N), Arl1(Q71L), ARF1T31N, and ARF1Q71L in pGBKT7 and assayed for interaction. The four GRIP mutants of Golgin-97: E696A, Y697A, K699A, and Q728A, were generated by standard PCR-mediated mutagenesis and cloned into pGADT7 vector between EcoRI and BamHI sites. These Golgin-97 GRIP domain mutants were transformed into yeast Y187 and tested for their interaction with active Arl1 by mating with Arl1(Q71L) containing yeast AH109. Arl1 mutants: ARF1(Q71L)(1–92), Arl1(93–181), Arl1(Q71L, T72D), Arl1(Q71L, S73K), Arl1(Q71L, Y77L), Arl1(Q71L, C80H), Arl1(Q71L, A87G), Arl1Q71L-ARF1swII (referred to as Arl1SIIm for short), and ARF1SIIm were generated by standard PCR-mediated mutagenesis and cloned into EcoRI and BamHI sites of pGBKT7. The resulting transformed AH109 yeast cells were mated to Y187 yeast cells containing Golgin-97 GRIP domain, Golgin-245 GRIP domain, or POR1. After yeast mating, diploid yeast cells containing DNA-BD and DNA-AD fusion constructs were selected on SD/-Trp/-Leu medium and then the selected cells were streaked on the SD/-Trp/-Leu/-His/-Ade (QDO) plate. The selected diploid yeast cells were subjected to ONPG (o-nitrophenol β-D-galactopyranoside, Sigma, St. Louis, MO) β-galactosidase assays were performed according to Yeast Protocols Handbook (Clontech).
In Vitro GST Pull-down Experiment
The coding sequence of human ARL4 (EST clones, GenBank Accession number T85847) were PCR-cloned into EcoRI/BamHI sites of pGEB. Arl1 wild type, Arl1(Q71L), Arl1(T31N), and ARF1 wild type in pGEB were described before (Lu et al., 2001). Recombinant GST fusion proteins were produced in Escherichia coli DH5α as described (Lowe et al., 1996). These GST fusion proteins immobilized on glutathione Sepharose 4B beads (Amersham-Pharmarcia, Upsala, Sweden) were exchanged for GTPγ S or GDP using the described methods (Randazzo et al., 1995; Christoforidis and Zerial, 2000). Briefly, GST fusion proteins equilibrated with NE buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 10 mM EDTA, 5 mM MgCl2, 1 mM DTT) were incubated with NE buffer with 1 mM GTPγ S (or GDP), 0.1% (wt/vol) sodium cholate, and 3 mM L-α -dimyristoylphosphatidylcholine (DMPC; 30 mM DMPC stock was freshly prepared by sonication in NE buffer) for 1.5 h at room temperature. The beads were then washed with NS buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT) with 10 μM GTPγ S or GDP. Hela cell cytosol was prepared in NS buffer and subsequently centrifuged at 55,000 rpm in TLA100.2 rotor for 1.5 h and precleared by GST immobilized beads with BSA added to a final concentration of 4 mg/ml. About 60 μg of fusion proteins on beads were first blocked for nonspecific binding sites by incubating with 4 mg/ml BSA in NS buffer at 4°C for 2 h before incubation with the precleared cytosol at 4°C overnight. For GST Golgin-245 GRIP domain pull-down experiment, 100 μM final concentration of GTPγ S was added to the Hela cytosol during the incubation. After washing with NS buffer, proteins retained on the beads were eluted in SDS sample buffer and analyzed by Western blot.
Immunoprecipitation
pSTAR (Zeng et al., 1998) constructs for expressing wild-type Arl1, Arl1(T31N), and Arl1(Q71L) with the C-terminal EGFP tag (Lu et al., 2001) were transfected into 293T cells using Effectene Transfection Reagent (QIAGEN, Hilden, Germany) according to manufacturer's manual. After 28 h of incubation at 37°C in the presence of 12 μg/ml doxycyclin (Clontech), cells were lysed in 25 mM HEPES, pH 7.4, 125 mM KAc, 1.0% Triton X-100 (TX-100), 1 mM DTT with Complete Protease Inhibitor (EDTA-Free; Roche, Basel, Switzerland) and 1 mM PMSF. The 14,000 rpm supernatant of each cell lysate was collected and precleared by human IgG Sepharose Fast Flow (Amersham Pharmacia) at 4°C for 1 h. BSA was added to each lysate to a final concentration of 2 mg/ml. Each cell lysate was then incubated with 2.5 μl of rabbit anti-EGFP polyclonal antibody (Clontech) at 4°C overnight and subsequently with 40 μl of Protein A Sepherose (Amersham Pharmacia; 50% slurry) at room temperature for 3 h. The beads were washed three times with 25 mM HEPES, pH 7.4, 125 mM KAc, 0.1% TX-100 followed by elution with SDS sample buffer and Western blotting analysis.
Mammalian Expression Plasmids
pSTAR construct containing Arl1SIIm was constructed by retrieving the region from pGBKT7 by EcoRI and BamHI and then cloned between EcoRI and BamHI sites of pSTAR vector. Arl1Q71L and Arl1T31N in pSTAR were described before (Lu et al., 2001). These constructs were used to transfect Hela cells, which were then incubated with 12 μg/ml doxycyclin at 37°C for 20 h to enhance the expression of proteins. Mouse SNX3 coding sequence was first PCR-cloned into EcoRI/XbaI sites of pDMyc-neo to express the N-terminal double myc-epitope–tagged SNX3. Rat Arl1Q71L or mouse ARF1Q71L coding sequence was PCR-cloned into XbaI/NotI sites of this pDMyc-neo/SNX3 for expressing chimeric proteins: Myc-SNX3-Arl1(Q71L) or Myc-SNX3-ARF1(Q71L). These constructs were transfected to A431 cells and incubated for 20 h before immunofluorescence analysis.
Antibodies
Wild-type Arl1 was cloned into NdeI/XhoI sites of pET37b vector and expressed as C-terminal His-tagged protein in BL21DE3 E. coli. The purified Arl1 was injected into New Zealand white rabbit to raise antibody according to described procedure (Lowe et al., 1996). The affinity-purified Arl1 antibody was shown to be specific for Arl1 because it did not react with recombinant mouse ARF1 in Western blot. This Arl1 antibody is very similar to our reported Arl1 antibody raised against E6P1 peptide (residues 130–150; Lowe et al., 1996) in that both antibodies react more strongly with rat than human Arl1 with about fivefold difference (unpublished results). Because rat and human Arl1 differ by only two amino acids within the E6P1 peptide region, it suggests that the major epitope recognized by this new antibody is within amino acids 130–150 region. Rabbit polyclonal antibody against β-1,4 galactosyltransferase (GT) was described previously (Subramaniam et al., 1992). Rabbit polyclonal antibody against Golgin-245 was a generous gift from Dr. Paul Gleeson. mAb (Mab), CDF4, against Golgin-97 was from Molecular Probes (Eugene, OR). Mabs against Golgin-245, p115, and GM130 were from BD Transduction Laboratories (San Diego, CA). Polyclonal anti-Myc antibody was from Upstate Biotechnology (Charlottesville, VA). Mab 1D9 (Cavenagh et al., 1996) against ARFs was from BioAffinity Reagents Inc (Golden, CO). Mabs against β-COP (Mad) and β-tubulin were from Sigma. Rabbit antibody against EGFP was from BD Clontech. Goat polyclonal antibodies against EEA1 were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
Cell Culture, Transfection, and Indirect Immunofluorescence Microscopy
Hela cells were cultured in RPMI with 10% FBS. A431, NRK, and 293T cells were maintained in DMEM supplemented with 10% FBS. Transient transfections were done using Effectene Transfection Reagent (QIAGEN) according to manufacturer's protocol. Indirect immunofluorescence microscopy was performed as described previously (Lu et al., 2001).
Knockdown of Endogenous Arl1 by Small Interference RNA
Small interference RNA (siRNA) targeting human Arl1 (5′-AAG AAG AGC UGA GAA AAG CCA-3′) and control siRNA (5′-AAG UCG GUG UGC UCU UGU UGG-3′) were synthesized at Dharmacon Research Inc (Lafayette, CO). Hela cells were transfected with siRNA using Oligofectamine transfection reagent (Invitrogen, Grand Island, NY) according to protocol provided by Dharmacon Research Inc. Cells were processed for immunofluorescence or immunoblotting after 48 h of incubation.
RESULTS
Interaction of GTP-restricted Arl1 with GRIP Domains of Golgin-97 and Golgin-245/p230
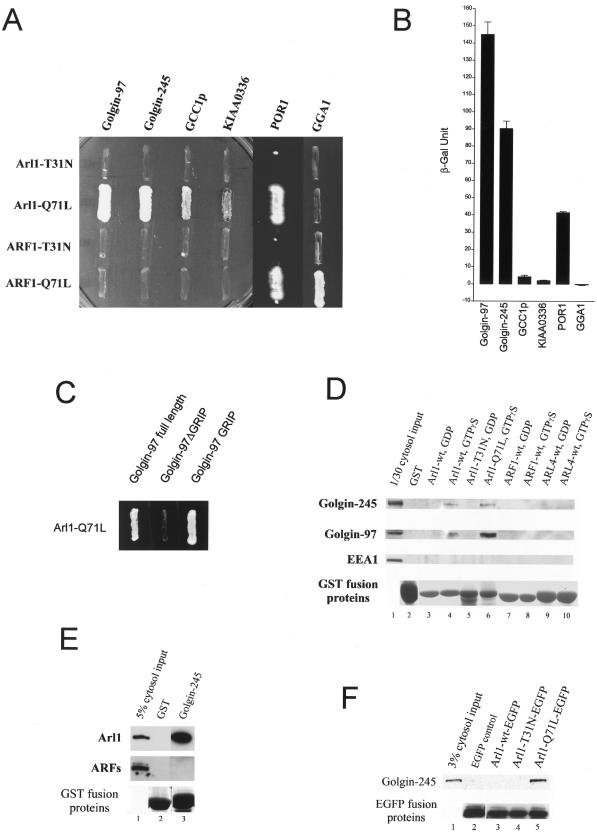
To gain insightful understanding about the function and mechanism of Arl1 action, we have earlier searched for potential effectors of Arl1 using yeast two-hybrid screens and have identified POR1, Golgin-97, and pericentrin as potential candidates for downstream effectors for GTP-restricted Arl1 (Lu et al., 2001). Because Golgin-245 was independently shown to interact with Arl1 by a yeast two-hybrid analysis (Van Valkenburgh et al., 2001) and both Golgin-97 and Golgin-245 interact specifically with Arl1 but not ARF1, we have first focused our studies on defining the nature and specificity of the interaction of Arl1 with Golgin-97 and found that GTP-restricted Arl1 interacts specifically with the C-terminal GRIP domain of Golgin-97 but not any other regions of this protein. Because GRIP domains were also present in Golgin-245 and two other proteins (GCC1p and KIAA0336), we have systematically analyzed the interaction of various forms of Arl1 with these GRIP domains by yeast two-hybrid analysis. As shown in Figure 1A, GTP-restricted (Arl1Q71L) but not GDP-restricted (Arl1T31N) of Arl1 interacted strongly with the GRIP domains of Golgin-97 and Golgin-245. Weaker interactions of Arl1Q71L with GRIP domains of GCC1p and KIAA0336 were also observed. Neither the GTP-restricted (ARF1Q71L) nor the GDP-restricted form (ARF1T31N) of ARF1 interacted with these GRIP domains. Under similar conditions, GGA1 interacted specifically with ARF1Q71L but not Arl1Q71L, ARF1T31N, or Arl1T31N, consistent with earlier results showing that GGA1 is a specific effector of ARF1 (Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000). POR1 interacted with GTP-restricted form of both Arl1 and ARF1 because it is an effector shared by Arl1, ARF1, and other small GTPases (such as ARF3, ARF6, and Rac1; D'Souza-Schorey et al., 1997; Lu et al., 2001; Van Valkenburgh et al., 2001). These results suggest that GTP-restricted Arl1 but not ARF1 can interact specifically and robustly with the GRIP domains of Golgin-97 and Golgin-245, whereas weaker interaction of Arl1 with GRIP domains of GCC1p and KIAA0336 was also observed.
Figure 1.
GTP-restricted Arl1 interacts with GRIP domains of Golgins. (A) Arl1Q71L, Arl1T31N, ARF1Q71L, and ARF1T31N were tested respectively in the yeast two-hybrid assay for interaction with the GRIP domain of Golgin-97, Golgin-245, GCC1p, KIAA0336, the full-length POR1, or GGA1. (B) ONPG β-galactosidase assay shows different interaction strengths of Arl1Q71L with various GRIP domains, POR1, or GGA. (C) Golgin-97 full-length but not Golgin-97Δ GRIP interacts with Arl1-Q71L in yeast two-hybrid assay, showing the interaction is only through the GRIP domain of Golgin-97. (D) Hela cell cytosol was incubated with 60 μg of various immobilized recombinant proteins as indicated. Proteins retained by the beads were eluted by SDS sample buffer and subjected to immunoblot analysis with the indicated antibodies. Significant amounts of Golgin-245 (first panel) and Golgin-97 (second panel) were retained on GTPγ S (lane 4 and 6) but not GDP (lane 3 and 5) charged forms of GST-Arl1. As negative controls, GST (lane 2), various form of GST-ARF1 (lane 7 and 8) or GST-Arl4 (lane 9 and 10) failed to retain Golgin-245 and Golgin-97. The endosomal EEA1 (third panel) was not retained by any of these beads. The bottom panel is the coomassie blue stained SDS-PAGE gel showing quantity of the respective GST fusion proteins. Lane 1 represents a 3% cytosol loading control for Golgin-245, Golgin-97 and EEA1. (E) Arl1 (top panel) but not ARFs (middle panel, blotted with Mab 1D9) in Hela cell cytosol was efficiently retained by immobilized GST-GRIP domain of Golgin-245 (lane 3) but not GST (lane 2). Lane 1 shows the 5% cytosol loading control. The bottom panel is the coomassie blue stained SDS-PAGE gel showing quantity of the recombinant GST or fusion protein. (F) Various forms of Arl1 tagged with EGFP, together with EGFP, were expressed in transfected 293T cells. The resulting cell lysates were immunoprecipitated with antibodies against EGFP, and the immunoprecipitates were analyzed by immunoblotting to detect Golgin-245. Golgin-245 was selectively coimmunoprecipited with GTP-restricted Arl1Q71L-EGFP (lane 5). Lane 1 represents 3% cytosol input used for the immunoprecipitation, and the bottom panel shows the 5% various EGFP fusion proteins recovered after immunoprecipitation.
This conclusion was strengthened by monitoring β-galactosidase activities (Figure 1B). High activities were observed when interaction was mediated by Arl1Q71L and the GRIP domain of Golgin-97 or Golgin-245. The weaker but significant interaction of Arl1Q71L with the GRIP domains of GCC1p and KIAA0336 was also verified by the β-galactosidase assay. Full-length Golgin-97, but not the GRIP domain deleted Golgin-97 (Golgin-97ΔGRIP), interacted with Arl1Q71L in yeast two-hybrid assays. These results suggest that Arl1 interacts specifically with the GRIP domain, but not the rest part of Golgin-97 (Figure 1C) and that this interaction can occur with either isolated GRIP domain or GRIP domain in the context of full-length polypeptide of Golgin-97.
To further demonstrate the biological relevance of this observed interaction of Arl1 with the GRIP domains, we investigated whether cellular Golgin-97 and Golgin-245 can interact with Arl1Q71L in vitro. Various forms of Arl1 and ARF1 were expressed as recombinant proteins fused to GST and these GST-fusion proteins, or GST alone as a negative control, were immobilized onto glutathione beads. Cytosols prepared from Hela cells were incubated with these beads and the amounts of Golgin-245 and Golgin-97 retained by the beads were analyzed by immunoblot analysis. As shown in Figure 1D, significant amounts of Golgin-245 and Golgin-97 in the cytosol were retained by immobilized wild-type Arl1 (lane 4) and Arl1Q71L charged with GTPγ S (lane 6). Conversely, wild-type Arl1 charged with GDP failed to retain either Golgin-245 or Golgin-97 (lane 3). Golgin-245 and Golgin-97 were not retained by immobilized Arl1T31N (lane 5) or any forms of ARF1 (lanes 7 and 8) or Arl4 (lane 9 and 10). These results suggest that, like isolated GRIP domains or full-length Golgin-97, intact Golgin-245, and Golgin-97 from cells can also interact specifically with GTP-restricted Arl1. Furthermore, when the GRIP domain of Golgin-245 was expressed as GST fusion protein and immobilized onto glutathione beads, it could achieve qualitative recovery of cytosolic Arl1 in the presence of GTPγS (Figure 1E), whereas ARFs were not retained, confirming the interaction of GTP-restricted Arl1 with the GRIP domain of Golgin-245. Finally, we have provided evidence that GTP-restricted Arl1 and Golgin-245 could interact in intact cells as revealed by coimmunoprecipitation experiment (Figure 1F). Hela cells were transiently transfected with wild-type (Arl1wt-EGFP), GDP-restricted (Arl1T31N-EGFP), or GTP-restricted (Arl1Q71L-EGFP) form of Arl1 tagged with EGPP at the C termini. The resulting cell lysates were processed for immunoprecipitation using antibodies against EGFP and analyzed by immunoblot analysis using antibodies against Golgin-245 (top panel) and EGFP (bottom panel). As shown, endogenous Golgin-245 was specifically coimmunoprecipitated with Arl1Q71L-EGFP (lane 5) but not Arl1T31N-EGFP (lane 4). Recombinant GRIP domain of Golgin-245 can also interact with recombinant Arl1 in the presence of GTPγ S (our unpublished results), suggesting that the Arl1 and GRIP interaction is direct. These results, taken together, establish that Arl1 (in its GTP-restricted status) can interact specifically and directly with the GRIP domain of Golgin-97 and Golgin-245, and this interaction occurs with intact proteins in vivo.
Arl1-GRIP Domain Interaction Depends on Conserved Residues of GRIP Domains That Are Important for Golgi Targeting
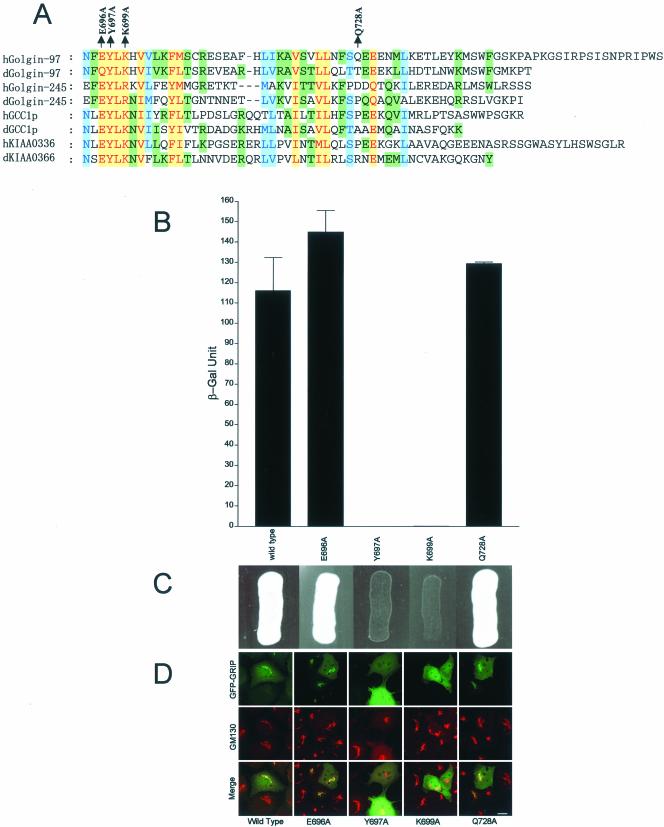
The GRIP domain is conserved evolutionally and is present in proteins from yeast, fly, worm and mammals (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999). The GRIP domains of four mammalian proteins (Golgin-97, Golgin-245, GCC1p, and KIAA0336) are able to mediate Golgi targeting by a saturable mechanism, and mutations of some conserved residues abolished Golgi targeting (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999). As shown in Figure 2A, several residues are conserved in the GRIP domains of all four mammalian proteins and their putative fly counterparts. To sustain the conclusion that GRIP domains of Golgin-97 and Golgin-245 interact with Arl1 and to establish the importance of conserved residues of GRIP domain in mediating this interaction, we constructed four different mutant versions of Golgin-97 GRIP domain: E696A, Y697A, K699A, and Q728A. The conserved residues E696, Y697, and K699 of Golgin-97 in the GRIP domain have been replaced by Ala in E696A, Y697A, and K699A, respectively. As a negative control, nonconserved Q728 of Golgin-97 was replaced by Ala in Q728A. As shown in Figure 2, B and C, individual mutation of conserved Y697 or K699 abolished interaction with Arl1Q71L, whereas mutation of the conserved E696 and the nonconserved Q728 did not affect its interaction with Arl1Q71L. These results firmly established the importance of two key conserved residues of Golgin-97 GRIP domain in mediating interaction with GTP-restricted form of Arl1. Furthermore, there is a strong correlation between interaction with Arl1 and Golgi targeting for these mutants (Figure 2D). The Arl1-interacting E696A and Q728A were targeted to the Golgi apparatus, whereas the Arl1 noninteracting Y697A and K699A were distributed in the cytosol and/or nucleus. This correlation suggests that interaction with GTP-restricted Arl1 is likely to be necessary for GRIP domains to mediate Golgi targeting.
Figure 2.
Conserved residues of the GRIP domain are important for interaction with Arl1 as well as for Golgi targeting. (A) Alignment of the amino acid sequences of four GRIP domains of human (prefix h) and Drosophila (prefix d) Golgins defines several conserved residues of the GRIP domains. The most conserved residues are indicated in red with a yellow background. The intermediate conserved residues are shown in blue under light blue background and the residues conserved in only some GRIP domains were highlighted with green background. The positions of point mutations analyzed in this study are indicated above the sequence of human Golgin-97. (B) Interaction of various forms of the Golgin-97 GRIP domain with Arl1Q71L as analyzed by ONPG β-galactosidase assay using the yeast two-hybrid system. (C) Interaction of various forms of Golgin-97 GRIP domain with Arl1Q71L as analyzed by cell growth in the yeast two-hybrid system. (D) The indicated forms of Golgin-97 GRIP domain mutants tagged with EGFP were expressed in transfected NRK cells (top row) and colabeled antibody against GM130 (Texas Red, middle row). There exists a strong correlation between interaction with Arl1Q71L and Golgi targeting of these mutant GRIP domains. Bar, 10 μm.
Arl1-GRIP Domain Interaction Depends on the Switch II Region of Arl1
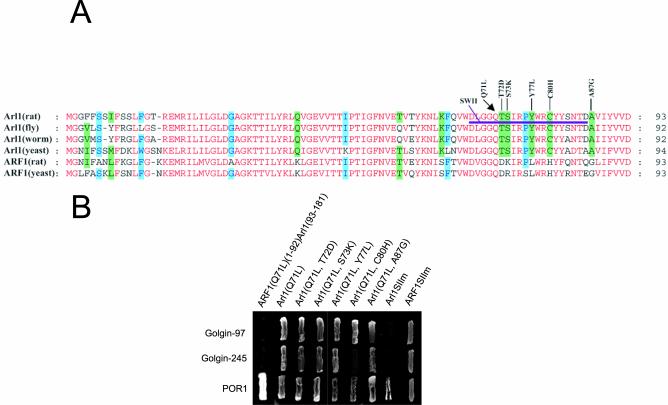
Because both Arl1 and ARF1share significant sequence homology as well as a common effector (POR1) and only Arl1 interacts specifically with the GRIP domains of Golgin-97 and Golgin-245, it is therefore of great interest to define the structural basis underlying the interaction of Arl1 with GRIP domains. Alignment of Arl1 from rat, fly, worm, and budding yeast with ARF1 of rat and budding yeast revealed that there are several residues that are selectively conserved in all Arl1 proteins but not in ARF1, including Q38, K62, T72, S73, Y77, C80, A87, D98, S159, and K162 (Figure 3A). We first constructed a chimeric protein, ARF1Q71L(1–92)-Arl1(93–181), consisting of the N-terminal portion (residue 1–92) of ARF1Q71L followed by the C-terminal region (residue 93–181) of Arl1. We have found that this chimeric protein failed to interact with GRIP domains of Golgin-97 and Golgin-245, although normal interaction with POR1 was observed (Figure 3B), suggesting that the region between residue 1 and 92 of Arl1 is important for its unique ability to interact with GRIP domains. Because the majority of Arl1-conserved residues (T72, S73, Y77, C80, and A87) are located in the switch II region of Arl1 and the switch II region of small GTPases is known to participate in interaction with specific effectors, we have focused our investigation on these residues. Replacement of each of these residues in the switch II region individually by the corresponding residue in ARF1 (Q71L-T72D, Q71L-S73K; Q71L-Y77L, Q71L-C80H, and Q71L-A87G) did not affect interaction with GRIP domains, although Q71L-C80H has significantly reduced its activity in interacting with the GRIP domain of Golgin-245. These results suggest that individual residues may not be responsible for the observed specific interaction of Arl1 with the GRIP domains. In this regard, we have created another mutant version of Arl1 (Arl1SIIm), in which the entire switch II region (residues 67–86) of Arl1Q71L was replaced by that of ARF1Q71L. As shown (Figure 3B), Arl1SIIm failed to interact with the GRIP domain of Golgin-97 and Golgin-245, although interaction with POR1 was still observed. Conversely, when the switch II region (residues 67–86) of ARF1Q71L was replaced by that of Arl1Q71L, the chimeric ARF1 (ARF1SIIm) showed interaction with the GRIP domain of Golgin-97 and Golgin-245. Taken together, these results suggest that the collective/concerted action of Arl1-specific residues in the switch II region is important for Arl1-specific interaction with the GRIP domain of Golgin-97 and Golgin-245.
Figure 3.
The switch II region of Arl1 is necessary for interaction with GRIP domains. (A) Alignment of N-terminal 93 residues of rat Arl1 with the corresponding regions of other Arl1 as well as rat and yeast ARF1. Identical residues are colored red. Residues conserved in four of the six proteins were highlighted with a green background. The point mutation mutants of Arl1Q71L are indicated above. The switch II region of Arl1 is underlined and Arl1SIIm has its switch II region replaced by that of ARF1, whereas ARF1SIIm has its switch II region replaced by that of Arl1. (B) The various Arl1Q71L mutants were tested for interaction with the GRIP domain of Golgin-97, Golgin-245, or the full-length POR1 by the yeast two-hybrid assay.
Concerted Responses of Arl1 and Golgin-97 to Brefeldin A Treatment
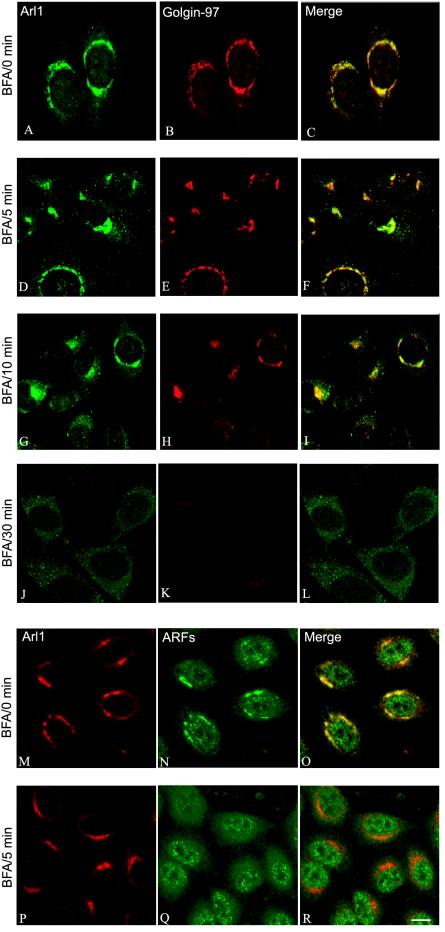
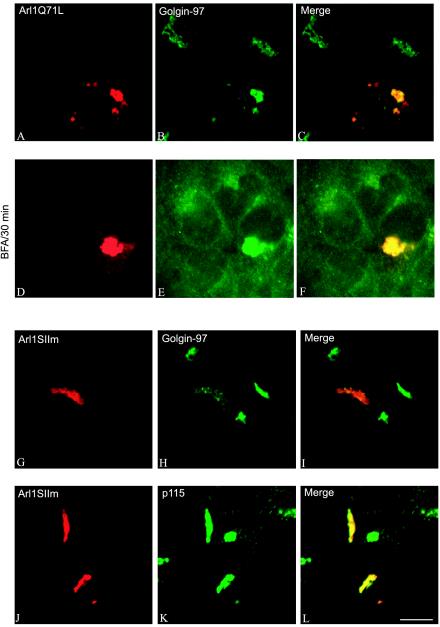
In contrast to Golgi ARFs and components of COPI, which dissociate from the Golgi apparatus within 2 min of brefeldin A treatment, Arl1 remains associated with the Golgi apparatus even after 5 min of treatment (Lowe et al., 1996). To gain additional understanding about the potential role of Arl1-GRIP domain interaction in mediating Golgi association of Golgin-97 and Golgin-245, we compared the kinetics of brefeldin A–induced dissociation of Arl1 and Golgin-97. As shown in Figure 4, both Arl1 and Golgin-97 remained Golgi-associated after 5 min of brefeldin A treatment. After 10-min treatment, Arl1 was dissociated from only a sub-population of the cells. Most intriguingly, it was observed that Golgin-97 remained Golgi-associated in cells that still have Arl1 in the Golgi apparatus, whereas Golgin-97 was dissociated from the Golgi apparatus in cells whose Arl1 has also been dissociated into the cytosol. As a control, ARFs (as detected by Mab1D9 antibody) were essentially dissociated in cells that have intact Golgi labeling of Arl1 after 5 min of brefeldin A treatment. Similar parallel response of Golgin-245 and Arl1 to brefeldin A treatment was also observed (our unpublished results). The similar kinetics of Golgi dissociation of Golgin-97 and Arl1 in response to brefeldin A treatment suggests that Golgi association of these two proteins are temporally coupled. In conjunction with the observation that residues within the GRIP domain important for Arl1 interaction are crucial for GRIP-domain–mediated Golgi targeting, it is tempting to suggest that interaction with activated Arl1 is an underlying mechanism for GRIP domain–driven Golgi recruitment.
Figure 4.
Golgi dissociation of Golgin-97 and Arl1 are temporally coupled in response to brefeldin A (BFA) treatment. Hela cells were treated with 10 μg/ml brefeldin A for 0 min (A–C and M–O), 5 min (D–F and P–R), 10 min (G–I), and 30 min (J–L). Cells were then processed for double-labeling using Arl1 antibody (A, D, G, and J) and Golgin-97 Mab (B, E, H, and K), or double-labeling using Arl1 antibody (M and P) and Mab anti-ARFs (N and Q). Bar, 10 μm.
Recruitment of Golgin-97 by Arl1Q71L but not by Arl1SIIm
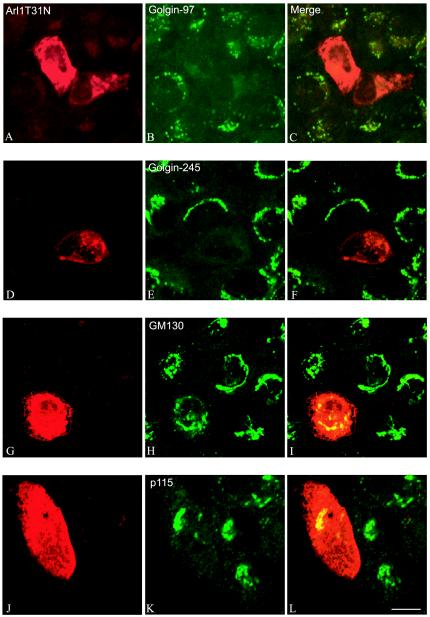
To directly examine whether Arl1 plays a role in the recruitment of Golgin-97 onto the Golgi, we first used a gain-offunction approach (Figure 5). As shown, when Arl1Q71L was overexpressed in transfected cells, it was massively recruited onto the Golgi apparatus (panel A). Interestingly, massive recruitment onto the Golgi apparatus was similarly observed for Golgin-97 (panel B), suggesting that Arl1 could effect a recruitment of Golgin-97 onto the Golgi membrane. Furthermore, the recruited Arl1Q71L and Golgin-97 became resistant to brefeldin A treatment (panels D–F). Although Arl1SIIm was recruited onto the Golgi apparatus in transfected cells (panel G), it exhibited no effect on recruitment of Golgin-97 (panel H). In fact, the amount of Golgi-associated Golgin-97 was reduced by the expression of Arl1SIIm, likely because of a competition of Arl1SIIm with endogenous Arl1 for regulatory factors. This result suggests that the switch II region of Arl1 is not only important for interaction with GRIP domains but also essential for Arl1-mediated Golgi recruitment of Golgin-97. Similar recruiting efforts on Golgin-245 were observed (our unpublished results). On the other hand (Figure 6), in cells expressing GDP-restricted Arl1 (Arl1T31N), both Golgin-97 (panel B) and Golgin-245 (panel E) became cytosolic, although GM130 (panel H) and p115 (panel K) remained with the Golgi structure. These results strengthen the notion that Arl1-mediated interaction with the GRIP domain of Golgin-97 and Golgin-245 may be responsible for Golgi recruitment of Golgin-97 and Golgin-245.
Figure 5.
GTP-restricted Arl1 recruits Golgin-97 onto the Golgi apparatus in a switch II region-dependent manner. A431 cells were transiently transfected with expressing plasmid for Arl1Q71L (A–F) or Arl1SIIm (G–L). The resulting cells were either not treated (A–C and G–L) or treated with 10 μg/ml brefeldin A for 60 min (D–F) and then processed for double-labeling using a limiting amount of Arl1 antibody that selectively detect overexpressed protein (A, D, G, and J) and Mab against Golgin-97 (B, E, and H) or p115 (K). Bar, 10 μm.
Figure 6.
GDP-restricted Arl1T31N abolishs Golgi association of Golgin-97 and Golgin-245 but not GM130 and p115. Arl1T31N was expressed transiently in A431 cells. Cells were then processed for the double-labeling with a limiting amount of Arl1 antibody (A, D, G, and J) and Mab against Golgin-97 (B), Golgin-245 (E), GM130 (H), or p115 (K). Bar, 10 μm.
Tethering of Active Arl1, but not ARF1, to the Endosome Results in Significant Recruitment of Golgin-97 onto the Endosomal Membrane
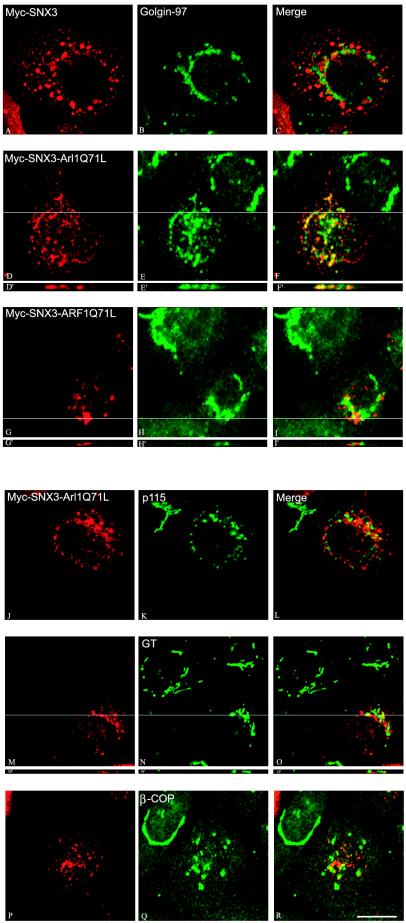
To directly examine whether Arl1 plays a sufficient and decisive role in mediating membrane recruitment of Golgin-97, we have engineered two chimeric proteins that will target the GTP-bound form of Arl1 [myc-SNX3-Arl1(Q71L)] or ARF1 [myc-SNX3-ARF1(Q71L)] onto the endosomal compartment. The chimeric proteins will be delivered to the early and recycling endosomes by PX domain of SNX3 (Xu et al., 2001). As shown in Figure 7, myc-SNX3 was delivered to the spotty endosomes (panel A), whereas Golgin-97 was detected in the Golgi apparatus (panel B). Both myc-SNX3-Arl1(Q71L) (panel D) and myc-SNX3-ARF1(Q71L) (panel G) were delivered to dotty endosomes. Interestingly, a significant amount of Golgin-97 was redistributed onto the endosomal structures marked by myc-SNX3-Arl1(Q71L) (panels D–F for x-y confocal section and panels D′–F′ for an indicated x-z section), establishing that activated Arl1 residing at endosomes could mediate recruitment of Golgin-97 onto this nonresidential new compartment. However, endosomal-targeted myc-SNX3-ARF1(Q71L) (panels G–I for x-y section and G′–I′ for an defined x-z section) had no effect on the Golgi distribution of Golgin-97. The same effects were observed for Golgin-245 (our unpublished results). These results suggest that Arl1 has a decisive and direct role in membrane recruitment of Golgin-97 and Golgin-245.
Figure 7.
Ectopic targeting of Arl1Q71L but not ARF1Q71L onto endosomes results in selective recruitment of Golgin-97 onto the endosomal membrane. A431 cells were transiently transfected with expressing construct for Myc-SNX3 (A–C), Myc-SNX3-Arl1Q71L (D–F′l and J–R), or Myc-SNX3-ARF1Q71L (G–I′). Cells were double-labeled with antibody against Myc to reveal the exogenously expressed SNX3 or its fusion proteins (left panels) and antibodies against Golgin-97 (B, E, E′, H, and H′), p115 (K), GT (N and N′), or β-COP (Q). Images D′–F′, G′–I′, and M′–O′ are the indicated x-z section of the x-y images of D–F, G–I, and M–O, respectively. Bar, 10 μm.
Interestingly, the Golgi apparatus marked by p115 (panel K), β1, 4-galactosyltransferase (GT; panel N), and β-COP (panel Q) was fragmented in cells expressing myc-SNX3-Arl1(Q71L). Unlike Golgin-97, p115, GT or β-COP was not detected in endosomal structures marked by myc-SNX3-Arl1(Q71L). The molecular basis for the observed Golgi fragmentation upon myc-SNX3-Arl1(Q71L) expression is likely due to mistargeting of Arl1 effectors to the endosomes, implying an essential role of its effectors in maintaining an intact Golgi structure. The selective delivery of Golgin-97 onto the endosomal membranes suggests that myc-SNX3-Arl1(Q71L) has a dominant role in determining the subcellular targeting of this Golgin.
Knockdown of Arl1 Dissociates Golgin-97 and Golgin-245 from the Golgi Apparatus
To provide complementary evidence for a role of Arl1 in recruiting Golgin-97 and Golgin-245 onto the Golgi apparatus, we used a loss-of-function approach by using the 21-base pair duplex siRNA technique (Elbashir et al., 2001) to knockdown the level of endogenous Arl1 and to see its consequence on Golgi association of Golgin-97 and Golgin-245. As revealed in Figure 8A, Arl1 level was specifically reduced in the entire population of cells transfected with Arl1 siRNA. When analyzed by indirect immunofluorescence microscopy, siRNA of Arl1 had a spectrum of knock-down effects, ranging from reduced levels of Arl1 in some cells to undetectable levels of Arl1 in other cells. This variability provided a useful internal control on the effect of Arl1 knockdown. As shown in Figure 8B, Golgin-97 (panels D–F) and Golgin-245 (panels J–L) became dissociated from the Golgi into the cytosol in cells with no detectable Arl1. However, reduced levels of Golgin-97 and Golgin-245 were clearly detected in the Golgi in cells with also reduced but detectable Golgi Arl1. In contrast to dissociation of Golgin-97 and Golgin-245, GM130 (panels M–O), another member of the Golgin family that does not contain a GRIP domain, was not dissociated into the cytosol but remained associated with the Golgi apparatus in cells with no detectable levels of Arl1. These results suggest that Arl1 is specifically required for Golgi association of Golgin-97 and Golgin-245.
Figure 8.
Knockdown of Arl1 levels by siRNA dissociates Golgin-97 and Golgin-245 from the Golgi apparatus. (A) Hela cells were transfected with indicated siRNA. After 48 h, the total cell lysates were subjected to SDS-PAGE and immunoblotting analysis to detect Arl1 and β-tubulin. (B) Hela cells were transfected with Arl1 siRNA (D–F and J–O) or control siRNA (A–C and G–I). After 48 h, cells were double-labeled with Arl1 antibody (left panels) and antibodies against Golgin-97 (B and E), Golgin-245 (H and K), or GM130 (panel N). Bar, 10 μm.
DISCUSSION
In our present study, we have addressed two important biological questions. The first relates to the cellular function and molecular mechanism of Arl1 action, and the second is the molecular mechanism underlying the process of GRIP domain-mediated Golgi targeting of autoantigens Golgin-97 and Golgin-245. Our results suggest that Golgin-97 and Golgin-245 are likely downstream effectors of Arl1, based on several lines of evidence. First, Golgin-97 and Golgin-245 uncovered by yeast two-hybrid screens using GTP-restricted form of Arl1 are shown to interact with Arl1 through their GRIP domains, which have been previously shown to mediate their Golgi targeting (Barr, 1999; Kjer-Nielsen et al., 1999; Munro and Nichols, 1999). Second, the importance of the GRIP domain in mediating interaction with Arl1 was further established by our demonstration that mutation of two conserved residues (Y697 and K699) of the GRIP domain of Golgin-97 abolished this interaction, demonstrating that Arl1 interacts with the intact GRIP domains. Because these two mutations also abolished the Golgi targeting property of the GRIP domain, it suggests that Golgi targeting of Golgin-97 (and likely Golgin-245 because they share the same conserved residues) is likely mediated through direct interaction of GRIP domains with activated form of Arl1. Third, the specificity of Arl1 interaction with GRIP domains was strengthened by the fact that its homologous Golgi-localized ARF1 does not interact with GRIP domains. Mutagenesis studies have uncovered the importance of switch II region of Arl1 in conferring this unique ability to Arl1. Because GRIP domains interact specifically with GTP-restricted forms of Arl1, interaction with GRIP domains of Golgins is likely triggered by guanine nucleotide exchange of GDP for GTP of Arl1. Finally, the biological relevance of GRIP domain-Arl1 interaction was demonstrated by our observation that Arl1 can interact with intact cellular Golgin-97 and Golgin-245, and this interaction is similarly depends on GTP-restricted status of Arl1. Furthermore, coimmunoprecipitation of endogenous Golgin-245 with transiently expressed Arl1 in a GTP-restricted manner strongly suggests that Golgi-245 and Arl1 could interact in intact cells.
The physiological relevance/function of the interaction of GRIP domains with Arl1 was further addressed by several experiments. First, because various proteins of the Golgi apparatus and endosomal pathway respond differently to brefeldin A treatment in mammalian cells (Klausner et al., 1992), we have compared the kinetics of Arl1, Golgin-97, and Golgin-245 redistribution during brefeldin A treatment and have found that they have a unique concerted dissociation kinetics. Unlike ARF1 and COPI components, which are dissociated from the Golgi apparatus even within 2 min of brefeldin A treatment, Arl1, Golgin-97, and Golgin-245 remain Golgi-associated after 5 min of brefeldin A treatment. At 10 min upon treatment, Arl1, Golgin-97, and Golgin-245 are dissociated from the Golgi in only some cells. Remarkably, there is a perfect correlation between the distribution of Arl1 with that of Golgin-97 and Golgin-245 in response to brefeldin A. This heterogeneity of cellular response to brefeldin A at 10 min treatment and the identical behavior of Arl1, Golgin-97, and Golgin-245 within the heterogeneous population provided sound evidence for a concerted kinetics of Golgi association/dissociation of these proteins.
Second, mutations of conserved residues in the GRIP domain of Golgin-97 not only abolished interaction with Arl1 but also abrogated their ability to mediate Golgi targeting, suggesting that interaction with Arl1 is likely necessary for GRIP domain to mediate Golgi targeting. Third, we have shown that overexpression of Arl1Q71L could elicit a massive recruitment of Golgin-97 onto the Golgi, suggesting that GTP-restricted Arl1 could mediate Golgi recruitment of Golgin-97. Because this effect depends on switch II region of Arl1, it is likely that the observed recruitment is a result of a direct interaction of Arl1Q71L with the GRIP domain. In support of this interpretation is the observation that overexpression of GDP-restricted form of Arl1 resulted in redistribution of Golgin-97 and Golgin-245 into the cytosol, whereas two other Golgins GM130 and p115 that do not contain the GRIP domain remained associated with Golgi-like structure.
Fourth, we have shown that Arl1 plays a direct and decisive role in membrane recruitment of Golgin-97. When Arl1(Q71L) but not ARF1(Q71L) was ectopically targeted to the endosomes by linking to SNX3, a significant amount of Golgin-97 but not p115, GT or β-COP was mistargeted to the endosomes. Furthermore, overexpression of Myc-SNX3-Arl1(Q71L) also caused fragmentation of the Golgi apparatus, likely because of mistargeting of most putative effectors of this small GTPase to the endosomes. Finally, a loss-of-functional approach has revealed an essential role of endogenous Arl1 in mediating Golgi targeting of Golgin-97 and Golgin-245, as revealed by data obtained from siRNA-mediated knockdown of Arl1 that had resulted in selective dissociation of Golgin-97 and Golgin-245 into the cytosol, whereas Golgi association of GM130 was not affected. These results collectively suggest that one of cellular function of Arl1 is to mediate Golgi targeting of Golgin-97 and Golgin-245, a process that is tightly linked to its guanine nucleotide exchange-driven interaction with the GRIP domains.
During the revision of our manuscript, the Golgi recruitment of Imh1p, the only GRIP domain containing protein in yeast, was shown to be regulated by Arl1p, the yeast homolog of Arl1 (Panic et al., 2003; Setty et al., 2003). These results suggest that Arl1-regulated mechanism of Golgi recruitment of GRIP domain proteins is evolutionally conserved. Our results also defined the switch II region of Arl1 confers the specificity and that conserved residues of the GRIP domains are key residues involved in interaction with Arl1-GTP.
Acknowledgments
The authors thank Drs. Tang Bor Luen and Paramjeet Singh for their careful reading of this manuscript, Drs. Marvin J. Fritzler and Edward K.L. Chan for the full-length cDNA of Golgin-97 and Dr. Paul Glesson for antibody against Golgin-245/p230. This work was support by a grant from Agency for Science, Technology, and Research (A*STAR), Singapore (to W.H.). W.H. is also a faculty member of the Department of Biochemistry, National University of Singapore.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0864. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0864.
References
- Aridor, M., Fish, K.N., Bannykh, S., Weissman, J., Roberts, T.H., Lippincott-Schwartz, J., and Balch, W.E. (2001). The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J. Cell Biol. 152, 213–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr, F.A. (1999). A novel Rab6-interacting domain defines a family of Golgi-targeted coiled-coil proteins. Curr. Biol. 9, 381–384. [DOI] [PubMed] [Google Scholar]
- Bar-Sagi, D., and Hall, A. (2000). Ras and Rho GTPases: a family reunion. Cell 103, 227–238. [DOI] [PubMed] [Google Scholar]
- Boman, A.L., and Kahn, R.A. (1995). ARF proteins: the membrane traffic police? Trends Biochem. Sci. 20, 147–150. [DOI] [PubMed] [Google Scholar]
- Boman, A.L., Zhang, C.J., Zhu, X., and Kahn, R.A. (2000). A family of ADP-ribosylation factor effectors that can alter membrane transport through the trans-Golgi. Mol. Biol. Cell 11, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, D.L., Heimann, K., Lock, J., Kjer-Nielsen, L., van Vliet, C., Stow, J.L., and Gleeson, P.A. (2001). The GRIP domain is a specific targeting sequence for a population of trans-Golgi network derived tubulo-vesicular carriers. Traffic 2, 336–344. [DOI] [PubMed] [Google Scholar]
- Cavenagh, M.M., Whitney, J.A., Carroll, K., Zhang, C-J, Boman, A.L., Rosenwaldi, A.G., Mellman, I., and Kahn, R.A. (1996). Intracellular distribution of ARF proteins in mammalian cells: ARF6 is uniquely localized to the plasma membrane. J. Biol. Chem. 271, 21767–21774. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., and Zerial, M. (2000). Purification of EEA1 from bovine brain cytosol using Rab5 affinity chromatography and activity assays. Methods 20, 403–410. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., Puertollano, R., Mullins, C., Aguilar, R.C., Vargas, J.D., Hartnell, L.M., and Bonifacino, J.S. (2000). GGAs: a family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Boshans, R.L., McDonough, M., Stahl, P.D., and Van Aelst, L. (1997). A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 16, 5445–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Erlich, R., Gleeson, P.A., Campbell, P., Dietzsch, E., and Toh, B.H. (1996). Molecular characterization of trans-Golgi p230. A human peripheral membrane protein encoded by a gene on chromosome 6p12–22 contains extensive coiled-coil alpha-helical domains and a granin motif. J. Biol. Chem. 271, 8328–8337. [DOI] [PubMed] [Google Scholar]
- Fritzler, M.J., Lung, C.C., Hamel, J.C., Griffith, K.J., and Chan, E.K. (1995). Molecular characterization of Golgin-245, a novel Golgi complex protein containing a granin signature. J. Biol. Chem. 270, 31262–31268. [DOI] [PubMed] [Google Scholar]
- Griffith, K.J., Chan, E.K., Lung, C.C., Hamel, J.C., Guo, X., Miyachi, K., and Fritzler, M.J. (1997). Molecular cloning of a novel 97-kd Golgi complex autoantigen associated with Sjogren's syndrome. Arthritis Rheum. 40, 1693–1702. [DOI] [PubMed] [Google Scholar]
- Hetzer, M., Gruss, O.J., and Mattaj, I.W. (2002). The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat. Cell Biol. 4, E177–E184. [DOI] [PubMed] [Google Scholar]
- Hirst, J., Lui, W.W., Bright, N.A., Totty, N., Seaman, M.N., and Robinson, M.S. (2000). A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J. Cell Biol. 149, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingley, E. et al. (1999). A novel ADP-ribosylation like factor (ARL-6), interacts with the protein-conducting channel SEC61beta subunit. FEBS Lett. 459, 69–74. [DOI] [PubMed] [Google Scholar]
- Jacobs, S., Schilf, C., Fliegert, F., Koling, S., Weber, Y., Schurmann, A., and Joost, H.G. (1999). ADP-ribosylation factor (ARF)-like 4, 6, and 7 represent a subgroup of the ARF family characterization by rapid nucleotide exchange and a nuclear localization signal. FEBS Lett. 456, 384–388. [DOI] [PubMed] [Google Scholar]
- Kjer-Nielsen, L., Teasdale, R.D., van Vliet, C., and Gleeson, P.A. (1999). A novel Golgi-localisation domain shared by a class of coiled-coil peripheral membrane proteins. Curr. Biol. 9, 385–388. [DOI] [PubMed] [Google Scholar]
- Klausner, R.D., Donaldson, J.G., and Lippincott-Schwartz, J. (1992). Brefeldin A: insights into the control of membrane traffic and organelle structure. J. Cell Biol. 116, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, S.L., Wong, S.H., and Hong, W. (1996). The mammalian ARF-like protein 1 (Arl1) is associated with the Golgi complex. J. Cell Sci. 109, 209–220. [DOI] [PubMed] [Google Scholar]
- Lu, L., Horstmann, H., Ng, C., and Hong, W.J. (2001). Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 114, 4543–4555. [DOI] [PubMed] [Google Scholar]
- Luke, M.R., Kjer-Nielsen, L., Brown, D.L., Stow, J.L., and Gleeson, P.L. (2002). GRIP domain-mediated targeting of two new coiled coil proteins, GCC88 and GCC185, to subcompartments of the trans-Golgi network. J. Biol. Chem. published November 20, 2002 as 10.1074/jbc.M210387200. [DOI] [PubMed]
- Munro, S., and Nichols, B.J. (1999). The GRIP domain—a novel Golgi-targeting domain found in several coiled-coil proteins. Curr. Biol. 9, 377–380. [DOI] [PubMed] [Google Scholar]
- Panic, B., Whyte, J.R.C., and Munro, S. (2003). The ARF-like GTPase Arl1p and Arl3p act in pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 13, 405–410. [DOI] [PubMed] [Google Scholar]
- Randazzo, P.A., Weiss, O., and Kahn, R.A. (1995). Preparation of recombinant ADP-ribosylation factor. Methods Enzymol. 219, 128–135. [DOI] [PubMed] [Google Scholar]
- Schurmann, A., Breiner, M., Becker, W., Huppertz, C., Kainulainen, H., Kentrup, H., and Joost, H.-G. (1994). Cloning of two novel ADP-ribosylation factor-like proteins and characterization of their differential expression in 3T3–L1 cells. J. Biol. Chem. 269, 15683–15688. [PubMed] [Google Scholar]
- Schurmann, A., Koling, S., Jacobs, S., Saftig, P., Krauss, S., Wennemuth, G., Kluge, R., and Joost, H.G. (2002). Reduced sperm count and normal fertility in male mice with targeted disruption of the ADP-ribosylation factor-like 4 (Arl4) gene. Mol. Cell. Biol. 22, 2761–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty, S.R.G., Shin, M., E., Yoshini, A., Marks, M.S., and Burd, C.G. (2003). Golgi Recruitment of GRIP domain proteins by Arf-like GTPase 1 is regulated by Arf-like GTPAse 3. Curr. Biol. 13, 401–404. [DOI] [PubMed] [Google Scholar]
- Sharer, J.D., Shern, J.F., Van Valkenburgh, H., Wallace, D.C., and Kahn, R.A. (2002). ARL2 and BART enter mitochondria and bind the adenine nucleotide transporter. Mol. Biol. Cell 13, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, V.N., bin Mohd Yusoff, A.R., Wong, S.H., Lim, G.B., Chew, M., and Hong, W. (1992). Biochemical fractionation and characterization of proteins from Golgi-enriched membranes. J. Biol. Chem. 267, 12016–12021. [PubMed] [Google Scholar]
- Tamkun, J.W., Kahn, R.A., Kissinger, M., Brizuela, B., Rulka, C., Scott, M.P., and Kennison, J.A. (1991). The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc. Natl. Acad. Sci. USA 88, 3120–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, C.E., and Brown, M.C. (2001). Cell motility: ARNO and ARF6 at the cutting edge. Curr. Biol. 11, R875–R877. [DOI] [PubMed] [Google Scholar]
- Van Valkenburgh, H., Shern, J.F., Sharer, J.D., Zhu, X., and Kahn, R.A. (2001). ADP-ribosylation factors (ARFs) and ARF-like 1 (ARL1) have both specific and shared effectors: characterizing ARL1-binding proteins. J. Biol. Chem. 276, 22826–22837. [DOI] [PubMed] [Google Scholar]
- Xu, Y., Hortsman, H., Seet, L., Wong, S.H., and Hong, W. (2001). SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 3, 658–666. [DOI] [PubMed] [Google Scholar]
- Zeng, Q., Tan, Y.H., and Hong, W. (1998). A single plasmid vector (pSTAR) mediating efficient tetracycline-induced gene expression. Anal. Biochem. 259, 187–194. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107–117. [DOI] [PubMed] [Google Scholar]