Abstract
In Saccharomyces cerevisiae, polarized morphogenesis is critical for bud site selection, bud development, and cell separation. The latter is mediated by Ace2p transcription factor, which controls the daughter cell-specific expression of cell separation genes. Recently, a set of proteins that include Cbk1p kinase, its binding partner Mob2p, Tao3p (Pag1p), and Hym1p were shown to regulate both Ace2p activity and cellular morphogenesis. These proteins seem to form a signaling network, which we designate RAM for regulation of Ace2p activity and cellular morphogenesis. To find additional RAM components, we conducted genetic screens for bilateral mating and cell separation mutants and identified alleles of the PAK-related kinase Kic1p in addition to Cbk1p, Mob2p, Tao3p, and Hym1p. Deletion of each RAM gene resulted in a loss of Ace2p function and caused cell polarity defects that were distinct from formin or polarisome mutants. Two-hybrid and coimmunoprecipitation experiments reveal a complex network of interactions among the RAM proteins, including Cbk1p–Cbk1p, Cbk1p–Kic1p, Kic1p–Tao3p, and Kic1p–Hym1p interactions, in addition to the previously documented Cbk1p–Mob2p and Cbk1p–Tao3p interactions. We also identified a novel leucine-rich repeat-containing protein Sog2p that interacts with Hym1p and Kic1p. Cells lacking Sog2p exhibited the characteristic cell separation and cell morphology defects associated with perturbation in RAM signaling. Each RAM protein localized to cortical sites of growth during both budding and mating pheromone response. Hym1p was Kic1p- and Sog2p-dependent and Sog2p and Kic1p were interdependent for localization, indicating a close functional relationship between these proteins. Only Mob2p and Cbk1p were detectable in the daughter cell nucleus at the end of mitosis. The nuclear localization and kinase activity of the Mob2p–Cbk1p complex were dependent on all other RAM proteins, suggesting that Mob2p–Cbk1p functions late in the RAM network. Our data suggest that the functional architecture of RAM signaling is similar to the S. cerevisiae mitotic exit network and Schizosaccharomyces pombe septation initiation network and is likely conserved among eukaryotes.
INTRODUCTION
The establishment and maintenance of cell polarity are critical for proper cellular function and development. Indeed, the importance of cell polarity is abundantly evident in a variety of cells, such as neurons and intestinal epithelial cells, for which polarized morphogenesis is essential for their specialized functions (Andersen and Bi, 2000). The temporal and spatial regulation of cell polarity and morphogenesis seems to be highly complex, involving the integration of multiple signaling networks, including those that regulate cell size and cell division (Knust, 2000; Rupes, 2002).
In the budding yeast Saccharomyces cerevisiae, multiple signaling networks coordinate bud emergence, polarized growth, and cell cycle progression (Pruyne and Bretscher, 2000a,b; Casamayor and Snyder, 2002). In response to favorable nutrient conditions, cell size, and initiation of cell cycle progression, a Ras-like GTPase signaling network becomes activated to specify the site of bud emergence. Coincident with initiation of DNA replication, the G1/S phase cyclin-dependent kinase (CDK) complex, Cln1/2p–Cdc28p, promotes bud emergence through activation of the Cdc42p GTPase cycle, which controls the assembly of macromolecular complexes containing multiple polarity determinant proteins (Lew and Reed, 1995; Pruyne and Bretscher, 2000a,b). In particular, the formin protein Bni1p controls the assembly of actin cables, which guide myosin-motor directed secretion (Evangelista et al., 2002; Pruyne et al., 2002; Sagot et al., 2002). During the initial stages of bud development, secretion is polarized to the bud tip, such that growth is focused to the bud apex. Later in the cell cycle, a switch is made from apical to isotropic growth, such that secreted materials are directed uniformly over the entire surface of the developing bud. This switch correlates with the coupling of CDK to the mitotic cyclins, Clb1/2p, and subsequent activation of a network involving the Nim1-like kinase Gin4p and the PAK-like kinase Cla4p, which terminate Cdc42p signaling (Gulli et al., 2000). At the end of mitosis, polarized cell growth is redirected from the bud cortex to the mother-daughter cell junction at the bud neck to facilitate septum formation and cell separation (Kilmartin and Adams, 1984).
The mechanisms involved in polarized growth in S. cerevisiae are also required for the formation of mating projections. Chemotropic growth of mating projections enables haploid cells of opposite mating types to make contact and subsequently form a diploid zygote through cell and nuclear fusion. Mating pheromone activates a cell surface G protein-coupled receptor, which stimulates a downstream mitogen-activated kinase kinase pathway and Cdc42p-signaling to induce the expression of mating genes, G1 cell cycle arrest, and polarized secretion, leading to the formation of a mating projection or shmoo (Leberer et al., 1997; Elion, 2000). Like polarized bud growth, formation of polarized mating projections requires Bni1p-mediated assembly of actin cables, which direct the secretion of plasma membrane and cell wall components to the growing tip of the cell (Evangelista et al., 2002; Pruyne et al., 2002; Sagot et al., 2002).
The S. cerevisiae Mob2p–Cbk1p kinase complex has been implicated in the regulation of polarized growth during budding and mating (Colman-Lerner et al., 2001; Weiss et al., 2002). Cbk1p kinase belongs to a conserved family of serinethreonine protein kinases, whose members include Dbf2p, a component of the S. cerevisiae mitotic exit network (MEN), S. pombe Orb6 and Sid2, and mammalian Ndr and LATS kinases (Verde et al., 1998; Racki et al., 2000; Bardin and Amon, 2001; Bidlingmaier et al., 2001). Cbk1p kinase activity peaks during periods of polarized bud growth and during late mitosis (Weiss et al., 2002). Cbk1p activity is dependent on Mob2p, a member of the Mob family of proteins that includes Mob1p, a Dbf2p binding protein and component of the S. cerevisiae MEN (Luca and Winey, 1998; Weiss et al., 2002). Cells deleted for either CBK1 or MOB2 are round, as are cells expressing a catalytically inactive form of Cbk1p (Racki et al., 2000; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002). Similarly, S. pombe mutant cells that lack CBK1 and MOB2 orthologs are round (Verde et al., 1998; Hou et al., 2003). The round cell morphology of cbk1Δ and mob2Δ mutants indicates a failure to establish or maintain apical bud growth. Consistent with a general role for the Mob2p–Cbk1p protein complex in polarized morphogenesis, mob2Δ and cbk1Δ mutants are defective in pheromone-induced projection formation (Bidlingmaier et al., 2001; Weiss et al., 2002). Moreover, the conditional inactivation of Cbk1p kinase results in the depolarization of the actin cytoskeleton at the growing tips of mating projections (Weiss et al., 2002).
In addition to their role in polarized growth, Cbk1p and Mob2p are critical for regulating the localization and activity of Ace2p transcription factor, which mediates mother-daughter cell separation at the end of mitosis (Colman-Lerner et al., 2001; Weiss et al., 2002). In wild-type cells, Ace2p localizes to daughter cell nuclei during mitotic exit and activates a set of genes that are essential for mother-daughter cell separation, such as CTS1 and SCW11, which encode proteins involved in septum degradation (Dohrmann et al., 1992; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Doolin et al., 2001; Weiss et al., 2002). Consistent with their dual roles in cell polarity and Ace2p regulation, Cbk1p and Mob2p localize to sites of cortical growth during G1 through mid-mitosis and to the bud neck region and the daughter cell nucleus at the end of mitosis (Colman-Lerner et al., 2001; Weiss et al., 2002). Cells deleted for ACE2, CBK1, or MOB2 display similar cell separation defects; however, unlike cbk1Δ and mob2Δ mutant cells, ace2Δ cells are not defective in polarized morphogenesis (Racki et al., 2000; Bidlingmaier et al., 2001). Thus, the role of the Mob2p–Cbk1p protein complex in Ace2p regulation is distinct from its role in polarized morphogenesis.
The mechanism for Mob2p–Cbk1p kinase activation remains to be elucidated. However, given the sequence similarities between Mob1p and Mob2p and between their respective kinase partners, Dbf2p and Cbk1p, it is likely that the Mob2p–Cbk1p complex is activated within a signaling pathway whose circuitry resembles that of the S. cerevisiae MEN and S. pombe septation initiation network (SIN) (Bardin and Amon, 2001). So far, Hym1p, an ortholog of the Aspergillus nidulans hyphal growth protein hymA and the mouse MO25 protein (Miyamoto et al., 1993; Karos and Fischer, 1999), and Tao3p (Pag1p), a 270 kDa protein conserved from yeast to humans of undefined molecular role, are the only known proteins that seem to function within the Mob2p-Cbk1p pathway (Dorland et al., 2000; Bidlingmaier et al., 2001; Du and Novick, 2002). However, Hym1p- and Tao3p-like proteins have not been implicated in MEN or SIN signaling.
Herein, we used a variety of approaches, including microarray analysis, protein–protein interaction methodologies, live cell microscopy, and in vitro kinase assays to investigate the functional relationships between Cbk1p, Mob2p, Tao3p, and Hym1p. We conducted genetic and two hybrid screens to identify additional genes involved in both polarized morphogenesis and cell separation. We present evidence that the serine-threonine kinase Kic1p (Sullivan etal., 1998) and a novel leucine-rich repeat (LRR)-containing protein Sog2p collaborate with Cbk1, Mob2p, Tao3p, and Hym1p to regulate Ace2p transcriptional activity and polarized morphogenesis. Each of these proteins localizes similarly to sites of polarized growth and displays a complex array of physical interactions and interdependencies for subcellular localization, indicating an intimate functional relationship. Notably, we establish that Mob2p–Cbk1p activity is dependent on Kic1p, Tao3p, Hym1p, and Sog2p, and we provide evidence that Hym1p, Sog2p, and Kic1p interact to form a functional unit. Given the sequence similarity of Kic1p to the MEN kinase Cdc15p, which activates the Mob1p–Dbf2p kinase complex directly (Mah et al., 2001), it is probable that Kic1p activates Mob2p-Cbk1p for regulating Ace2p and cellular morphogenesis.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
The yeast strains used in this study (Table 1) are derived from W303 (Evangelista et al., 1997) or S288C (Roberts et al., 2000; Weiss et al., 2002) and were cultured as described previously. Strains deleted for BNI1 (Y3314), CBK1 (Y1747), MOB2 (Y3424), TAO3 (Y3152), HYM1 (Y1560), KIC1 (Y3323), and SOG2 (FLY1492), were created using a polymerase chain reaction (PCR)-based targeted gene replacement strategy, as described previously (Longtine et al., 1998).
Table 1.
Strain list
| Name | Relevant Genotype | Source |
|---|---|---|
| SY2585 | MATa his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,—112 trp 1-1 ade2-1 can 1-100 | Sprague lab |
| SY2625 | MATa bar 1Δhis3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | Sprague lab |
| W303-1A | MATa ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | |
| Y282 | MATa lys2Δ::FUS1-pADE8hoΔ::GAL1-α2 ade8Δ his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y483 | MATa bnilΔ::URA3 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y580 | MATa spa2Δ::LEU2 trp1 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 ade2-1 can1-100 | This study |
| Y581 | MATa bnilΔ::LEU2 trp1 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 ade2-1 can1-100 | This study |
| Y587 | MATa bnilΔ::URA3 bar1::LEU2 trp1 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 ade2-1 can1-100 | This study |
| Y773 | MATa bud6Δ::URA3 bar1 his3::FUSI-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y837 | MATa bud6Δ::LEU2 bar1 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y860 | MATα 8 LEXA operator-ADE2::URA3 ade2-1 his3-11,15 leu2-3,117 trp1-1 | Evangelista et al. (2000) |
| Y871 | MATa hym1 bar1Δ::LEU2 lys2Δ::FUS1-pADE8 hoΔ::GAL1-α2 ade8Δhis3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y1026 | MATa ura3::URA3-LexAop-LacZ ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | Evangelista et al. (2000) |
| Y1028 | MATa/α ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y1560 | MATa hym1Δ::URA3 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y1603 | MATa hym1Δ::HIS3 ura3-1 leu2-3, 112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y1614 | MATa hym1Δ::LEU2 ura3-1 leu2-3, 112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y1622 | MATa/α hym1Δ::LEU2/hym1Δ::LEU2 ura3-1/ura3-1 leu2-3, 112/leu2-3, 112 his3-11,-15/his3-11,-15 trp1-1/trp1-1 ade2-1/ade2-1 can1-100/can1-100 | This study |
| Y1726 | MATa cbk1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 can1-100 | R. Geyer |
| Y1744 | MATa hym1Δ::URA3 bar1Δ::LEU2 ura3-1 leu2-3,-112 can1-100 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ | This study |
| Y1747 | MATa cbk1Δ::kanMX6 ura3-1 leu2-3,-112 can1-100 his3::FUS1-HIS3 ade2-1 mfa2Δ::FUS1 lacZ | This study |
| Y1749 | MATa hym1::URA3 cbk1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y2619 | MATa/α bni1::kanMX6/bni1Δ:LEU2 trp1/trp1 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ ura3-1/ura3-1 leu2-3,-112/leu2-3,-112 ade2-1/ade2-1 can1-100/can1-100 | This study |
| Y2623 | MATa/α cbk1Δ::kanMX6/cbk1Δ::kanMX6 ura3-1/ura3-1 leu2-3,-112/leu2-3,-112 can1-100/can1-100 his3::FUS1-HIS3 mfa2Δ::FUS1-lacZ | This study |
| Y2862 | MATa his3::OCH1-HIS3 mfa2-Δ1::OCH1-lacZ ura3-1 leu2-3,-112 his3-11,-15 trp1-ΔFA ade2-1 can1-100 | This study |
| Y3152 | MATa tao3Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y3280 | MATa CBK1-13myc::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y3320 | MATa ACE2-GFP::HIS3MX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y3323 | MATa kic1Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y3382 | MATa KIC1-GFP::HIS3MX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y3424 | MATa mob2Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| Y3481 | MATa tao3Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 | This study |
| Y3482 | MATa mob2Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 lys2 | This study |
| Y3487 | MATa kic1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 lys2 | This study |
| Y3527 | MATa/αmob2Δ::kanMX6/mob2Δ::kanMX6 bar1Δ::LEU2/BAR1 ura3-1/ura3-1 leu2-3,-112/leu2-3,-112 his3-11,-15/his3-11,-15 trp1-1/trp1-1 ade2-1/ade2-1 can1-100/can1-100 | This study |
| Y3529 | MATa/α kic1Δ::kanMX6/kic1Δ::kanMX6 bar1Δ::LEU2/BAR1 ura3-1/ura3-1 leu2-3,-112/leu2-3,-112 his3-11,-15/his3-11,-15 trp1-1/trp1-1 ade2-1/ade2-1 can1-100/can1-100 lys2/LYS2 | This study |
| Y3565 | MATa/α tao3Δ::kanMX6/tao3Δ::kanMX6 bar1Δ::LEU2/BAR1 ura3-1/ura3-1 leu2-3,-112/leu2-3,-112 his3-11,-15/his3-11,-15 trp1-1/trp1-1 ade2-1/ade2-1 can1-100/can1-100 lys2/LYS2 | This study |
| Y3606 | MATa kic1Δ::kanMX6 cbk1Δ::kanMX6 bar 1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 lys2 | This study |
| Y3608 | MATa hym1Δ::URA3 mob2Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y3609 | MATa hym1Δ::URA3 tao3Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y3615 | MATa KIC1 - HAx3::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-10 | This study |
| Y3625 | MATa kic1Δ::kanMX6 mob2Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 lys2 | This study |
| Y3626 | MATa mob2Δ::kanMX6 cbk1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 lys2 | This study |
| Y3627 | MATa tao3Δ::kanMX6 cbk1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 lys2 | This study |
| Y3628 | MATa hym1Δ::URA3 kic1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y3644 | MATa tao3Δ::kanMX6 kic1Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 | This study |
| Y3645 | MATa tao3Δ::kanMX6 mob2Δ::kanMX6 bar1Δ::LEU2 ura3-1 leu2-3,-112 trp1-1 ade2-1 can1-100 lys2 | This study |
| Y4032 | MATa CBK1-13myc::HIS3MX6 hym1Δ::URA3 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 trp1-1 | This study |
| Y4033 | MATa CBK1-13myc::HIS3MX6 tao3Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 trp1-1 | This study |
| Y4034 | MATa CBK1-13myc::HIS3MX6 kic1Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 trp1-1 | This study |
| Y4035 | MATa CBK1-13myc::HIS3MX6 mob2Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 trp1-1 | This study |
| Y4089 | MATa HϒM1-13myc::kanMX6 KIC1 - HAx3::kanMX6 ura3-52 leu2-3, 112 trp1Δ1 his3Δ200 | This study |
| Y4122 | MATa ACE2 - GFP::HIS3MX6 cbk1Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 lys2 | This study |
| FLY811 | MATa ACE2-GFP::kanMX6 ura3-52 leu2-3,-112 trp1Δ1 his3Δ200 | Weiss et al. (2002) S |
| FLY849 | MATa ACE2-GFP::kanMX6 mob2Δ::HIS3 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | Weiss et al. (2002) S |
| FLY853 | MATa ACE2-GFP::kanMX6 cbk1Δ::HIS3MX6 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | Weiss et al. (2002) S |
| FLY891 | MATa HϒM1-GFP::kanMX6 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | Weiss et al. (2002) S |
| FLY893 | MATa/α MOB2-GFP::kanMX6/MOB2-GFP::HIS3MX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200/ | Weiss et al. (2002) S |
| FLY895 | MATa/α CBK1-GFP::KANMX/CBK1-GFP::kanMX6 ura3-52/ura3-52 leu2-3, 112/leu2-3, 112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | Weiss et al. (2002) S |
| FLY947 | MATa KIC1-GFP::kanMX6 ura3 leu2-3,112 trp1 his3 | This study |
| FLY1081 | MATa ACE2-GFP::kanMX6 hym1Δ::URA3 ura3 leu2-3,112 trp1 his3 | This study |
| FLY1124 | MATa ACE2-GFP::kanMX6 tao3Δ::kanMX6 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | This study |
| FLY1134 | MATa ACE2-GFP::kanMX6 kic1Δ::kanMX6 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | This study |
| FLY1244 | MATa/α HϒM1-GFP::kanMX6/HϒM1-GFP::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200 | This study |
| FLY1245 | MATa/αHϒM1-GFP::kanMX6/HϒM1-GFP::kanMX6 mob2Δ::HIS3/mob2Δ::HIS3 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1246 | MATa/α HϒM1-GFP::kanMX6/HϒM1-GFP::kanMX6 cbk1Δ::HIS3MX6/cbk1Δ::HIS3MX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1247 | MATa/α KIC1-GFP::kanMX6 mob2Δ::HIS3 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1248 | MATa/αKIC1-GFP::kanMX6/KIC1-GFP::kanMX6 cbk1Δ::HIS3MX6/cbk1Δ::HIS3MX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1249 | MATa/α KIC1-GFP::kanMX6/KIC1-GFP::kanMX6 tao3Δ::kanMX6/tao3Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1258 | MATa/α KIC1-GFP::kanMX6/KICI-GFP::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1259 | MATa/α CBK1-GFP::kanMX6/CBK1-GFP::kanMX6 tao3Δ::kanMX6/tao3Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1260 | MATa/α MOB2-GFP::HIS3MX6/MOB2-GFP::HIS3MX6 tao3Δ::kanMX6/tao3Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1261 | MATa/α HϒMI-GFP::kanMX6/HϒM1-GFP::kanMX6 kic1Δ::kanMX6/kic1Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1262 | MATa/α HϒM1-GFP::kanMX6/HϒM1-GFP::kanMX6 tao3Δ::kanMX6/tao3Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1263 | MATa TAO3-GFP::kanMX6 ura3-52 leu2-3, 112 trp1Δ1 his3Δ200 | This study |
| FLY1267 | MATa/α TAO3-GFP::HIS3MX6/TAO3-GFP::HIS3MX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1268 | MATa CBK1-13myc::HIS3MX6 ace2Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 trp1-1 ade2-1 can1-100 | This study |
| FLY1278 | MATa/α KIC1-GFP::kanMX6/KIC1-GFP::kanMX6 hym1Δ::URA3/hym1Δ::URA3 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1324 | MATa/α TAO3-GFP::HIS3MX6/TAO3-GFP::HIS3MX6 cbk1Δ::kanMX6/cbk1Δ::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1325 | MATa/α TAO3-GFP::HIS3MX6/TAO3-GFP::HIS3MX6 kic1Δ::kanMX6/kic1Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1326 | MATa/α TAO3-GFP::HIS3MX6/TAO3-GFP::HIS3MX6 hym1Δ::URA3/hym1Δ::URA3 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1342 | MATa/α MOB2-GFP::HIS3MX6/MOB2-GFP::HIS3MX6 hym1Δ::URA3/hym1Δ::URA3 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1343 | MATa/α MOB2-GFP::HIS3MX6/MOB2-GFP::HIS3MX6 kic1Δ::kanMX6/kic1Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1344 | MATa/α CBK1-GFP::kanMX6 kic1Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1347 | MATa SOG2-GFP::kanMX6 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | This study |
| FLY1382 | MATa/α SOG2-GFP::kanMX6/SOG2-GFP::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1383 | MATa/α CBK1-GFP::kanMX6/CBK1-GFP::kanMX6 hym1Δ::URA3/hym1Δ::URA3 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1386 | MATa/α TAO3-GFP::HIS3MX6/TAO3-GFP::HIS3MX6 mob2Δ::HIS3/mob2Δ::HIS3 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1465 | MATa/α KIC1-GFP::kanMX6/KIC1-GFP::kanMX6 sog2Δ::kanMX6/sog2Δ::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1470 | MATα Ace2-GFP::kanMX6 sog2Δ::kanMX6 ura3-52 leu2-3,112 trp1Δ1 his3Δ200 | This study |
| FLY1471 | MATa/α SOG2-GFP::kanMX6/SOG2-GFP::kanMX6 kic1Δ::kanMX6/kic1Δ::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1472 | MATa/α SOG2-GFP::kanMX6/SOG2-GFP::kanMX6 hym1Δ::URA3/hym1Δ::URA3 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1473 | MATa/α SOG2-GFP::kanMX6/SOG2-GFP::kanMX6 tao3Δ::kanMX6/tao3Δ::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1474 | MATa/α SOG2-GFP::kanMX6/SOG2-GFP::kanMX6 mob2Δ::HIS3/mob2Δ::HIS3 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1475 | MATa/α SOG2-GFP::kanMX6/SOG2-GFP::kanMX6 cbk1Δ::HIS3 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1492 | MATa CBK1 CBK1-13myc::HIS3MX6 sog2Δ::kanMX6 ura3-1 leu2-3,-112 his3-11,-15 ade2-1 can1-100 trp1-1 | This study |
| FLY1499 | MATa/α MOB2-GFP::HIS3MX6/MOB2-GFP::HIS3MX6 sog2Δ::kanMX6/sog2Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1500 | MATa/α TAO3-GFP::HIS3MX6/TAO3-GFP::HIS3MX6 sog2Δ::kanMX6/sog2Δ::kanMX6 ura3/ura3 leu2-3,-112/leu2-3,112 trp1/trp1 his3/his3 | This study |
| FLY1514 | MATa/α HYM1-GFP::kanMX6/HYM1-GFP::kanMX6 sog2Δ::kanMX6/sog2Δ::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
| FLY1515 | MATa/α CBK1-GFP::kanMX6/CBK1-GFP::kanMX6 sog2Δ::kanMX6/sog2Δ::kanMX6 ura3-52/ura3-52 leu2-3,112/leu2-3,112 trp1Δ1/trp1Δ1 his3Δ200/his3Δ200 | This study |
Cells expressing C-terminal–tagged proteins (3 × hemagglutinin [HA], 13 × Myc, or green fluorescent protein [GFP]) were constructed via PCR-based gene fusion, as described previously (Longtine et al., 1998). These alleles did not cause any observable phenotypes, indicating that the tags did not interfere with protein function. Double mutant combinations were constructed through standard genetic crossing methods.
Identification of Bilateral Mating Mutants
Cells were engineered to select for mutants with reduced mating efficiency. Mutant ade2Δ cells grown in medium containing low levels of adenine are red, due to the accumulation of a red biosynthetic precursor of adenine (Ugolini and Bruschi, 1996). ade2Δ ade8Δ double mutants fail to accumulate the red pigment due to a blockage at an earlier step in biosynthesis and therefore grow as white colonies. In haploid cells, α2 encodes a transcriptional repressor of MATa-specific genes. In diploid cells, a1-α2 represses haploid-specific genes, such as FUS1 and HO. FUS1pr-ADE8 (ADE8 behind the FUS1 promoter) was used as a haploid-specific pheromone response reporter. When integrated into the genome of a haploid ade2Δ ade8Δ cells, the basal activation of FUS1 expression is strong enough to produce adequate amounts of Ade8p, thus yielding red colonies. Colonies that successfully mate will produce diploid cells that will repress the production of FUS1pr-ADE8 (due to a1-α2 repression of haploid-specific genes) and therefore look white, due to a lack of production of Ade8p. Mutations in the pheromone response cascade repress the basal signaling of FUS1 and thus yield white colonies. We selected for more subtle bilateral mating mutants that cannot mate efficiently and therefore continue to express FUS1pr-ADE8. Colonies of such mutants are red or contain red and white sectors when grown on adenine-deficient plates.
Strain Y282 (MATa ade2Δ ade8Δ GAL1pr-MATα2, FUS1pr-ADE8) was transformed with p1240, a YCp50 (CEN, URA3) plasmid containing HO, which encodes an endonuclease that catalyzes mating type conversion. Cells were grown in synthetic medium lacking uracil and containing 2% galactose and plated onto rich medium containing minimal amounts of adenine and 2% glucose. They were then immediately mutagenized with UV light to a 10% survival rate. Of the 20,000 colonies screened, 262 were chosen for further analysis based on a red sectoring quality. Colonies were streaked onto 5-fluoroorotic acid medium to counterselect for HO plasmid and assayed for mating type. MATa isolates were assayed for G1 arrest, by using a modified protocol from (Fink and Styles, 1972) and for pheromone production and response as described previously (Boone et al., 1993). Of 262 colonies, 75 were found to be normal for these assays and subjected to further analysis. FAR1 was transformed into these strains and found to complement 11, which were subsequently removed from further analysis. One of the mutants that showed a round cell morphology, and a cell separation defect (Y871) was transformed with a YCp50 (CEN, URA3)-based genomic library. Approximately 14,000 colonies were screened for restored mating ability. Plasmids were recovered and sequenced from cells exhibiting an increased mating efficiency. A single complementing plasmid clone was isolated (p1319) that contained the HYM1 gene and flanking sequences. HYM1 was subcloned and confirmed to complement the mutant phenotypes of Y871.
Isolation of Cell Separation Mutants
Y2862, which contains an integrated copy of OCH1pr-HIS3 and OCH1pr-lacZ, was transformed with an mTn-3 × HA/GFP mutagenized library (Burns et al., 1994) and plated onto synthetic medium lacking uracil and histidine and containing 15 mM 3-amino triazole. Colonies (340,000) were screened for growth on medium containing 15 mM 3-amino triazole, and 59 colonies were selected. Eight colonies of the 59 also exhibited a cell separation phenotype and round cell morphologies. These mutants were placed into four complementation groups and direct genomic sequencing was used to determine the identity and transposon insertion site of each of the mutants (Horecka and Jigami, 2000). Four alleles of TAO3, two alleles of KIC1, and single alleles of MOB2 and CBK1 were recovered. All mutations showed elevated OCH1 expression (approximately twofold above wild type) by lacZ measurement (Horecka, unpublished data). Only cbk1Δ cells showed elevated levels of OCH1 expression (twofold) by DNA microarray analysis, which illustrates the greater sensitivity of the lacZ-reporter for analysis of gene expression.
Suppression of Cell Separation Mutants
Y1614 (hym1Δ::LEU2), which is defective for cell separation, was transformed with the YEp24-based (URA3, 2 μ) genomic library. Approximately 18,000 colonies were pooled and placed vertically at room temperature for 40 min and allowed to sediment. A small volume (0.25 ml) of liquid was transferred from the culture and transferred to 50 ml of fresh SD-URA and grown at 30°C to saturation. This enrichment for nonsedimenting cells was repeated for two additional rounds. Liquid was taken from the surface of the culture and plated onto SD-URA at a density of ∼200 colony-forming units/plate. Forty-nine round, smooth colonies were chosen for microscopic analysis to confirm cell separation. Plasmids were extracted and examined by restriction analysis, one was sequenced and found to contain a 10.8-kb insert containing ACE2 (p3679).
DNA Microarrays
Yeast cultures were grown for three generations to mid-logarithmic phase in synthetic complete medium. Cells were pelleted and frozen in liquid nitrogen. RNA extractions and hybridizations were performed as described previously (Roberts et al., 2000).
Assays for Development of Mating Projections
Logarithmically growing MATa cells were exposed to 50 nM synthetic α-factor for 2 h and stained with rhodamine-phalloidin to visualize filamentous actin. Cells were scored for ability to polarize filamentous actin patches to discrete sites at the tips of cells.
Analysis of Budding Patterns
Strains expressing a BUD4-containing plasmid (p2698; a gift from Silvia Sanders, Massachusetts Institute of Technology, Cambridge, MA) or the vector pRS316 were grown in medium lacking uracil to mid-logarithmic phase. Cells were sonicated for cell separation and Calcofluor White was added to 2 μg/ml concentration. Bud scars were visualized by fluorescence microscopy and bud patterns were defined as described previously (Chant and Pringle, 1995). Only cells containing three or more bud scars were counted. Bud scars residing on opposite thirds of the cells were considered to be bipolar. Bud patterns were considered axial only if scars resided immediately adjacent to one another. Cells were considered to have random patterns if bud scars resided in the middle third of the cell.
Two-Hybrid Screens and Assays
Two-hybrid bait and prey plasmids are listed in Table 2 and were based on pEG202, which encodes the LexA-DNA binding domain, and pJG4-5, which encodes the B42 transcriptional activation domain (Gyuris et al., 1993). DNA fragments of various genes were amplified by the PCR with primers that incorporated 5′-BamHI and 3′-NotI restriction sites (unless otherwise noted in Table 2) for insertion into the two-hybrid vectors.
Table 2.
Two-hybrid plasmids
| Plasmid name | Insert | Vector |
|---|---|---|
| p2139 | Hym1p (1-300) Xho1 | pEG202 Xho1 |
| p3411 | Cbk1p (1-347) Xho1 | pEG202 Xho1 |
| p3412, | Cbk1p (1-112) Xho1 | pEG202 Xho1 |
| p4424 | Kic1p (1-282) BgIII | pEG202 BamHI |
| p4429 | Tao3p (1-510) Xho1 | pEG202 Xho1 |
| p4434 | Tao3p (1141-1775) Xho1 | pEG202 Xho1 |
| p4437 | Tao3p (1776-2377) Xho1 | pEG202 Xho1 |
| p4445 | Tao3p (1776-2377) Xho1 | pEG202 Xho1 |
| p2872 | Cbk1p (1-756) | pJG4-5 |
| p3413 | Cbk1p (346-756) | pJG4-5 |
| p3415 | Cbk1p (1-347) | pJG4-5 |
| p3416 | Cbk1p (1-117) | pJG4-5 |
| p4422 | Mob2p (1-326) | pJG4-5 |
| p4423 | Kic1p (1-282) BgIII | pJG4-5 BamHI |
| p4426 | Kic1p (283-1081) BgIII | pJG4-5 BamHI |
| p4428 | Tao3p (1-510) | pJG4-5 |
| p4431 | Tao3p (511-1140) | pJG4-5 |
| p4433 | Tao3p (1141-1775) | pJG4-5 |
| p4436 | Tao3p (1776-2377) | pJG4-5 |
Two-hybrid assays were performed as described previously (Phizicky and Fields, 1995) by mating Y1026, which contains a LexA DNA-binding domain plasmid, and Y860, which contains a prey plasmid. Diploids were grown on selective media and subjected to β-galactosidase expression analysis (Sheu et al., 2000). Strains qualitatively more reactive than the vector control were classified as positive. p2139 was used as bait to identify genes encoding Hym1p-binding proteins from a yeast cDNA library derived from pACT (Durfee et al., 1993). Plasmids encoding positive interactors were recovered and sequenced. One plasmid, designated p3153, contained a fragment of YOR353c (SOG2).
Plasmid Construction
To construct yeast strain Y282, several integrating plasmids (p40, p65 and p89) were created. The plasmid p40 introduces a frame-shift mutation at codon 45 of ADE8 and was created in a three-step process. First, a 2-kb EcoRI-HindIII ADE8 fragment was cloned into YIplac211 (URA3; Gietz and Sugino, 1988) to create p23. p23 was cut with EcoRI and SnaBI, treated with Klenow and dNTPs, and religated to remove an MscI site. The resultant plasmid, designated p38, was cut with Eco47III-HpaI and ligated to a BamHI linker (5′-CGGGATCCCG-3′) to create an ADE8 frame-shift mutation. This plasmid (p40) was cut with MscI to target integration to the ADE8 locus. p65 was used to integrate GAL1-α2 at the HO locus, thereby disrupting HO after codon 83. It was constructed by cutting YI-plac211 with PvuII to remove the multicloning site. A HindIII linker (5′-CCCAAGCTTGGG-3′) was then ligated to create p41, which was subsequently cut with HindIII and ligated to a HindIII fragment carrying HO and its promoter, creating p53. p53 was cut with PstI-BamHI and ligated with an oligonucleotide linker containing EcoRI-SstI sites; the resultant plasmid was cut with EcoRI and SstI and ligated to a EcoRI-SstI fragment containing the GAL1 promoter (p113, Boone laboratory collection) and a BamHI-SstI fragment containing a MATα2 PCR product. The MATα2 fragment was PCR amplified from genomic DNA by using primers AAAGGATCCA AAATGAGAAC GGCCGTA and AAAGAGCTCT TGGAAAAATC CATTAACT, which incorporate BamHI and SstI sites at the 5′ and 3′ ends, respectively.
p89 was used to introduce the FUS1pr-ADE8 reporter was constructed as follows. A YIp plasmid containing a 4.8 Kb LYS2 genomic fragment (pCP6; provided by Eric Foss, Fred Hutchinson Cancer Research Center, Seattle, WA), was digested with HpaI and ligated to linkers to introduce HindIII-SrfI-XbaI sites within the LYS2 gene. The resultant plasmid (p72) was cut with HindIII and XbaI and ligated to the HindIII-NheI fragment of FUS1pr-ADE8 from a pUC13-based plasmid (p51), to create p89.
The human MST3 gene was cloned from a pool of infant brain, placenta, spinal chord, and colon total RNAs (BD Biosciences Clonetech, Palo Alto, CA) by reverse transcription-PCR by using the primers CGCGGATATC ACCATGGCTC ACTCCCCGGT GCA and CGCGGTCGAC GTGGGATGAA GTTCCTCCAC CACT. The PCR fragment was ligated into EcoRV and SalI-digested pCMV-TAG 4A (Stratagene, La Jolla, CA), which resulted in a C-terminal FLAG-tagged MST3 cDNA under the control of the cytomegalovirus promoter, creating pMST3-FLAG.
Yeast Immunoprecipitation and Protein Kinase Assays
For coprecipitation experiments, 1-liter culture of yeast cells was grown to early log phase in rich media, and extracts were prepared by grinding frozen cell pellets in lysis buffer (0.1% Triton X-100, 50 mM Tris-HCl, 100 mM NaCl, 10 mM EDTA) containing a protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Immunoprecipitations were carried out with 1.5-ml extracts, containing 20–25 mg/ml total protein, monoclonal antibody HA.11 (Babco, Richmond, CA), or monoclonal antibody c-Myc (Babco) and G-Sepharose beads (Pharmacia, Peapack, NJ). Total yeast extract (25 μg) was analyzed on immunoblots, and 10 and 90% of the immunoprecipitated material was analyzed for detection of the immunoprecipitated and coprecipitating proteins, respectively. Immunoblot analysis was performed as described previously (Peter et al., 1993) and probed using rabbit polyclonal HA antibody (Babco) and rabbit polyclonal c-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, CA).
For Cbk1p protein kinase assays, cells were grown to mid-log phase and synchronized in G1 phase with mating pheromone. Cell extracts were prepared by glass bead lysis as described previously (Weiss et al., 2002). Cell extracts were normalized to 5 mg/ml and immunoprecipitated by incubating in 4 μg of anti-Myc antibody (9E10) for 45 min at 4°C followed by 25 μl of Gamma-bind G-Sepharose (Pharmacia) for 45 min at 4°C. Immunoprecipitated material was washed four times in lysis buffer and three times in kinase buffer (50 mM HEPES pH 7.4, 60 mM sodium acetate, 10 mM MgCl2, 1 mM dithiothreitol). Half the immunoprecipitated protein was processed for SDS-PAGE and immunoblotted. The other half was incubated in kinase buffer containing 5 μg of histone H1, 10 μM ATP, and 10 μCi of 32P-ATP for 30 min at 30°C. Kinase reactions were terminated by addition of protein sample buffer and were processed for PAGE and autoradiography. Relative kinase activity was determined using a GS-525 PhosphorImager (Bio-Rad, Hercules, CA) and Multianalyst software.
Fluorescence Microscopy
Live cell microscopy was performed as described previously (Luca et al., 2001; Weiss et al., 2002).
RESULTS
CBK1, MOB2, TAO3, HYM1, and KIC1 Function in the Same Signaling Network
To identify S. cerevisiae genes that are required for polarized morphogenesis during both budding and mating, we performed two different genetic screens. First, we used a screen designed to uncover mutations that caused decreased efficiency in bilateral mating and defects in the formation of mating projections (see MATERIALS AND METHODS). From this screen, we identified an allele of HYM1 that exhibited cell separation and round cell morphology defects, similar to that reported for hym1Δ cells (Dorland et al., 2000; Bidlingmaier et al., 2001). Second, we screened for other mutations that cause cell separation and morphogenesis defects and identified alleles of CBK1, MOB2, TAO3, and KIC1. KIC1 encodes a protein kinase that interacts with centrin-related Cdc31p and controls polarized growth and cell wall integrity (Sullivan et al., 1998; Vink et al., 2002). HYM1, CBK1, MOB2, TAO3, and KIC1 are all essential for viability in the S288C-derived strain that was used by the yeast deletion consortium (Winzeler et al., 1999). However, in our laboratory isolates of S288C (Luca laboratory) and W303 yeast strains (Luca and Boone laboratories), none of the genes is essential, which is likely due to a mutation in the SSD1 gene (Du and Novick, 2002; Jorgensen et al., 2002). We found that viable hym1Δ, cbk1Δ, mob2Δ, tao3Δ, and kic1Δ deletion cells exhibit indistinguishable round cell morphology and cell separation defects (Figure 1) and fail to mate efficiently (our unpublished data).
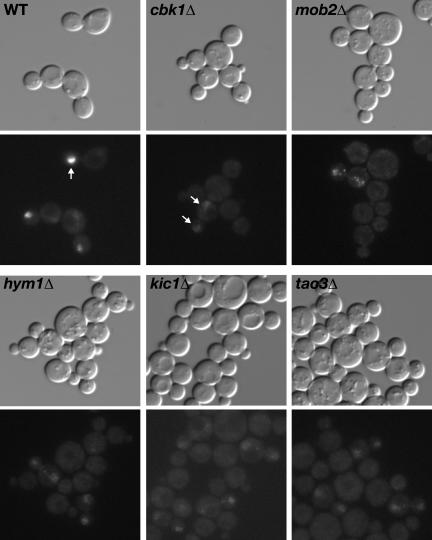
Figure 1.
RAM deletion mutants are round in morphology and are defective for cell separation and Ace2p localization. Logarithmically growing wild-type (FLY811), hym1Δ (FLY1081), cbk1Δ (FLY853), tao3Δ (FLY1124), kic1Δ (FLY1134), and mob2Δ (FLY849) cells expressing Ace2-GFP were visualized by differential interference contrast microscopy (top) and fluorescence microscopy (bottom). Arrows point to Ace2-GFP in the nuclei of representative cells. Note that the relative fluorescence of Ace2-GFP in the nuclei of RAM mutants is weaker than in the daughter cell nuclei of wild-type cells. Morphological and cell separation phenotypes were not affected by Ace2-GFP expression (our unpublished data). All images were captured and processed identically.
Yeast cell separation requires the activation of Ace2p transcription factor, which controls the daughter cell-specific expression of genes required for septum degradation, such as CTS1 and SCW11 (Dohrmann et al., 1992; Racki et al., 2000; Colman-Lerner et al., 2001; Doolin et al., 2001). Ace2p localizes to the daughter cell nucleus at the end of mitosis (Colman-Lerner et al., 2001; Weiss et al., 2002), and its activation and daughter-specific localization are dependent on Mob2p and Cbk1p (Colman-Lerner et al., 2001; Weiss et al., 2002). In tao3Δ, hym1Δ and kic1Δ cells, we found that Ace2p-GFP was not restricted to the daughter cell nucleus at the end of mitosis, but localized weakly to both mother and daughter cell nuclei (Figure 1), as observed in mob2Δ and cbk1Δ cells (Weiss et al., 2002; Figure 1). These findings suggest that Mob2p, Cbk1p, Tao3p, Hym1p, and Kic1p function within the same signaling network, which we have designated the RAM network for regulation of Ace2p activity and cellular morphogenesis.
CBK1, MOB2, TAO3, HYM1, and KIC1 Are Essential for Ace2p-dependent Transcription
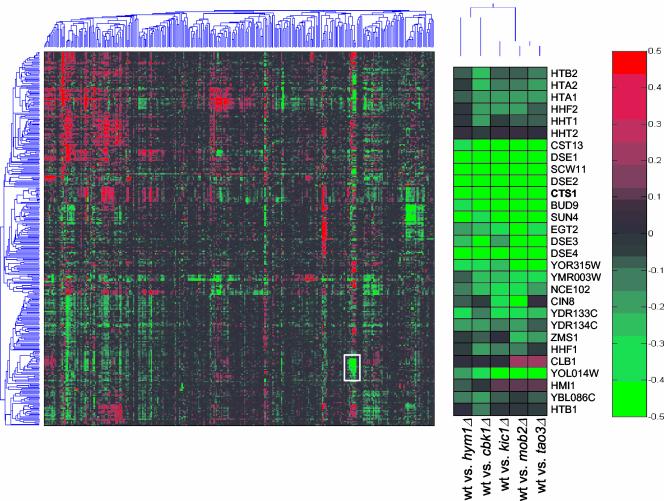
Ace2p is required for the daughter cell-specific transcription of a specific subset of genes (Colman-Lerner et al., 2001). The gene expression profiles of cbk1Δ cells were shown to be similar to ace2Δ cells (Bidlingmaier et al., 2001). To determine whether Mob2p, Tao3p, Hym1p, and Kic1p control the expression of a similar set of genes, we monitored global changes in gene expression for cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells during vegetative growth by microarray analysis (DeRisi et al., 1997; Roberts et al., 2000). We compared the gene expression profiles of the RAM network deletion mutants to a compendium of >300 reference gene expression profiles (Hughes et al., 2000). We found that the expression profiles associated with cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ were highly similar and formed a unique cluster (Figure 2). In particular, several Ace2p-regulated genes, such as CTS1, SCW11, and DSE1, DSE2, DSE3, and DSE4 (Colman-Lerner et al., 2001; Doolin et al., 2001) were coregulated across numerous experiments and were dependent on Cbk1p, Mob2p Tao3p, Hym1p, and Kic1p for normal expression (Figure 2). Thus, all of these proteins function in a common signaling pathway that controls Ace2p function.
Figure 2.
DNA microarray analysis of RAM deletion mutants. DNA microarray analysis of hym1Δ (Y1614), cbk1Δ (Y1726), tao3Δ (Y3152), kic1Δ (Y3323), and mob2Δ (Y3424) cells show overlapping expression profiles. Logarithmically growing cells were harvested or treated with 50 nM α-factor for 2 h before harvesting. Cells growing for vegetative profiling were grown in synthetic media containing all amino acids, cells treated with α-factor were grown in rich media. Two-dimensional clustering of DNA microarray profiles is presented. Gene expression is measured on a color scale (log10) with increased gene induction (red) or repression (green) corresponding to increased intensity. A gene cluster corresponding to Ace2p regulated genes is enlarged from the two-dimensional clustering for detailed examination (middle).
CBK1, MOB2, TAO3, HYM1, and KIC1 Are Required for Polarized Growth
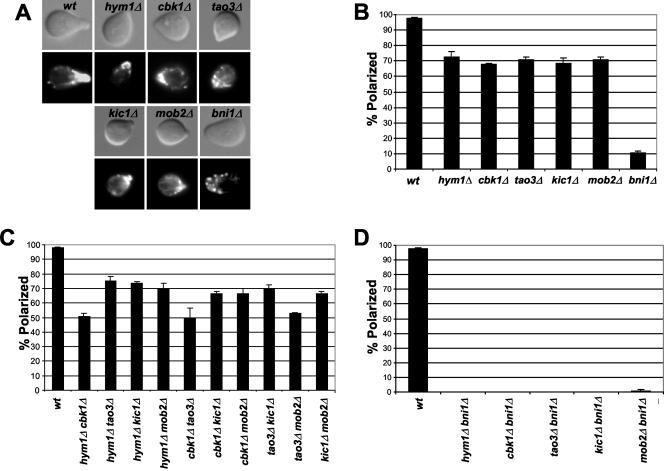
The round cell morphologies of cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells suggest that, in addition to Ace2p regulation, Cbk1p, Mob2p, Tao3p, Hym1p, and Kic1p have roles in polarized morphogenesis. To examine the roles of these proteins in pheromone-induced morphogenesis, we exposed cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells to saturating levels of pheromone for 2 h and stained with rhodaminephalloidin to visualize the filamentous actin cytoskeleton. Each of the five mutants formed broader and less pronounced mating projections than wild-type cells (Figure 3A), as observed for cbk1Δ and mob2 Δ cells (Bidlingmaier et al., 2001; Weiss et al., 2002). Only ∼70% of the mutant cells, in contrast to ∼98% of the wild-type cells, displayed a polarized distribution of cortical actin patches (Figure 3, A and B). Double mutant cells carrying different combinations of cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ displayed defects in mating projection formation that were comparable to single mutants (Figure 3C). In contrast, similarly treated cells lacking the formin gene BNI1 exhibited more severe defects in mating projection formation than cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ single mutants or double mutants (Figure 3B). Significantly, cells lacking one of the RAM genes in combination with bni1Δ exhibited more severe defects in mating projection formation than single mutants (Figure 3D). Thus, Cbk1p, Mob2p, Tao3p, Hym1p, and Kic1p seem to collaborate to control cell polarity and likely function independently from Bni1p, which is required for actin cable assembly during polarized morphogenesis.
Figure 3.
HYM1, CBK1, TAO3, KIC1, and MOB2 are necessary for formation of mating projections. Logarithmically growing cells were treated with 50 nM α-factor for 2 h and stained with rhodamine-phalloidin to visualize filamentous actin. (A) Representative micrographs are shown of cells that successfully formed mating projections. Cells were treated with rhodamine-phalloidin for detecting F-actin. Top, differential interference contrast microscopy; bottom, fluorescence microscopy. (B) Wild-type, hym1Δ, cbk1Δ, tao3Δ, kic1Δ, and mob2Δ and bni1Δ cells were scored for their ability to form mating projections. (C) RAM double mutants were scored for their ability to form mating projections. (D) Cells harboring RAM gene deletions in combination with bni1Δ were scored for their ability to form mating projections. More than 100 cells were scored in three separate counts. Strains used were wild type (SY2625), hym1Δ (Y1744), cbk1Δ (1726), tao3Δ (3481), kic1Δ (3487), mob2Δ (3482), bni1Δ (Y587), hym1Δcbk1Δ (Y1749), hym1Δtao3Δ (Y3609), hym1Δkic1Δ (Y3628), hym1Δmob2Δ (Y3608), cbk1Δtao3Δ (Y3627), cbk1Δkic1Δ (Y3606), cbk1Δmob2Δ (Y3626), tao3Δkic1Δ (Y3644), tao3Δmob2Δ (Y3645), and kic1Δmob2Δ (Y3625).
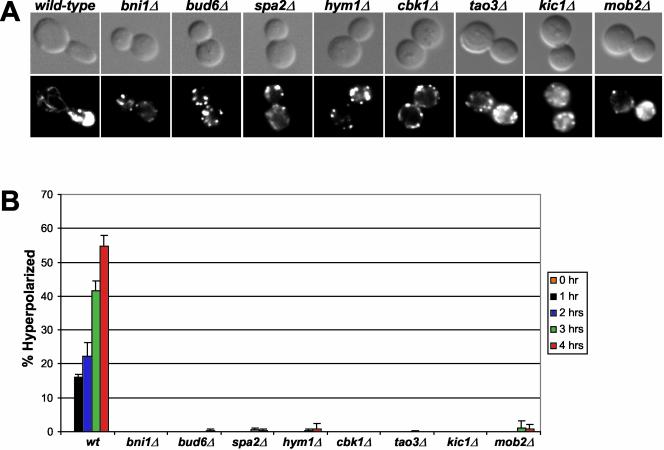
During budding, the cyclin-dependent kinase Cdc28p promotes polarized apical growth when coupled to the G1 cyclins and isotropic growth when coupled to mitotic cyclins (Lew and Reed, 1995). The apical growth phase can be prolonged by G1 cyclin overexpression, resulting in hyperelongated buds. Cells deleted for genes encoding cell polarity proteins, such as Bni1p, Bud6p and Spa2p (Chenevert et al., 1994; Amberg et al., 1997; Evangelista et al., 1997; Sheu et al., 1998), do not form hyperelongated buds in response to overexpression of the G1 cyclin CLN1, presumably due to a defect in actin-dependent polarized secretion to the bud tip (Sheu et al., 2000). To test Cbk1p, Mob2p, Tao3p, Hym1p, and Kic1p for a role in apical growth during budding, we overexpressed CLN1 in deletion strains and scored for the presence of hyperelongated buds. With prolonged CLN1 expression, wild-type cells showed a steady increase in the percentage of hyperelongated buds, reaching 55% of the total population after 4 h (Figure 4). In contrast, large budded cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells resembled bni1Δ, bud6Δ, or spa2Δ cells, showing no apparent hyperelongation of buds and displayed a random distribution of actin patches. Immunoblot analysis revealed that the mutants and wild-type cells expressed comparable levels of galactose-induced Cln1p (our unpublished data). Thus, Cbk1p, Mob2p, Tao3p, Hym1p, and Kic1p are all required for the establishment or maintenance of apical bud growth.
Figure 4.
HYM1, CBK1, TAO3, KIC1, and MOB2 are important for apical growth. (A) Cells overexpressing GAL1pr-CLN1 are shown. Top, differential interference contrast microscopy; bottom, fluorescence microscopy. Strains containing GAL1-CLN1 were induced by addition of galactose and stained with rhodamine-phalloidin to visualize filamentous actin. (B) Cells were scored for bud hyperelongation over time. Scoring consisted of counting >100 cells three independent times. The strains used were wild type (W3031A), bni1Δ (Y581), bud6Δ (Y837), spa2Δ (Y580), hym1Δ (Y1603), cbk1Δ (Y1747), tao3Δ (Y3152), kic1Δ (Y3323), and mob2Δ (Y3424). pMT485, containing GAL1pr-CLN1 with the epitope tag HAx3 was provided by Mike Tyers (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada).
HYM1, CBK1, TAO3, KIC1, and MOB2 Affect Budding Patterns
Cells that are defective in actin-based polarized morphogenesis often display errors in bipolar bud site selection (Casamayor and Snyder, 2002). We examined the budding patterns of diploid cbk1Δ, mob2Δ, tao3Δ, hym1Δ, or kic1Δ cells that contained three or more calcofluor-stained bud scars. For wild-type diploids, 76% of the cells displayed bipolar budding patterns, whereas only 5–9% of diploid cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells showed a bipolar pattern (Table 3). We also examined the budding patterns of haploid cells. The yeast strain W303 exhibits an axial budding defect, which can be rescued by introducing a plasmid carrying BUD4 (our unpublished data). Approximately 83% of haploid W303 cells that were transformed with pBUD4 showed an axial pattern (Table 3). In contrast, all five RAM deletion mutants exhibited reduced axial budding (Table 3). Only 55, 44, 56, 64, and 40% of cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells, respectively, displayed axial budding patterns. Thus, Cbk1p, Mob2p, Tao3p, Hym1p, and Kic1p are not essential for axial budding but seem to participate in this process.
Table 3.
Mutations in RAM genes result in axial and bipolar budding defects
| Axial, % | Bipolar, % | Random, % | |
|---|---|---|---|
| wt (vector) | 3.0 ± 2.6 | 68.4 ± 1.4 | 28.6 ± 3.9 |
| wt (pBUD4) | 82.7 ± 2.0 | 4.5 ± 1.3 | 12.8 ± 3.0 |
| hym1 | 59.2 ± 0.5 | 3.1 ± 1.6 | 38.1 ± 1.2 |
| cbk1 | 54.6 ± 2.1 | 1.0 ± 0.1 | 44.4 ± 2.0 |
| tao3 | 56.1 ± 2.9 | 3.1 ± 0.6 | 40.8 ± 2.3 |
| kic1 | 40.1 | 2.3 | 57.7 |
| mob2 | 43.6 | 3.9 | 52.5 |
| bni1 | 85.9 ± 4.0 | 0.6 ± 0.5 | 13.6 ± 3.6 |
| wt (vector) | 0.0 ± 0.0 | 70.1 ± 2.6 | 29.9 ± 2.6 |
| wt (pBUD4) | 0.0 ± 0.0 | 75.8 ± 3.7 | 24.2 ± 3.7 |
| hym1/hym1 | 1.0 ± 1.7 | 7.0 ± 2.6 | 92.0 ± 4.0 |
| cbk1/cbk1 | 0.0 ± 0.0 | 8.6 ± 1.9 | 91.3 ± 2.0 |
| tao3/tao3 | 0.3 ± 0.6 | 12.9 ± 3.0 | 86.8 ± 3.3 |
| bni1/bni1 | 0.0 ± 0.0 | 9.6 ± 3.2 | 90.4 ± 3.2 |
Strains containing plasmid p2698 with BUD4 (pBUD4) or pRS316 (vector) were grown to midlogarithmic phase, sonicated for cell separation and stained with Calcofluor to visualize bud scars. Strains contain pBUD4 unless otherwise indicated. Upper rows show haploid strains, lower rows show diploid strains. For each sample, a minimum of 100 cells was counted from at least three independent fields. The strains used were wild-type (W303-1A and Y1028), hym1Δ (Y1614 and Y1622), cbk1Δ (Y1747 and Y2623), tao3Δ (Y3152 and Y3565), kic1Δ (Y3323 and Y3529), mob2Δ (Y3424 and Y3527), and bni1Δ (Y581 and Y2619).
Cbk1p Kinase Activity Is Dependent on Mob2p, Tao3p, Hym1p, and Kic1p
Cbk1p kinase activity is critical for cell separation and maintenance of polarized growth (Racki et al., 2000; Colman-Lerner et al., 2001; Weiss et al., 2002) and thus may be a marker for RAM network activation. In agreement, Cbk1p kinase activity is stimulated during periods of polarized growth and peaks during late mitosis (Weiss et al., 2002). Moreover, Cbk1p activation requires Mob2p (Weiss et al., 2002). To test whether other RAM proteins are important for Cbk1p kinase activation, we immunoprecipitated Cbk1-Myc from pheromone-treated ace2Δ, mob2Δ, tao3Δ, hym1Δ, kic1Δ, and wild-type cells and performed in vitro kinase reactions by using histone H1 as substrate. Immunoblots revealed that similar amounts of Cbk1-Myc immunoprecipitated from ace2Δ, tao3Δ, hym1Δ, kic1Δ, and wild-type cells, whereas the amount of Cbk1-Myc immunoprecipitated from mob2Δ cells was more variable, ranging from equivalent to twofold less than that from wild-type cells (Figure 5). In vitro kinase assays showed that there was ∼5- to 20-fold less kinase activity associated with immunoprecipitated Cbk1-Myc from either mob2Δ, tao3Δ, hym1Δ, or kic1Δ cells than from ace2Δ and wild-type cells (Figure 5). We obtained similar data from immunoprecipitants of asynchronous cells (our unpublished data). Thus, Mob2p, Tao3p, Hym1p, and Kic1p are essential for maximal Cbk1p kinase activity, suggesting that they function in concert with Cbk1p or before Cbk1p in RAM network signaling. In contrast, Cbk1p kinase activation does not require Ace2p, which is consistent with Ace2p being a downstream target of RAM signaling.
Figure 5.
Cbk1p kinase activity is dependent on Mob2p, Hym1p, Kic1p, and Tao3p. Top, immunoblot of Cbk1-Myc immunoprecipitated from mating pheromone-treated wild-type (Y3280), mob2Δ (Y4035), hym1Δ (Y4032), kic1Δ (Y4034), tao3Δ (Y4033), ace2Δ (FLY1268), or untagged cells (W303-1A). The immunoblot was probed with anti-Myc antibody (9E10). Middle, an autoradiograph of the corresponding histone H1 kinase assays. The bottom panel shows graph of relative kinase activity. The Cbk1p-dependent histone H1 kinase activity from untagged cells was normalized to zero. The data from one of five experiments are presented; each experiment showed similar results. The maximal Cbk1p kinase activities in wild-type and ace2Δ cells are variable from experiment to experiment; thus, the apparent difference between kinase activities of wild-type and ace2Δ cells in the presented experiment is not significant.
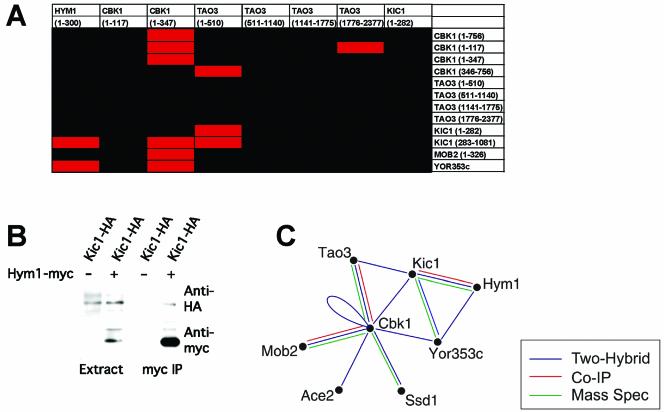
Network of Hym1p, Cbk1p, Tao3p, Kic1p, and Mob2p Protein–Protein Interactions
To test for pairwise protein interactions between RAM network components, we performed two-hybrid assays on various fragments of these proteins (Figure 6A). We confirmed previously demonstrated Mob2p–Cbk1p and Tao3p–Cbk1p interactions (Racki et al., 2000; Du and Novick, 2002; Weiss et al., 2002) and observed Cbk1p–Cbk1p, Cbk1p–Kic1p, Tao3p–Kic1p, and Hym1p–Kic1p interactions (Figure 6A). We corroborated the Hym1p–Kic1p two-hybrid interactions by coimmunoprecipitating epitope-tagged proteins from asynchronous cells (Figure 6B).
Figure 6.
RAM protein interaction network. (A) Pairwise two-hybrid qualitative LacZ assays were performed among the various RAM proteins. The corresponding amino acid regions of each protein are noted in parentheses. Red boxes indicate a positive interaction; black boxes indicate no interaction. (B) Coimmunoprecipitation of Kic1-HA with Hym1-Myc. Extracts prepared from cells expressing Hym1-Myc or untagged Hym1p were immunoprecipitated with anti-Myc. Immunoprecipitated proteins were detected by immunoblot analysis with antibodies directed against HA (top) or Myc (bottom). Strains used were Kic1-HA Hym1-Myc (Y4089) and Kic1-HA (Y3615). (C) Summary of the RAM protein interaction network based on our data and that of others (Racki et al., 2000; Ito et al., 2001; Du and Novick, 2002; Ho et al., 2002; Weiss et al., 2002). YOR353c = SOG2.
Additional protein interactions were identified in a parallel study (Ho et al., 2002). In these experiments, overproduced FLAG epitope-tagged Hym1p, Cbk1p, and Mob2p were immunoprecipitated from yeast extracts and the coprecipitated proteins were identified by mass spectroscopy. Tao3p, Mob2p, Ace2p, and Ssd1p, a protein implicated in Sit4p phosphatase function and polarized morphogenesis (Sutton et al., 1991), copurified with Cbk1p, and Cbk1p copurified with Mob2p. The Cbk1p–Ssd1p interaction is likely to be functionally relevant, because deletion of the SSD1 gene suppresses the lethality associated with disruption of RAM network signaling in certain strain backgrounds (Du and Novick, 2002; Jorgensen et al., 2002). The various interaction data are summarized in Figure 6C. This complex network of interactions is reminiscent of other signal transduction pathways, such as the yeast pheromone response pathway (Schultz et al., 1995; Elion, 2000; Dohlman and Thorner, 2001).
Sog2p Is a Novel RAM Component
To identify additional RAM-associated proteins, we conducted a two-hybrid screen by using Hym1p as bait and identified YOR353c as a gene encoding a Hym1p-interacting protein. YOR353c is registered as SOG2 in the Saccharomyces Genome Database (http://www.yeastgenome.org) and is designated as such throughout this manuscript. SOG2 encodes an 87-kDa protein that contains leucine-rich repeats and shares weak similarity with the NH2-terminal region of S. cerevisiae adenylate cyclase. In parallel studies, Sog2p was found to coprecipitate with Cbk1p and Hym1p by large-scale precipitation methods (Ho et al., 2002) and to interact with Cbk1p in independent two-hybrid screens (Uetz et al., 2000; Ito et al., 2001).
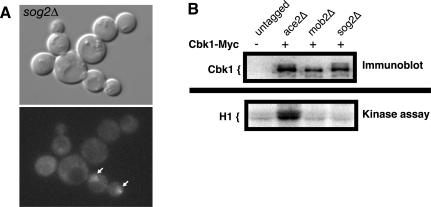
To investigate whether Sog2p functions similarly to other RAM proteins, we analyzed the phenotypes of sog2Δ cells. SOG2 and the RAM genes are essential for viability in the S288C-derived strains used for the Saccharomyces genome deletion project (Winzeler et al., 1999). However, SOG2 and the RAM genes were not essential for viability in our laboratory isolates of S288C and W303 strains, which carry the defective ssd1-d allele (Figure 7; our unpublished data). Using these strains, we found that cells lacking SOG2 exhibited round morphologies and failed to separate upon completion of mitosis (Figure 7A). Moreover, sog2Δ cells were unable to restrict Ace2p to the daughter cell nucleus (Figure 7A). To determine whether Sog2p is important for Cbk1p kinase activity, as are Mob2p, Hym1p, Tao3p, and Kic1p (Figure 5), we conducted in vitro kinase assays of immunoprecipitated Cbk1p derived from sog2Δ cells. We found that Cbk1p kinase activity was greatly diminished in the sog2Δ cells, indicating that Mob2p–Cbk1p function was dependent on Sog2p (Figure 7B). These data indicate that Sog2p is an essential component of the RAM network.
Figure 7.
Sog2p is required for proper Ace2p localization, cell morphogenesis, and Cbk1p kinase activity. (A) Logarithmically growing sog2Δ (FLY1470) cells expressing Ace2-GFP were visualized by differential interference contrast microscopy (top) and fluorescence microscopy (bottom). Arrows point to Ace2-GFP in the nuclei of representative cells. (B) Top, immunoblot of Cbk1-Myc immunoprecipitated from mating pheromone-treated untagged cells (W303-1A), ace2Δ (FLY1268), mob2Δ (Y4035), and sog2Δ cells (FLY1492). The immunoblot was probed with anti-Myc antibody (9E10). Bottom, an autoradiograph of the corresponding histone H1 kinase assays. The relative kinase activity of immunoprecipitated Cbk1p from sog2Δ and mob2Δ cells was as low as immunoprecipitated kinase activity from untagged cells and was ∼10-fold less than from ace2Δ cells. Similar data was obtained from asynchronous cells (our unpublished data).
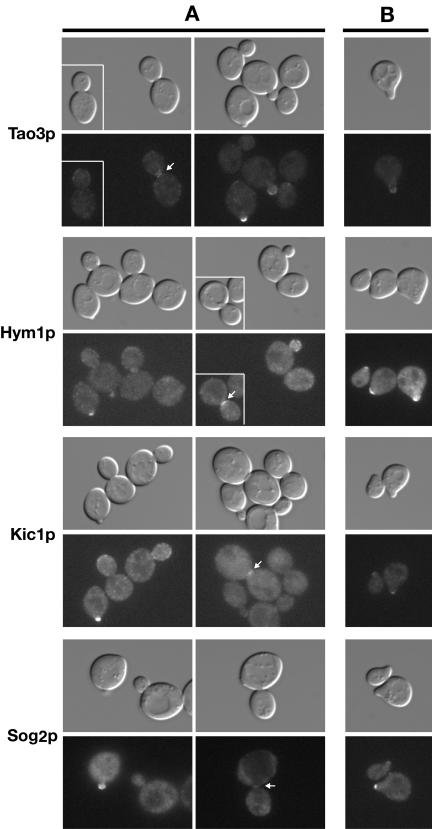
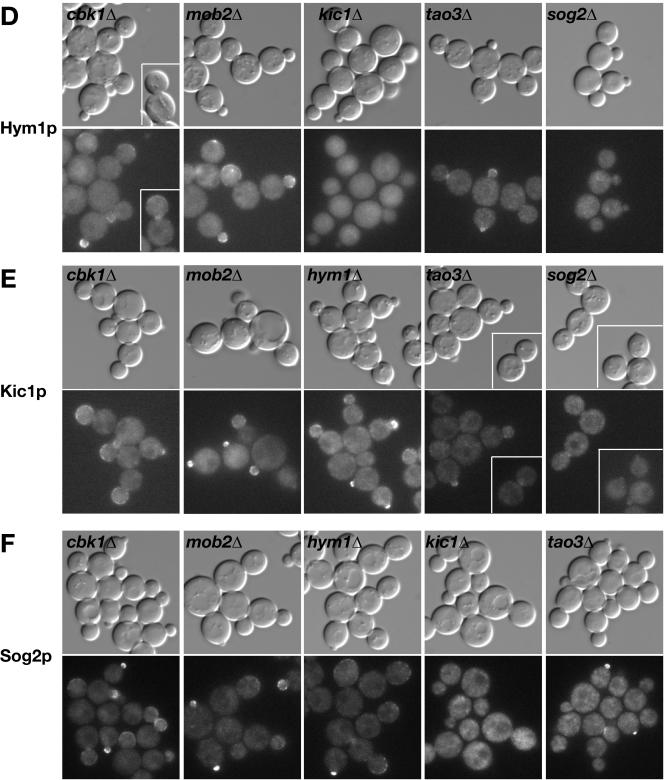
Tao3p, Hym1p, Sog2p, and Kic1p Localize to Sites of Polarized Growth and Control Mob2p–Cbk1p Nuclear Localization
We examined the subcellular localizations of GFP-tagged RAM proteins to gain insight to their functional specificity. Indeed, consistent with roles in both polarized growth and Ace2p regulation, Cbk1p and Mob2p localize to the cortex during apical bud growth and to the bud neck and daughter cell nucleus during late mitosis (Colman-Lerner et al., 2001; Weiss et al., 2002). Tao3p was also shown to localize to sites of polarized growth but was not observed in the daughter nucleus (Du and Novick, 2002). We confirmed these observations by monitoring cells expressing Tao3-GFP (Figure 8A). Similarly, Kic1-GFP, Hym1-GFP, and Sog2-GFP localized prominently to incipient bud sites and small buds and weakly to the cortex and bud necks of large budded cells (Figure 8A). Tao3p, Hym1p, Sog2p, and Kic1p also localized to the growing tips of mating projections (Figure 8B), as shown for Mob2p and Cbk1p (Weiss et al., 2002). We did not detect Tao3p, Hym1p, Sog2p, or Kic1p in the nucleus. Thus, because Mob2p and Cbk1p are the only proteins of the network that are detected in the daughter cell nucleus, it is likely that Mob2p and Cbk1p function downstream of the other RAM proteins with respect to Ace2p regulation.
Figure 8.
Tao3p, Hym1p, Kic1p, and Sog2p localize to sites of polarized growth. (A) Logarithmically growing homozygous diploid strains expressing Tao3-GFP (FLY1267), Hym1-GFP (FLY1244), Kic1-GFP (FLY1258), and Sog2-GFP (FLY1382) were visualized by differential interference contrast or fluorescence microscopy. Arrows point to bud neck localizations. (B) Tao3-GFP, Hym1-GFP, Kic1-GFP, and Sog2-GFP proteins localize to the tips of the mating projections upon pheromone treatment. MATa cells (FLY1263, FLY891, FLY947, and FLY1347) were treated with 5 μM mating pheromone for 2 h before visualization. All images were captured and processed identically.
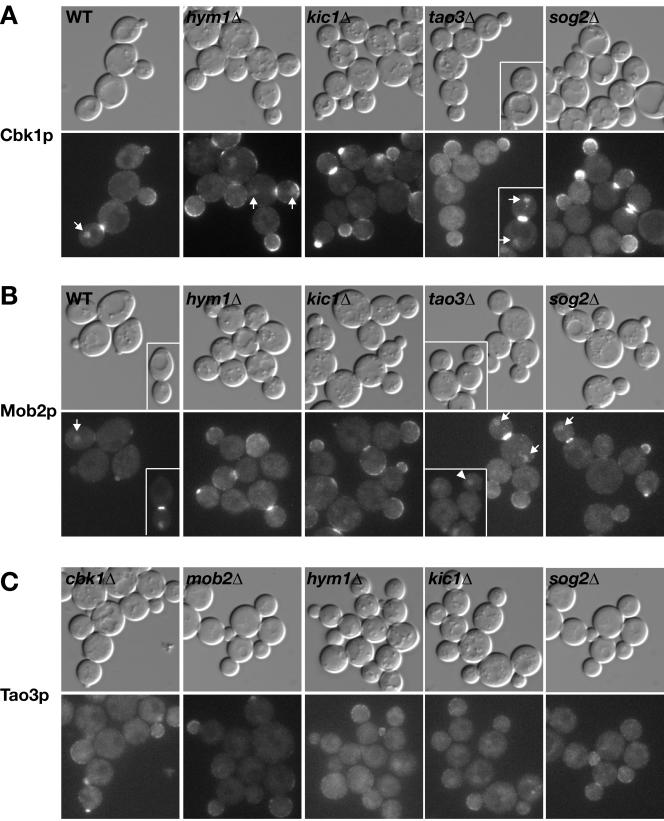
Interdependency of RAM Protein Localization
To determine whether Cbk1p, Mob2p, Tao3p, Hym1p, Sog2p, and Kic1p are interdependent for localization, we systematically examined the localization of each protein in various RAM deletion strains. The results are summarized in Table 4.
Table 4.
Summary of RAM protein localization in wild type and mutant cells
| Cbk1p | Mob2p | Tao3p | Hym1p | Kic1p | Sog2p | |
|---|---|---|---|---|---|---|
| WT | B N DN | B N DN | B N | B N | B N | B N |
| cbk1Δ | cytoplasma | B+ N | B+ N | B+ | B+ N | |
| mob2Δ | M+DNa | B+ N | B+ N | B+ | B+ N | |
| tao3Δ | B N M+DN | B N M+DN | B N | B- N- | B N | |
| hym1Δ | B N M+DN | B N | B N | B N | B+ N | |
| kic1Δ | B N | B N | B N | cytoplasm | cytoplasm | |
| sog2Δ | B N | B N M+DN | B N | cytoplasm | cytoplasmb |
B, bud cortex; B+, B-, stronger or weaker cortical localization than in wild type cells; DN, daughter cell nucleus; M+DN, mother and daughter cell nuclei. N, bud neck during late mitosis; WT, wild type cells; See results for detailed description.
A greatly diminished amount of Kic1p can be detected on the cortex in some sog2Δ cells.
We asked whether Mob2p and Cbk1p localization was dependent on other RAM proteins. As observed previously (Weiss et al., 2002), Cbk1p and Mob2p localized interdependently at the cortex, bud neck, and daughter cell nucleus. In contrast, both Cbk1p and Mob2p were able to localize to the bud cortex and bud neck in tao3Δ, hym1Δ, sog2Δ, and kic1Δ cells (Figure 9, A and B). Nevertheless, the nuclear localization of Mob2p and Cbk1p was distinctly aberrant in these cells. Mob2p was not detected in any kic1Δ or hym1Δ cell nucleus but was weakly detectable in (∼9%) daughter nuclei or in (∼9%) both mother and daughter nuclei of large budded sog2Δ cells (Figure 9B, arrows). Cbk1p was also undetectable in kic1Δ and sog2Δ cell nuclei but localized weakly to the daughter cell nucleus (∼7%) or to both mother and daughter cell nuclei (∼15%; see arrows Figure 9A) in some large budded hym1Δ cells. Cbk1p and Mob2p also localized to both mother and daughter cell nuclei in ∼25–46% of the large budded tao3Δ cells (Figure 2A, inset) and to the daughter nucleus in only ∼15% of the large budded tao3Δ cells. In particular, Mob2p was detectable in some late G1 tao3Δ cell nuclei (see arrowhead, Figure 9B, inset), which is in contrast to its localization in wild-type cells where Mob2p disappears from the daughter nucleus in early G1, well before bud emergence (Weiss et al., 2002). Together, these findings support a model in which Kic1p, Sog2p, Hym1p, and Tao3p function upstream of Mob2p-Cbk1p to control the catalytic activity and proper nuclear localization of the Mob2p–Cbk1p complex.
Figure 9.
Relationship between Cbk1p, Mob2p, Tao3p, Hym1p, Kic1p, and Sog2p for protein localization. Logarithmically growing cells expressing GFP-tagged proteins were visualized by differential interference contrast and fluorescence microscopy. (A) Localization of Cbk1-GFP in wild-type (FLY895), hym1Δ (FLY1383), kic1Δ (FLY1344), tao3Δ (FLY1259), and sog2Δ cells (FLY1515). Arrows point to nuclear localization of Cbk1-GFP in late mitotic cells. (B) Localization of Mob2-GFP in wild-type (FLY893), hym1Δ (FLY1342), kic1Δ (FLY1343), tao3Δ (FLY1260) and sog2Δ (FLY1499) cells. Arrows point to nuclear localization of Mob2-GFP. The arrowhead points to a G1 tao3Δ cell (inset) with Mob2-GFP in the nucleus. (C) Localization of Tao3-GFP in cbk1Δ (FLY1324), mob2Δ (FLY1386), hym1Δ (FLY1326), kic1Δ (FLY1325), and sog2Δ (FLY1500) cells. (D) Localization of Hym1-GFP in cbk1Δ (FLY1246), mob2Δ (FLY1245), kic1Δ (FLY1261), tao3Δ (FLY1262), and sog2Δ (FLY1514) cells. (E) Localization of Kic1-GFP in cbk1Δ (FLY1248), mob2Δ (FLY1247), hym1Δ (FLY1278), tao3Δ (FLY1249), and sog2Δ (FLY1465) cells. (F) Localization of Sog2-GFP in cbk1Δ (FLY1475), mob2Δ (FLY1474), hym1Δ (FLY1472), kic1Δ (FLY1471), and tao3Δ (FLY1473) cells. Immunoblots of the deletion strains reveal no significant differences in RAM protein levels (our unpublished data). Each image was captured and processed identically.
There were subtle, but significant changes in RAM protein localization in mob2Δ and cbk1Δ cells. In these cells, Tao3-GFP, Hym1-GFP, Sog2-GFP, and Kic1-GFP seemed brighter at the cortexes of small and large buds than in wild-type cells (Figure 9, C,Figure 9, D–F). Moreover, there were significantly fewer large budded mob2Δ and cbk1Δ cells with Kic1-GFP (0%), Hym1-GFP (11–14%), or Sog2-GFP (4–6%) localized to bud necks than in wild-type cells (49%, n ≥ 50 large budded cells). Immunoblots of the RAM deletion strains reveal no significant difference in protein levels of the remaining RAM proteins (our unpublished data). Thus, these data suggest that Mob2p and Cbk1p influence the localized concentration or abundance of RAM proteins at the sites of polarized growth.
Figure 9.
Relationship between Cbk1p, Mob2p, Tao3p, Hym1p, Kic1p, and Sog2p for protein localization. Logarithmically growing cells expressing GFP-tagged proteins were visualized by differential interference contrast and fluorescence microscopy. (A) Localization of Cbk1-GFP in wild-type (FLY895), hym1Δ (FLY1383), kic1Δ (FLY1344), tao3Δ (FLY1259), and sog2Δ cells (FLY1515). Arrows point to nuclear localization of Cbk1-GFP in late mitotic cells. (B) Localization of Mob2-GFP in wild-type (FLY893), hym1Δ (FLY1342), kic1Δ (FLY1343), tao3Δ (FLY1260) and sog2Δ (FLY1499) cells. Arrows point to nuclear localization of Mob2-GFP. The arrowhead points to a G1 tao3Δ cell (inset) with Mob2-GFP in the nucleus. (C) Localization of Tao3-GFP in cbk1Δ (FLY1324), mob2Δ (FLY1386), hym1Δ (FLY1326), kic1Δ (FLY1325), and sog2Δ (FLY1500) cells. (D) Localization of Hym1-GFP in cbk1Δ (FLY1246), mob2Δ (FLY1245), kic1Δ (FLY1261), tao3Δ (FLY1262), and sog2Δ (FLY1514) cells. (E) Localization of Kic1-GFP in cbk1Δ (FLY1248), mob2Δ (FLY1247), hym1Δ (FLY1278), tao3Δ (FLY1249), and sog2Δ (FLY1465) cells. (F) Localization of Sog2-GFP in cbk1Δ (FLY1475), mob2Δ (FLY1474), hym1Δ (FLY1472), kic1Δ (FLY1471), and tao3Δ (FLY1473) cells. Immunoblots of the deletion strains reveal no significant differences in RAM protein levels (our unpublished data). Each image was captured and processed identically.
If Hym1p, Sog2p, and Kic1p are functionally linked as suggested by their binding interactions, then they may influence each other's subcellular distributions, as shown for Mob2p and Cbk1p (Weiss et al., 2002). Indeed, Hym1-GFP and Sog2-GFP mislocalized from the cortex and bud neck in kic1Δ cells and instead localized diffusely to the cytoplasm (Figure 9, D and F). Hym1p similarly mislocalized to the cytoplasm in sog2Δ cells, but localized normally in tao3Δ cells. Strikingly, Kic1p localization was severely diminished at the cortex or bud neck in sog2Δ cells, whereas Kic1p localization was normal in hym1Δ cells. In tao3Δ cells, the pattern of Kic1p localization was normal but seemed slightly diminished (Figure 9E). Sog2p distribution was normal in hym1Δ and tao3Δ cells, as was Tao3p in hym1Δ, sog2Δ, and kic1Δ cells (Figure 9, C, and Figure 9F). These data indicate that Hym1p localization requires both Sog2p and Kic1p and that Sog2p and Kic1p are interdependent for localization.
DISCUSSION
Cbk1p, Mob2p, Tao3p, Hym1p, Sog2p, and Kic1p Function in a Common Signaling Network
We present several lines of evidence that Cbk1p, Mob2p, Hym1p, Tao3p, Sog2p, and Kic1p function in a common signaling network, which we refer to as the RAM network. Cells deleted for each RAM gene exhibit dramatic cell separation defects, reduced mating efficiency and are rounder in morphology than wild-type cells. In addition cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ mutants display indistinguishable defects in bud site selection and pheromone-induced polarized morphogenesis. There are no additive effects of RAM mutations, which further suggests the RAM proteins function in a common pathway (Figure 3; our unpublished data). Moreover, the RAM proteins form a complex network of protein–protein interactions and localize similarly to cortical sites of polarized growth during budding and cell separation, with some proteins dependent upon one another for localization. Although the combined results of previous studies regarding Mob2p, Cbk1p, Tao3p, and Hym1p anticipated the existence of a common signaling network that controls morphogenesis and cell separation (Dorland et al., 2000; Racki et al., 2000; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Du and Novick, 2002; Weiss et al., 2002), this study firmly establishes and defines the RAM network. Significantly, we have elucidated the in vivo relationships between the RAM proteins and have demonstrated that Kic1p kinase and the LRR-containing Sog2p are key components of the RAM network.
The Role of RAM Network in Regulating Ace2p Activity
It is apparent that the RAM network controls cell separation via regulation of the Ace2p transcription factor. Ace2p was previously shown to localize to the daughter nucleus at the end of mitosis and to activate daughter-specific transcription of a subset of genes important for septum degradation (Colman-Lerner et al., 2001; Doolin et al., 2001; Weiss et al., 2002). The RAM proteins Mob2p and Cbk1p also localize to the daughter cell nucleus at the end of mitosis and were shown to influence Ace2p function (Racki et al., 2000; Bidlingmaier et al., 2001; Colman-Lerner et al., 2001; Weiss et al., 2002). Herein, we demonstrate that deletion of the RAM genes CBK1, MOB2, TAO3, HYM1, SOG2, or KIC1 cause indistinguishable defects in Ace2p localization and function. Specifically, in the deletion mutants, Ace2p is not restricted to the daughter cell nucleus at the end of mitosis, but instead enters both nuclei (Figures 1 and 7). Previous data suggest that Mob2p–Cbk1p kinase activity is necessary for inhibition of Ace2p nuclear export from the daughter cell nucleus (Weiss et al., 2002). Our microarray analyses reveal that RAM deletion strains are markedly reduced for expression of Ace2-regulated genes (Figure 2). Thus, RAM signaling is required to regulate both Ace2p localization and activity.
Ace2p seems to be regulated at multiple levels. Previous studies have shown that Ace2p nuclear import is inhibited by CDK phosphorylation (O'Conallain et al., 1999), but activated by MEN-dependent signaling (Weiss et al., 2002). Nevertheless, Ace2p activation cannot be solely regulated by nuclear import, because Ace2p can enter nuclei in RAM deletion mutants and yet remains inactive as a transcription factor. It has been suggested that Mob2p-Cbk1p phosphorylates Ace2p, or an associated protein, to prevent its nuclear export (Weiss et al., 2002). Our data are consistent with this possibility. Of the six RAM network proteins, Mob2p and Cbk1p are the only ones detected in the daughter nucleus. Thus, Mob2p-Cbk1p may function late in the network, with respect to Ace2p activation, and may phosphorylate Ace2p (or an associated protein), thereby inhibiting Ace2p nuclear export and, at the same time, activating Ace2p.
RAM Signaling and Cell Polarity
The role of the RAM network in Ace2p regulation is distinct from its role in polarized growth. Notably, ace2Δ cells are not defective in polarized growth (Racki et al., 2000; Bidlingmaier et al., 2001), and moderate overexpression of ACE2 on high copy plasmids suppresses the cell separation but not the polarized growth defects of cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells (Racki et al., 2000; Colman-Lerner et al., 2001; our unpublished data). The RAM network likely contributes to polarized growth by regulating actin-based secretion. Polarized actin cables seem to act as tracks that guide myosin motors carrying cargo of secretory vesicles containing materials necessary for cell surface expansion. The yeast formin, Bni1p controls the assembly and polarization of actin cables (Burns et al., 1994; Evangelista et al., 2002), and a bni1Δ deletion mutation results in defects in cable assembly and polarization, leading to general defects in cell polarization (Evangelista et al., 2002). In contrast to bni1Δ cells, the actin cables seem relatively normal in cbk1Δ, mob2Δ, tao3Δ, hym1Δ, and kic1Δ cells (Bidlingmaier et al., 2001; Du and Novick, 2002; Weiss et al., 2002; our unpublished data). Furthermore, the role of RAM signaling in maintenance of cell polarity seems distinct from that of Bni1p, because deletion of RAM genes in combination with bni1Δ results in a synergistic defect in mating projection formation. Perhaps the RAM network operates at the level of vesicle transport or exocyst function (TerBush et al., 1996; Lipschutz and Mostov, 2002). Consistent with this idea, Cbk1p was shown to bind Sec2p, a guanine nucleotide exchange factor required for vesicle transport and exocytosis, in a two-hybrid assay (Racki et al., 2000).
Order of RAM Protein Function
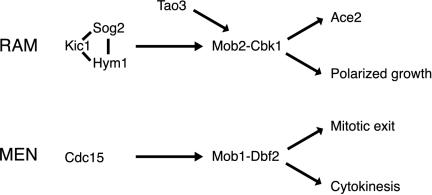
By analogy to the S. cerevisiae MEN (Bardin and Amon, 2001), it seems plausible that Kic1p kinase activates the Mob2p–Cbk1p complex directly (Figure 10). Recombinant Kic1p-related Cdc15p kinase was shown to phosphorylate and activate the Mob2p–Cbk1p-related Mob1p–Dbf2p kinase complex (Mah et al., 2001). The presence of Kic1p in the RAM network suggests a similar mechanism for the Mob2p–Cbk1p kinase activation. Consistent with this hypothesis, we have shown that Mob2p-Cbk1p kinase activity is dependent on Kic1p.
Figure 10.
Models of conserved regulatory MEN and RAM networks in yeast. The Mob2p–Cbk1p kinase complex likely functions late in the RAM network because 1) Cbk1p kinase activity is dependent on all known RAM proteins, and 2) Mob2p and Cbk1p are the only RAM proteins detectable in the daughter cell nucleus at the end of mitosis. Kic1p, Sog2p, and Hym1p may function together as a complex because Hym1p binds Sog2p and Kic1p and is Sog2p- and Kic1p-dependent for localization. By analogy to the MEN protein Cdc15p, which activates the Mob1p–Dbf2p kinase complex in vitro, Kic1p may phosphorylate and activate the Mob2p–Cbk1p complex. Tao3p is a 270-kDa protein of unknown function that interacts with Cbk1p and Kic1p. Thus, Tao3p may serve as a scaffold that facilitates activation of Mob2p–Cbk1p kinase by Kic1p.
Hym1p, Sog2p, and Tao3p are not related to known MEN components, yet are also required for Mob2–Cbk1p kinase activation. What might be their molecular functions? Our data indicate that Hym1p, Sog2p and Kic1p are functionally linked. Hym1p coimmunoprecipitates with Kic1p and mislocalizes in the absence of Kic1p or Sog2p. Moreover, Sog2p, which associates with Hym1p, Cbk1p, and Kic1p (Figure 6) (Ho et al., 2002), is interdependent with Kic1p for localization. Curiously, Kic1p and Sog2p localize to the bud cortex independently of Hym1p, suggesting that Kic1p and Sog2p function upstream of Hym1p or that Sog2p mediates the association of Hym1p with Kic1p. Hym1p and Sog2p may regulate Kic1p kinase activity or mediate interactions between Kic1p kinase and its substrates. Along these lines, Hym1p and Sog2p might stabilize Kic1p and Mob2p–Cbk1p interactions for Cbk1p kinase activation.
Tao3p is a large 270-kDa protein that interacts with Cbk1p and Kic1p (Figure 6; Du and Novick, 2002) that may also facilitate Mob2p–Cbk1p kinase activation by Kic1p. Moreover, Tao3p may be important for regulating the timing or duration of Mob2p-Cbk1p nuclear localization, because Mob2p and Cbk1p seem to remain localized to the nuclei for a longer duration in some tao3Δ cells than in wild-type cells (arrows Figure 9, A and B).
Although Tao3p, Hym1p, Sog2p, and Kic1p all seem to act upstream of Mob2p-Cbk1p with respect to Cbk1p kinase activation, it is not yet possible to definitively order their functions with respect to each other. Indeed, it is probable that the RAM network is not linear. In support, we have found that Hym1p, Sog2p, Kic1p, and Tao3p are more prominent at the bud cortex in the absence of Mob2p or Cbk1p. Thus, in addition to acting late in the RAM network, Mob2-Cbk1p may regulate Hym1p, Sog2p, Kic1p, and Tao3p localization or stability, perhaps as part of a feedback mechanism.
Other RAM Proteins and Associated Proteins
It is likely that other RAM network components or associated proteins will be identified. True RAM components will be required for cell morphogenesis and Ace2p regulation, although some effectors or targets of RAM signaling may have additional or separable functions. Indeed, the nonessential protein Ssd1p was also shown to bind Cbk1p, suggesting that it may have a role in RAM regulation or may function as an effector of the RAM pathway (Racki et al., 2000). Ssd1p was originally identified as a suppressor of mutation in Sit4p phosphatase (Sutton et al., 1991). Recently, ssd1Δ mutations were shown to suppress the lethality associated with deletion of RAM genes (Du and Novick, 2002; Jorgensen et al., 2002). These data suggest that Ssd1p negatively regulates a function that is essential in the absence of RAM signaling, and thus Ssd1p activity may be coordinated with RAM signaling.
RAM-like Signaling Networks Are Conserved
All of the components of the RAM network are conserved. In Drosophila, Cbk1p-related and Tao3p-like proteins were shown to be required to maintain the integrity of cellular extensions during morphogenesis (Geng et al., 2000; Cong et al., 2001). Similarly, S. pombe orthologs to Cbk1p, Mob2p, and Tao3p (designated Orb6p, Mob2p, and Mor2p) were shown to be essential for cell polarity and morphogenesis (Verde et al., 1998; Hirata et al., 2002; Hou et al., 2003). Deletion of S. pombe orb6, mob2, or mor2 genes cause a depolarization of the actin cytoskeleton resulting in striking spherical cell morphologies. Like their S. cerevisiae counterparts, the proteins encoded by S. pombe orb6, mob2, or mor2 genes localize to the sites of polarized growth. However, unlike S. cerevisiae Mob2p and Cbk1p, S. pombe Orb6p and Mob2p were not observed to localize to the nucleus or to regulate a transcription factor.
We suspect that future work will demonstrate that the overall organization of the RAM-related signaling networks is conserved in all eukaryotes. Thus, in an effort to map a mammalian RAM-like signaling network, we identified several proteins associated with the human Kic1p-like kinase Mst3 (Schinkmann and Blenis, 1997) (see Supplement). We found that Mst3 coimmunoprecipitates with phocein (Luca and Winey, 1998; Baillat et al., 2001; Moreno et al., 2001), a distantly related Mob1p/Mob2p-related protein that is highly conserved in animal cells (see Supplement Figure S1). This work supports the idea that RAM-like signaling modules are conserved in mammalian cells; however, more work is necessary to elucidate the function of Mst3 kinase complex and to identify other components of its signaling network.
Current evidence suggest that eukaryotic RAM and MEN signaling modules are important for regulating and coordinating a variety of important cellular processes, including mitotic exit, cytokinesis, cell polarity, gene transcription, and vesicle transport. In this sense, different RAM and MEN signaling networks can be likened to the different MAP kinase-signaling networks, which are compromised of conserved protein modules that affect different downstream processes (Posas et al., 1998; Wilkinson and Millar, 2000). Further work is necessary regarding the identity, function, and targets of these novel classes of regulatory networks.
Supplementary Material
Acknowledgments
We thank Christopher Roberts and Matthew Marton for help with the microarray analysis, and S. Kang for help with characterization of RAM pathway mutants. We also thank Eric Weiss, Chris Herbert, David Stillman, and members of the Luca and Boone laboratories for insightful discussions. This work was funded by grants from the National Institutes of Health (R01 GM-60575) to F.C.L.; Natural Sciences and Engineering Research Council, the National Cancer Institute of Canada to C.B.; and an Natural Sciences and Engineering Research Council graduate student fellowship to B.N.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0018. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0018.
Online version of this article contains supplementary data for some figures. Online version is available at http://www.molbiolcell.org.
References
- Amberg, D.C., Zahner, J.E., Mulholland, J.W., Pringle, J.R., and Botstein, D. (1997). Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell 8, 729–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, S.S., and Bi, G.Q. (2000). Axon formation: a molecular model for the generation of neuronal polarity. Bioessays 22, 172–179. [DOI] [PubMed] [Google Scholar]
- Baillat, G., Moqrich, A., Castets, F., Baude, A., Bailly, Y., Benmerah, A., and Monneron, A. (2001). Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell 12, 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin, A.J., and Amon, A. (2001). Men and sin: what's the difference? Nat. Rev. Mol. Cell. Biol. 2, 815–826. [DOI] [PubMed] [Google Scholar]
- Bidlingmaier, S., Weiss, E.L., Seidel, C., Drubin, D.G., and Snyder, M. (2001). The Cbk1p pathway is important for polarized cell growth and cell separation in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 2449–2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone, C., Davis, N.G., and Sprague, G.F., Jr. (1993). Mutations that alter the third cytoplasmic loop of the a-factor receptor lead to a constitutive and hypersensitive phenotype. Proc. Natl. Acad. Sci. USA 90, 9921–9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, N., Grimwade, B., Ross-Macdonald, P.B., Choi, E.Y., Finberg, K., Roeder, G.S., and Snyder, M. (1994). Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8, 1087–1105. [DOI] [PubMed] [Google Scholar]
- Casamayor, A., and Snyder, M. (2002). Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 5, 179–186. [DOI] [PubMed] [Google Scholar]
- Chant, J., and Pringle, J.R. (1995). Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert, J., Valtz, N., and Herskowitz, I. (1994). Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics 136, 1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner, A., Chin, T.E., and Brent, R. (2001). Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107, 739–750. [DOI] [PubMed] [Google Scholar]
- Cong, J., Geng, W., He, B., Liu, J., Charlton, J., and Adler, P.N. (2001). The furry gene of Drosophila is important for maintaining the integrity of cellular extensions during morphogenesis. Development 128, 2793–2802. [DOI] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Dohlman, H.G., and Thorner, J.W. (2001). Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70, 703–754. [DOI] [PubMed] [Google Scholar]
- Dohrmann, P.R., Butler, G., Tamai, K., Dorland, S., Greene, J.R., Thiele, D.J., and Stillman, D.J. (1992). Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 6, 93–104. [DOI] [PubMed] [Google Scholar]
- Doolin, M.T., Johnson, A.L., Johnston, L.H., and Butler, G. (2001). Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40, 422–432. [DOI] [PubMed] [Google Scholar]
- Dorland, S., Deegenaars, M.L., and Stillman, D.J. (2000). Roles for the Saccharomyces cerevisiae SDS3, CBK1 and HYM1 genes in transcriptional repression by SIN3. Genetics 154, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L.L., and Novick, P. (2002). Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee, T., Becherer, K., Chen, P.L., Yeh, S.H., Yang, Y., Kilburn, A.E., Lee, W.H., and Elledge, S.J. (1993). The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7, 555–569. [DOI] [PubMed] [Google Scholar]
- Elion, E.A. (2000). Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3, 573–581. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Blundell, K., Longtine, M.S., Chow, C.J., Adames, N., Pringle, J.R., Peter, M., and Boone, C. (1997). Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science 276, 118–122. [DOI] [PubMed] [Google Scholar]
- Evangelista, M., Klebl, B.M., Tong, A.H., Webb, B.A., Leeuw, T., Leberer, E., Whiteway, M., Thomas, D.Y., and Boone, C. (2000). A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148, 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., Pruyne, D., Amberg, D.C., Boone, C., and Bretscher, A. (2002). Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 4, 260–269. [DOI] [PubMed] [Google Scholar]
- Fink, G.R., and Styles, C.A. (1972). Curing of a killer factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 69, 2846–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, W., He, B., Wang, M., and Adler, P.N. (2000). The tricornered gene, which is required for the integrity of epidermal cell extensions, encodes the Drosophila nuclear DBF2-related kinase. Genetics 156, 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., and Sugino, A. (1988). New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534. [DOI] [PubMed] [Google Scholar]
- Gulli, M.P., Jaquenoud, M., Shimada, Y., Niederhauser, G., Wiget, P., and Peter, M. (2000). Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell 6, 1155–1167. [DOI] [PubMed] [Google Scholar]
- Gyuris, J., Golemis, E., Chertkov, H., and Brent, R. (1993). Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803. [DOI] [PubMed] [Google Scholar]
- Hirata, D., et al. (2002). Fission yeast Mor2/Cps12, a protein similar to Drosophila Furry, is essential for cell morphogenesis and its mutation induces Wee1-dependent G(2) delay. EMBO J. 21, 4863–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, Y., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Horecka, J., and Jigami, Y. (2000). Identifying tagged transposon insertion sites in yeast by direct genomic sequencing. Yeast 16, 967–970. [DOI] [PubMed] [Google Scholar]
- Hou, M.C., Wiley, D.J., Verde, F., and McCollum, D. (2003). Mob2p interacts with the protein kinase Orb6p to promote coordination of cell polarity with cell cycle progression. J. Cell Sci. 116, 125–135. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., et al. (2000). Functional discovery via a compendium of expression profiles. Cell 102, 109–126. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., Nelson, B., Robinson, M.D., Chen, Y., Andrews, B., Tyers, M., and Boone, C. (2002). High-resolution genetic mapping with ordered arrays of Saccharomyces cerevisiae deletion mutants. Genetics 162, 1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karos, M., and Fischer, R. (1999). Molecular characterization of HymA, an evolutionarily highly conserved and highly expressed protein of Aspergillus nidulans. Mol. Gen. Genet. 260, 510–521. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J.V., and Adams, A.E. (1984). Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98, 922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knust, E. (2000). Control of epithelial cell shape and polarity. Curr. Opin. Genet. Dev. 10, 471–475. [DOI] [PubMed] [Google Scholar]
- Leberer, E., Thomas, D.Y., and Whiteway, M. (1997). Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7, 59–66. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and Reed, S.I. (1995). Cell cycle control of morphogenesis in budding yeast. Curr. Opin. Genet. Dev. 5, 17–23. [DOI] [PubMed] [Google Scholar]
- Lipschutz, J.H., and Mostov, K.E. (2002). Exocytosis: the many masters of the exocyst. Curr. Biol. 12, R212–R214. [DOI] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Luca, F.C., Mody, M., Kurischko, C., Roof, D.M., Giddings, T.H., and Winey, M. (2001). Saccharomyces cerevisiae Mob1p is required for cytokinesis and mitotic exit. Mol. Cell. Biol. 21, 6972–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca, F.C., and Winey, M. (1998). MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell 9, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah, A.S., Jang, J., and Deshaies, R.J. (2001). Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA 98, 7325–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, H., Matsushiro, A., and Nozaki, M. (1993). Molecular cloning of a novel mRNA sequence expressed in cleavage stage mouse embryos. Reprod. Dev. 34, 1–7. [DOI] [PubMed] [Google Scholar]
- Moreno, C.S., Lane, W.S., and Pallas, D.C. (2001). A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin-family/protein phosphatase 2A complexes. J. Biol. Chem. 23, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Conallain, C., Doolin, M.T., Taggart, C., Thornton, F., and Butler, G. (1999). Regulated nuclear localisation of the yeast transcription factor Ace2p controls expression of chitinase (CTS1) in Saccharomyces cerevisiae. Mol. Gen. Genet. 262, 275–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, M., Gartner, A., Horecka, J., Ammerer, G., and Herskowitz, I. (1993). FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73, 747–760. [DOI] [PubMed] [Google Scholar]
- Phizicky, E.M., and Fields, S. (1995). Protein-protein interactions: methods for detection and analysis. Microbiol. Rev. 59, 94–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas, F., Takekawa, M., and Saito, H. (1998). Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1, 175–182. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000a). Polarization of cell growth in yeast. J. Cell Sci. 113, 571–585. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000b). Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113, 365–375. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A., and Boone, C. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- Racki, W.J., Becam, A.M., Nasr, F., and Herbert, C.J. (2000). Cbk1p, a protein similar to the human myotonic dystrophy kinase, is essential for normal morphogenesis in Saccharomyces cerevisiae. EMBO J. 19, 4524–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C.J., et al. (2000). Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287, 873–880. [DOI] [PubMed] [Google Scholar]
- Rupes, I. (2002). Checking cell size in yeast. Trends Genet. 18, 479–485. [DOI] [PubMed] [Google Scholar]
- Sagot, I., Klee, S.K., and Pellman, D. (2002). Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat. Cell Biol. 4, 42–50. [DOI] [PubMed] [Google Scholar]
- Schinkmann, K., and Blenis, J. (1997). Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J. Biol. Chem. 272, 28695–28703. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Ferguson, B., and Sprague, G.F., Jr. (1995). Signal transduction and growth control in yeast. Curr. Opin. Genet. Dev. 5, 31–37. [DOI] [PubMed] [Google Scholar]
- Sheu, Y.J., Barral, Y., and Snyder, M. (2000). Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol. Cell. Biol. 20, 5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y.J., Santos, B., Fortin, N., Costigan, C., and Snyder, M. (1998). Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18, 4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, D.S., Biggins, S., and Rose, M.D. (1998). The yeast centrin, cdc31p, and the interacting protein kinase, Kic1p, are required for cell integrity. J. Cell Biol. 143, 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton, A., Immanuel, D., and Arndt, K.T. (1991). The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol. Cell. Biol. 11, 2133–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush, D.R., Maurice, T., Roth, D., and Novick, P. (1996). The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 15, 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Ugolini, S., and Bruschi, C.V. (1996). The red/white colony color assay in the yeast Saccharomyces cerevisiae: epistatic growth advantage of white ade8–18, ade2 cells over red ade2 cells. Curr. Genet. 30, 485–492. [DOI] [PubMed] [Google Scholar]
- Verde, F., Wiley, D.J., and Nurse, P. (1998). Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA 95, 7526–7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink, E., Vossen, J.H., Ram, A.F., Van Den Ende, H., Brekelmans, S., De Nobel, H., and Klis, F.M. (2002). The protein kinase Kic1 affects 1,6-beta-glucan levels in the cell wall of Saccharomyces cerevisiae. Microbiology 148, 4035–4048. [DOI] [PubMed] [Google Scholar]
- Weiss, E.L., Kurischko, C., Zhang, C., Shokat, K., Drubin, D.G., and Luca, F.C. (2002). The Saccharomyces cerevisiae Mob2p-Cbk1p kinase complex promotes polarized growth and acts with the mitotic exit network to facilitate daughter cell-specific localization of Ace2p transcription factor. J. Cell Biol. 158, 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, M.G., and Millar, J.B. (2000). Control of the eukaryotic cell cycle by MAP kinase signaling pathways. FASEB J. 14, 2147–2157. [DOI] [PubMed] [Google Scholar]
- Winzeler, et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.