Abstract
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor, until now described only in vertebrates, that mediates many of the carcinogenic and teratogenic effects of certain environmental pollutants. Here, we describe orthologs of AHR and its dimerization partner AHR nuclear translocator (ARNT) in the nematode Caenorhabditis elegans, encoded by the genes ahr-1 and aha-1, respectively. The corresponding proteins, AHR-1 and AHA-1, share biochemical properties with their mammalian cognates. Specifically, AHR-1 forms a tight association with HSP90, and AHR-1 and AHA-1 interact to bind DNA fragments containing the mammalian xenobiotic response element with sequence specificity. Yeast expression studies indicate that C. elegans AHR-1, like vertebrate AHR, requires some form of post-translational activation. Moreover, this requirement depends on the presence of the domains predicted to mediate binding of HSP90 and ligand. Preliminary experiments suggest that if AHR-1 is ligand-activated, its spectrum of ligands is different from that of the mammalian receptor: C. elegans AHR-1 is not photoaffinity labeled by a dioxin analog, and it is not activated by β-naphthoflavone in the yeast system. The discovery of these genes in a simple, genetically tractable invertebrate should allow elucidation of AHR-1 function and identification of its endogenous regulators.
The mammalian aryl hydrocarbon receptor (AHR) is a ligand-inducible transcription factor that can be activated by environmental pollutants such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and benzo[a]pyrene, and AHR mediates the carcinogenic, teratogenic, hepatotoxic, and immunotoxic effects of these compounds (1, 2). Unliganded AHR resides in the cytoplasm in a complex with the 90-kDa heat shock protein (HSP90) and the immunophilin homolog ARA9 (3). Upon binding ligand, AHR translocates to the nucleus, dissociates from HSP90, and complexes with another transcription factor termed “AHR nuclear translocator” (ARNT) (4, 5). Both AHR and ARNT are members of a family of regulatory proteins that contain an N-terminal basic helix–loop–helix DNA-binding motif and a PAS domain named for its discovery in Drosophila Period, mammalian AHR and ARNT, and Drosophila Singleminded (6–10). The PAS domain in AHR has been shown to mediate interaction with HSP90, binding to ligand, and heterodimerization with ARNT (11–13). The AHR:ARNT heterodimer binds a specific DNA sequence, the xenobiotic response element (XRE; also known as the dioxin response element or AhRE), to regulate the transcription of target genes.
Current evidence suggests that there are at least two classes of AHR agonists, which initiate different cellular responses. In the first class, certain polycyclic aromatic hydrocarbons such as β-naphthoflavone (βNF), as well as the carcinogen benzo[a]pyrene produced by cigarette smoking and other combustion processes, initiate a limited response including the induction of at least three enzymes of the cytochrome P450 pathway and other drug metabolizing enzymes which then metabolize the inducing ligands (4). Some of the resultant electrophilic intermediates can mutate DNA (14). The second class of ligands, including halogenated aromatic hydrocarbons such as TCDD, are poorly metabolized and do not interact directly with DNA (15, 16). However, these agonists typically have greater potency than the first class because they initiate a broader pleiotropic response that includes induction of both the genes mentioned above and other less well characterized genes that are thought to be responsible for the toxic, teratogenic, and carcinogenic effects of these compounds.
Although an endogenous ligand for AHR has not yet been described, the embryonic expression of AHR (17) and the teratogenic effects of TCDD suggest that AHR and its presumptive endogenous ligand(s) play a role in embryonic development and/or homeostasis (18, 19). Disruption of the murine Ahr gene by homologous recombination has been reported by two groups whose results differ in several respects (20, 21). However, both studies conclude that Ahr−/− mice suffer from defects in liver development and exhibit decreased constitutive expression of xenobiotic metabolizing enzymes such as CYP1A2 and the glucuronosyl transferase UGT1A6.
Among the many questions that remain about the function and regulation of AHR are: Do endogenous ligands exist? Does AHR have ligand-independent or ARNT-independent functions? What other proteins interact with AHR or ARNT to influence the activity of the signaling complex? These questions have proven difficult to address in mammalian experimental systems, and until recently, it has not been possible to pursue them in a simple model organism amenable to genetic analysis because no invertebrate AHR ortholog had been described.¶ Here, we present molecular and biochemical characterization of AHR and ARNT orthologs in the nematode Caenorhabditis elegans. We have named the corresponding genes ahr-1 (AHR-related protein) and aha-1 (AHR-associated protein), respectively.
MATERIALS AND METHODS
Isolation of cDNAs.
We sequenced part of the C. elegans cosmid F52F3 and found a region predicted to encode a protein with similarity to the PAS domain of AHR. We designed primers 3gp16 (5′-ATTTCGGTGCTTATTGGATAAC) and 3gp23 (5′-GACTAGATTATAGAGTGGCATTGGTAG) and amplified a partial cDNA from a mixed stage cDNA library (22). We used the PCR product to screen 8 × 105 plaque-forming units of this library and isolated a single cDNA clone (96GA), which lacked the first exon of ahr-1. We used this cDNA to probe a blot of electrophoretically separated C. elegans RNA and found that it hybridized to RNA of a single size. We amplified the 5′ end from other cDNA libraries, first using a λgt11-specific primer (5′-ATATGGGGATTGGTGGCGACGAC or 5′-ATTATTTTTGACACCAGACCAACTG) and an ahr-1-specific primer, 3gp38 (5′-AGCTGACAGGAACTGAGAGTTGTGTAG).
We diluted and reamplified this product using a second λgt11-specific primer, λFX (5′-ACAAGATCTAGAGGTGGCGACGACTCCTGGAGCCCG) or λRX (5′-GTCAGATCTAGATTTGACACCAGACCAACTGGTAATG) or alternatively an SL1 trans-splice leader primer (23) (5′-GTTTAATTACCCAAGTTTGAG) and a primer specific to the ahr-1 helix–loop–helix domain, 3gp39 (5′-AAGAAAGCTGACTGCAAGTCGGAGC). Both strategies amplified the same cDNA sequence. An SL2 primer (24) and 3gp39 amplified no product.
To find a C. elegans ARNT homolog, we searched data from the Genomic Sequencing Consortium (http://eatworms.swmed.edu/) (25) by using the tblastn algorithm (26). In the summer of 1996, these researchers sequenced the cosmid C25A1, which includes aha-1. We designed primers NTp1 (5′-AATGGAAGATGAAGATATGGGCATGC) and NTp5 (5′-AACCACGTATTCGAACTGTTCCGAG) to predicted aha-1 coding sequence and amplified a cDNA fragment from existing libraries. We used this fragment to screen a mixed-stage λgt11 library and isolated cDNAs, some of which included part of the SL1 splice leader.
Construction of Expression Vectors.
We amplified the 96GA ahr-1 cDNA insert by using PCR with λFX and λRX primers, cut with BamHI (in ahr-1 exon 2) and BglII (in λ primer), and subcloned into pSP72 (Promega). This construct was cut with BamHI and ligated to the linker oligonucleotides 5′-AGCTTGCCACCATGGCTCACCATCATCATCACCACGTACGG and 5′-GATCCCGTACGTGGTGATGATGATGGTGAGCCA, which added a Kozak consensus translational start site (27). We created four further constructs as follows.
(i) pJ345: We cut the above plasmid with NcoI and BamHI and ligated to two linker oligonucleotides: 5′-CATGTATGCCAGCAAACGTCGCCAGCGGAACTTCAAAAGGGTACGG and 5′-GATCCCGTACCCTTTTGAAGTTCCGCTGGCGACGTTTGCTGGCATA, which added the ahr-1 sequences lacking in the 96GA isolate; (ii) pJ343: We PCR-amplified aha-1 cDNA from a λgt11 clone by using the primers λFX and NTATG: 5′-CTGAAGCTTGCCACCATGGCTCAGGATATATTTATGGA, which replaced the SL1 splice leader with a Kozak translational start consensus sequence. We digested the resulting product with BglII and HindIII and cloned into pSP72; (iii) pJ350Y: We digested pJ345 with NsiI and BglII and purified the ahr-1 3′ sequences. We then cut BTM116 (28) with EcoRI and BamHI and ligated to the ahr-1 sequences and linker oligonucleotides 5′-AATTCATGAGAGGGCATGCA and 5′TGCCCTCTCATG, and (iv) pJ353Y: We used 3gp30 (5′-CAATCAACAGCTGCACTTCCATTACC) and λFX to PCR-amplify 3′ ahr-1 sequences from 96GA phage. We subcloned the resulting fragment into the EcoRV site of pSP72, cut it out with EcoRI and BglII, and cloned it into BTM116 that had been digested with EcoRI and BamHI.
In Vitro Translation.
We expressed AHA-1, AHR-1, AHR, and ARNT in TNT rabbit reticulocyte lysates as recommended by the supplier (Promega), using the plasmid templates pJ343, pJ345, pSPORTM’AHR (12), and pSPORTARNT (12), respectively.
DNA Binding Assays.
We combined approximately equimolar amounts of each protein in the presence of 1 μM β-naphthoflavone (βNF; Aldrich) in 0.2% Me2SO or Me2SO alone and coincubated the proteins for 90 min at room temperature. We added 1 μg of poly (dIdC) and binding buffer (25 mM Hepes, pH 7.5/200 mM KCl/10 mM DTT/10% glycerol/5 mM EDTA) and incubated the proteins 10 min on ice in a total reaction volume of 16 μl. We then added a labeled probe (0.4 ng) and an unlabeled competitor, and after 10 min of additional incubation on ice, we loaded the reactions onto 0.5X TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) 4.5% acrylamide gels at 4°C. The XRE probe was 5′-TCGAGGGGCATTGCGTGACACC, annealed to 5′-TCGAGGTGTCACGCAATGCCCC. The XREmut2 competitor was 5′-TCGAGGGGCATTACGTGACACC annealed to 5′-TCGAGGTGTCACGTAATGCCCC (the point mutation is underlined).
Coimmunoprecipitation with HSP90.
We produced proteins by in vitro translation in the presence of [35S]methionine. We then used rabbit HSP90 antibody (generously provided by Alan Poland, NIOSH, Morgantown, WV) or preimmune sera to immunoprecipitate proteins from these reactions (13).
Photoaffinity Labeling.
We produced proteins as above, incubated them with 2-azido-3-[125I]iodo-7,8-dibromodibenzo-p-dioxin (125I-N3Br2DD), irradiated them for photoaffinity labeling, and separated them by SDS/PAGE (12).
Assays for Receptor Activation in Yeast.
We introduced pJ350Y, pJ353Y, and pLexA-AHRNΔ166 into Saccharomyces cerevisiae carrying the reporter plasmid pSH18–34 in which the GAL1 promoter is fused to the bacterial lacZ gene and the upstream activating sequence (UASG) has been replaced with eight LexA binding sites (29). We assayed β-galactosidase activity in triplicate for each strain.
RESULTS
Discovery of AHR and ARNT Homologs in C.
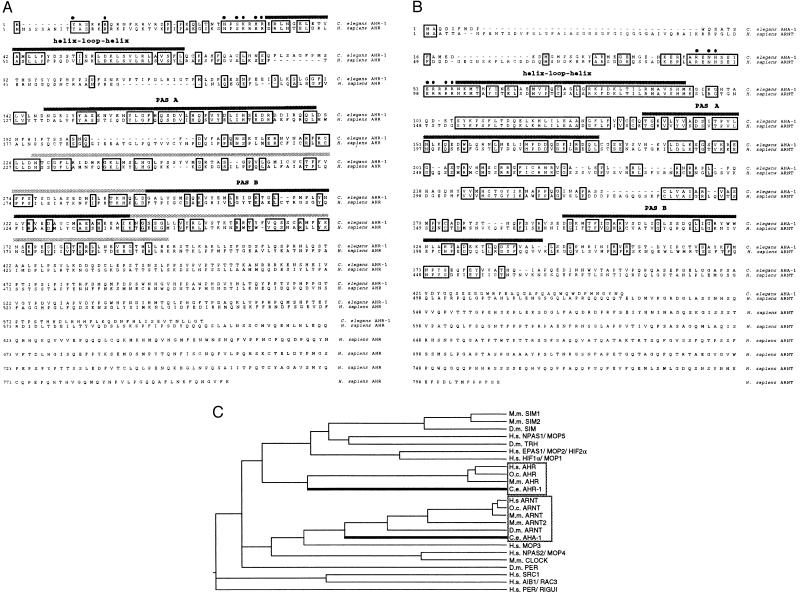
elegans. We discovered a genomic region predicted to encode a peptide with similarity to the PAS domain of mammalian AHR while sequencing a portion of cosmid F52F3 from C. elegans linkage group I in connection with another project. We designed primers to predicted coding sequences and used PCR to amplify a DNA fragment from a mixed-stage cDNA library (22), confirming that these sequences are transcribed. We labeled the fragment, probed the same library, and isolated a single phage clone (see Materials and Methods). The genomic sequence since has been confirmed by the C. elegans Genome Sequencing Consortium (25). We have named the gene ahr-1 (aryl hydrocarbon receptor-related). The predicted AHR-1 protein shares 38% identity with human AHR over a region of 395 amino acids (Fig. 1A), and sequence comparisons indicate that AHR-1 is related most closely to AHR among PAS-domain proteins (Fig. 1C).
Figure 1.
Predicted amino acid sequences of AHR-1 and AHA-1 and their comparison with other PAS domain proteins. (A and B) C. elegans AHR-1 and AHA-1 are aligned with their human homologs. Identical residues are boxed. There are only minimal sequence similarities to the human proteins beyond residues 400 and 375 for AHR-1 and AHA-1, respectively. The regions beyond the PAS domain are not aligned. Filled circles are placed over the residues in the basic domains of the mammalian proteins that have been shown to be important for DNA binding (30, 31); dark bars identify the PAS repeats, and in A, a lighter bar extends the PAS B repeat to include the sequences sufficient for AHR ligand binding (13). The sequences of AHR-1 and AHA-1 have been submitted to the GenBank database (accession nos. AFO39570 and AF039569). (C) N termini through the PAS domains (as defined in ref. 37) are aligned by using the clustal method (megalign) with a PAM250 residue weight table (dnastar, Madison, WI). Species names are abbreviated as follows: Homo sapiens (H.s.), Drosophila melanogaster (D.m.), Oryctolagus cuniculus (O.c.; rabbit), Mus musculus (M.m.), and C. elegans (C.e.). The accession numbers for the proteins included in this analysis are: U40575 (M.m. SIM1); U40576 (M.m. SIM2); P05709 (D.m. SIM); U42699 (D.m. TRH); U77968 (H.s. NPAS1, also termed MOP5 U51628); U81984 (H.s. EPAS1, also called MOP2 U51626); D89787 (M.m. HLF); PIR 138972 (H.s. HIFα); P35869 (H.s. AHR); D38226 (O.c. AHR); A46266 (M.m. AHR); A26588 (D.m. PER); P27540 (H.s. ARNT); A56241 (M.m. ARNT); D63644 (M.m. ARNT2); U51627 (H.s. MOP3); U77970 (H.s. NPAS2, also called MOP4 U51625); AF000998 (M.m. CLOCK); U59302 (H.s. SRC1); D45239 (O.c. ARNT); AF016053 (D.m. ARNT); AF012108 (H.s. AIB1); and AB002107 (H.s. PER, also called RUGUI AF022991).
To ask whether C. elegans also has an ARNT ortholog, we searched data from the sequencing consortium for sequences that could encode ARNT-like proteins and found such a sequence in cosmid C25A1, also from linkage group I in the vicinity of ahr-1. We isolated a cDNA for this gene that included an SL1 splice leader, indicating that it is full length. The amino-terminal sequence of 377 residues of the predicted protein is 45% identical to human ARNT (Fig. 1B) and is more similar to ARNT than to other PAS domain proteins (Fig. 1C). Based on this information and the results of experiments described below, we have named this gene aha-1 (AHR-1 associated).
Binding of AHR-1 and AHA-1 to the Xenobiotic Response Element.
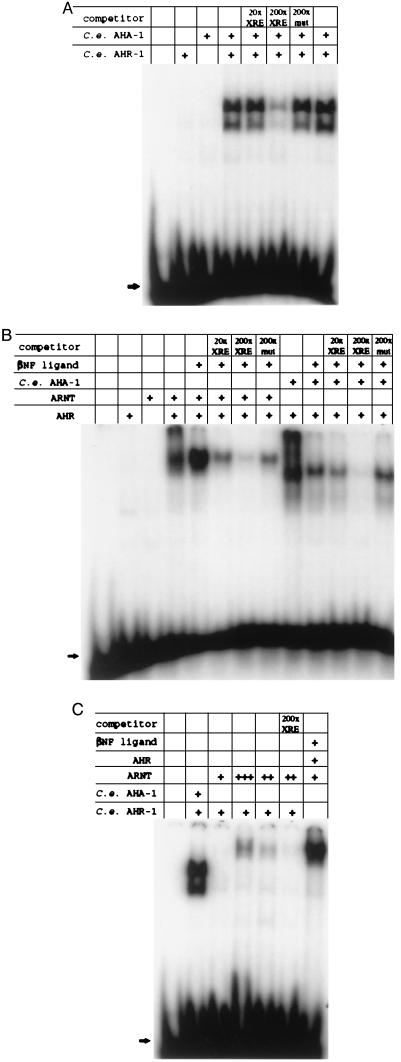
The AHR and ARNT amino acid residues demonstrated to be important for binding to the XRE are conserved in C. elegans AHR-1 and AHA-1, respectively (Figs. 1 A and B) (30, 31). Conservation of these residues distinguishes AHR-1 from PAS domain-containing proteins other than AHR and also suggests that an AHR-1:AHA-1 heterodimer might bind specifically to the XRE. To test this possibility, we transcribed ahr-1 and aha-1 in vitro, translated the resulting mRNAs in rabbit reticulocyte lysates, incubated aliquots of the reaction mixture with a labeled XRE probe, and then carried out electrophoretic gel mobility-shift assays. As shown in Fig. 2A, neither AHR-1 nor AHA-1 alone binds the probe, but when the proteins are coincubated, they interact to bind the XRE and decrease its mobility. At least two complexes are evident. This binding is sequence-specific: The labeled probe can be displaced by a 200-fold excess of unlabeled XRE but not by an unlabeled competitor having a single point mutation in the core binding domain (5′-TGCGTG mutated to 5′-TACGTG). Complex formation is not affected detectably by addition of the AHR ligands βNF or TCDD to the reaction (data not shown; see below).
Figure 2.
Gel mobility-shift assays for DNA binding. C. elegans AHR-1 and AHA-1 and mammalian AHR and ARNT were transcribed and translated in vitro in independent reactions, combined as indicated in the presence or absence of β-naphthoflavone (βNF), and incubated with a labeled XRE probe. The reactions were then electrophoresed on nondenaturing polyacrylamide gels. Unlabeled XRE or XREmut2 DNA was added as competitor to some reactions to assay the sequence specificity of binding. In (C), an additional 2 μl (+++) and 0.5 μl (++) of baculovirus-expressed ARNT (42) was added as indicated to the binding reactions. Arrows indicate the position of free probe.
We tested the ability of the C. elegans and mammalian proteins to form interspecies heterodimers in similar gel-shift assays. As shown in Fig. 2B, murine AHR can complex with C. elegans AHA-1 to bind the XRE sequence specifically. In the converse experiment, C. elegans AHR-1 can be made to complex with human ARNT but only when a large molar excess of ARNT is added to the reaction (Fig. 2C).
AHR-1 Binding to HSP90.
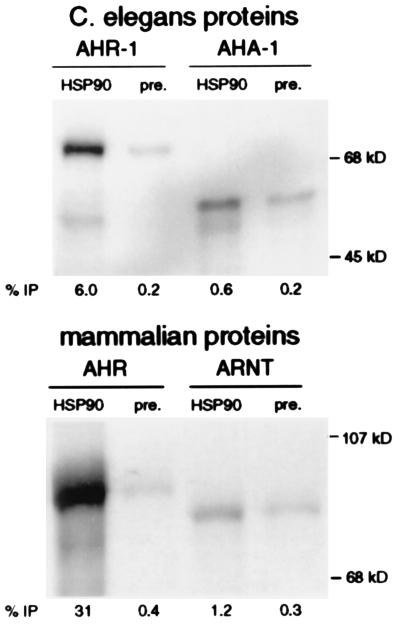
To determine whether C. elegans AHR-1 and AHA-1 share additional biochemical properties of their mammalian cognates, we assayed for coimmunoprecipitation with the HSP90 present in rabbit reticulocyte lysates. The results shown in Fig. 3 demonstrate that rabbit HSP90 binds AHR-1, although less effectively than it binds murine AHR. Like ARNT, AHA-1 does not bind HSP90 well; only threefold more AHA-1 is immunoprecipitated by the HSP90 antibodies than by the preimmune serum. We conclude that C. elegans AHR-1 and AHA-1 have HSP90 binding properties similar to those of mammalian AHR and ARNT, respectively.
Figure 3.
Coimmunoprecipitation with HSP90. C. elegans AHR-1 and AHA-1 and mammalian AHR and ARNT were independently translated and labeled with [35S]methionine in rabbit reticulocyte lysates. Rabbit HSP90 antibody or preimmune sera were used to immunoprecipitate (IP) proteins from these reactions, and the HSP90-associated proteins were resolved by SDS/PAGE. The percentage of total protein translated that immunoprecipitated was calculated, and the average of two experiments was recorded as “% IP”.
Dioxin Binding Assays.
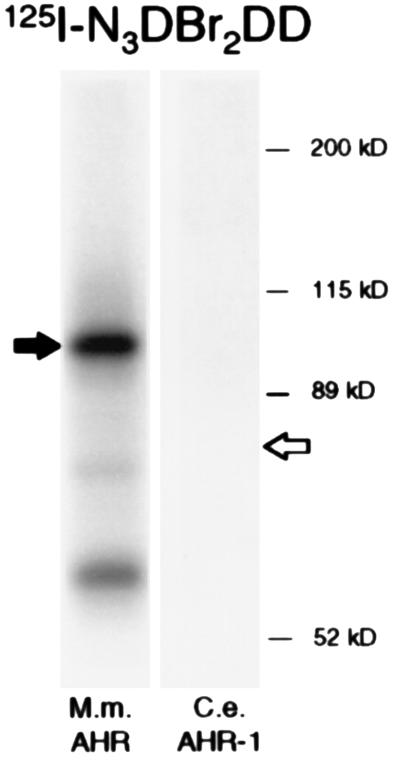
Dioxins such as TCDD are high affinity ligands for mammalian AHR, but no dioxin-binding proteins so far have been identified in invertebrates (32). To test whether AHR-1 could bind a dioxin, we produced the protein in vitro as above and attempted photoaffinity labeling with the dioxin analog 125I-N3Br2DD (12). As shown in Fig. 4, the photoactivated ligand covalently labeled murine AHR in a parallel control reaction but failed to label AHR-1.
Figure 4.
Photoaffinity labeling of AHR-1 and AHR with a dioxin analog. C. elegans AHR-1 and mammalian AHR were expressed in rabbit reticulocyte lysates. The proteins were incubated with 125I-N3Br2DD, photoaffinity labeled, separated by SDS/PAGE, and visualized by autoradiography. The sizes of murine AHR and C. elegans AHR-1 are indicated by filled and open arrows, respectively. No labeling of AHR-1 could be detected.
Assay for AHR-1 Activation in Yeast.
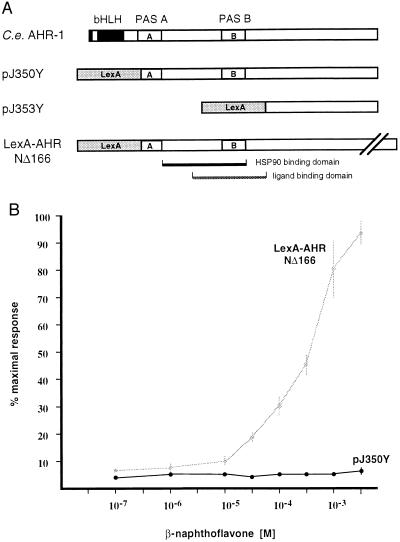
Three possible explanations for the inability of C. elegans AHR-1 to bind 125I-N3Br2DD could be: (i) AHR-1 activity is regulated by lipophilic ligands that do not include dioxins; (ii) AHR-1 does not bind a ligand, but the activation of AHR-1 is regulated by other post-translational events; and (iii) AHR-1 is localized always to the nucleus and is constitutively active. To help distinguish between these possibilities, we used a yeast expression system and two plasmid constructs to assay AHR-1 activity. In one expression construct (pJ350Y), we replaced the basic and helix–loop–helix domains of AHR-1 with the DNA-binding and dimerization domains of LexA (Fig. 5A). In the other (pJ353Y), we also removed the putative HSP90- and ligand-binding domains from AHR-1 to generate a constitutively active mutant. We assayed these expression vectors in cells containing a lacZ reporter driven by eight LexA binding sites upstream of a minimal promoter (29). pJ353Y activated transcription of the lacZ reporter, indicating that the carboxy-terminal region of AHR-1 contains a transcriptional activation domain. However, pJ350Y failed to activate expression of the reporter (see below). These data indicate that the PAS domain of AHR-1, which aligns with the HSP90- and ligand-binding regions of AHR, exerts a repressive function that inhibits nuclear translocation or transcriptional activation.
Figure 5.
Receptor activation assays in yeast. (A) Schematic representation of C. elegans AHR-1 and of the LexA protein fusions expressed by pJ350Y, pJ353Y, and the LexA-AHRNΔ166 control construct (29). In pJ350Y, the LexA DNA binding and dimerization domains replace the AHR-1 bHLH domain. pJ353Y lacks the PAS domains. The control construct LexA-AHRNΔ166 is a fusion of LexA and human AHR as diagrammed. The regions of the AHR PAS domain required for binding to HSP90 and to ligand (13) are indicated. (B) β-naphthoflavone (βNF) dose-response curves for the proteins encoded by pJ350Y and LexA-AHRNΔ166 in the yeast expression system. Immunoblot analyses (not shown) using anti-LexA antibody showed that similar amounts of the fusion proteins were synthesized in cells containing pJ350Y or LexA-AHRNΔ116.
To determine whether a known AHR ligand could activate AHR-1 in this system, we added βNF to yeast containing the pJ350Y construct. βNF activated a LexA–AHR fusion protein in a parallel control experiment, but it failed to activate AHR-1 (Fig. 5B). Immunoblot analyses (not shown) with LexA-specific antibodies indicated that approximately equal amounts of the AHR-1 and control murine AHR fusion proteins were produced.
DISCUSSION
Sequence analysis of ahr-1 and aha-1 and biochemical characterization of their gene products provide strong evidence that these genes are the C. elegans orthologs of Ahr and Arnt. AHR-1 shares 38% identity with human AHR over the regions of AHR shown to mediate DNA binding, dimerization with ARNT, interaction with HSP90, and ligand binding. Moreover, AHR-1 shares important biochemical properties with mammalian AHR. AHR-1 binds HSP90; it appears to require some form of post-translational activation in a yeast expression system, and it interacts with either human ARNT or C. elegans AHA-1 to bind the xenobiotic response element XRE in vitro. However, in our assays, AHR-1 is not activated by the AHR ligand βNF, and it does not bind the dioxin analog 125I-N3Br2DD.
Amino acid residues in AHR and ARNT that have been proven important for recognition and binding to the XRE (30, 31) are conserved in their respective C. elegans orthologs. The consensus XRE sequence for the AHR:ARNT complex, 5′-(T/G)NGCGTG-3′, is asymmetrical (33). ARNT recognizes the 3′ sequence GTG, and AHR binds the 5′ portion of the XRE (34–36). Two other human PAS domain proteins, HIF1α and MOP2/EPAS1/HIF2α, have been shown to prefer the 5′ half sites CAC or TAC over the XRE sequence NGC (37). In this study, we have shown that the binding of C. elegans AHR-1:AHA-1 to the XRE is not competed effectively by a DNA fragment containing the core sequence 5′-TACGTG-3′, demonstrating that C. elegans AHR-1 and mammalian AHR have similar half-site sequence preferences.
Whereas ARNT was known to dimerize with other PAS domain proteins, AHR had not been shown to form a DNA-binding complex with proteins other than ARNT and its closely related paralog ARNT2. We have demonstrated that mammalian AHR can interact with C. elegans AHA-1 to form a ligand-responsive complex that recognizes the XRE. C. elegans AHR-1 also can form an XRE-binding complex with mammalian ARNT, although this interaction requires excess amounts of baculovirus-expressed ARNT.
Activation of C. elegans AHR-1.
The data presented here are consistent with a model in which AHR-1 requires some form of post-translational activation to enter the nucleus or to unmask the transcriptional activation domain. This activating event may or may not include binding to a lipophilic ligand, but if AHR-1 ligands exist, they do not include 125I-N3Br2DD or βNF. When the basic helix–loop–helix motif in AHR-1 was replaced with the DNA-binding and dimerization domains of LexA (construct pJ350Y; Fig. 5), the fusion protein was unable to activate transcription of a reporter gene under the control of a LexA enhancer in yeast. Rather, it behaved like the similarly constructed AHR fusion in the absence of ligand, suggesting that the LexA–AHR-1 fusion is localized to the cytoplasm and repressed. Addition of saturating levels of βNF to the pJ350Y-containing cells did not increase expression of the reporter gene. When the PAS domain was deleted from this construct (in pJ353Y; Fig. 5), the resulting protein was able to activate transcription. These data indicate that the carboxy-terminal region of AHR-1 includes a transcriptional activation domain and that the PAS domain, which aligns with the HSP90- and ligand-binding regions of AHR, has a repressive function that inhibits nuclear translocation or transcriptional activation.
In gel-shift assays, we detect at least two presumed AHR-1:AHA-1 complexes. The presence of multiple complexes may reflect efficient binding by a rare truncation product or post-translational modification of one or both proteins. These DNA-binding complexes form in the absence of any exogenous ligands. Because we do not yet understand the molecular nature of AHR-1 activation, we cannot distinguish between two possible explanations for this finding: (i) that the absence of a nuclear barrier, or a low affinity of rabbit HSP90 for AHR-1, or both, in reticulocyte lysates allows the AHR-1:AHA-1 complex to form and bind the XRE without activation and (ii) that one or more of these complexes includes AHR-1 that has been “fully activated” by an AHR-1 ligand in the reticulocyte lysate or by some other mechanism. Whatever the explanation, the efficiency with which these complexes form suggests that AHA-1 can compete successfully with rabbit HSP90 for binding to AHR-1 under these assay conditions.
Other cellular signaling pathways may play a role in the regulation of AHR-1 activity. Protein kinase C-dependent phosphorylation is necessary for AHR signaling in some mammalian cell lines (38, 39). Specifically, dephosphorylation of AHR inhibits DNA binding, and phosphatase treatment of ARNT inhibits heterodimerization with AHR (40). AHR-1 may be phosphorylated differentially in vivo, and its phosphorylation state in the reticulocyte lysates or in yeast, or both, may have affected its behavior in these assay systems.
Implications of an Invertebrate AHR Complex.
The discovery of AHR and ARNT orthologs in C. elegans indicates that the AHR signaling complex evolved before nematodes and mammals diverged >500 million years ago and that orthologs of these molecules are likely to exist in other phyla. Indeed, an ARNT homolog recently has been identified in Drosophila (41). The sequence information presented here should facilitate the isolation and characterization of other such homologs. Further experimentation will reveal whether developmental functions of the AHR signaling complex also have been conserved. It will be of interest from environmental and toxicological standpoints to determine whether any of these gene products are activated by dioxins and other man-made pollutants.
We currently are screening for mutations in ahr-1 and aha-1 to allow functional analysis as well as generating reporter constructs and antibodies with which to examine the expression patterns of these gene products in C. elegans. We anticipate that genetic and phenotypic analysis of ahr-1 and aha-1 will clarify their roles in C. elegans development and lead to identification of genes that regulate AHR-1 signaling.
Acknowledgments
C.A.B. acknowledges the technical assistance of Lucy Carver and Kristen Carr. We thank Wendy Hanna-Rose, Weiqing Li, Clark Coffman, David Fay, and three reviewers for helpful comments on the manuscript. This work was supported by Grants GM-16294 (to J.A.P.C.), ES-05703 (to C.A.B.), and HD-11762 and HD-14958 (to W.B.W.). J.A.P.C. is a Special Fellow of the Leukemia Society of America; C.A.B. is a Burroughs–Wellcome Scholar in Toxicology, and W.B.W. is a member of the University of Colorado Cancer Center, U. of C. Health Sciences Center, Denver, CO 80262.
ABBREVIATIONS
- AHR
aryl hydrocarbon receptor
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- ARNT
AHR nuclear translocator
- XRE
xenobiotic response element, also known as the dioxin response element or AhRE
- βNF
β-naphthoflavone
- 125I-N3Br2DD
2-azido-3-[125I]iodo-7,8,-dibromodibenzo-p-dioxin
- PAS
period-AHR-singleminded
Note Added in Proof
While this article was in press, an independent analysis of data generated by the C. elegans genome project concluded that the genes we have termed ahr-1 and aha-1 are possible homologs of AHR and ARNT genes, respectively (43). In addition, an AHR homolog is encoded by the spineless-aristapedia gene in Drosophila (44).
Footnotes
References
- 1.Birnbaum, L. S. (1994) Environ. Health Perspect. 102 (Suppl. 9), 157–167. [DOI] [PMC free article] [PubMed]
- 2.Bock K W. Rev Physiol Biochem Pharmacol. 1994;125:1–42. doi: 10.1007/BFb0030908. [DOI] [PubMed] [Google Scholar]
- 3.Carver L A, Bradfield C A. J Biol Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 4.Hankinson O. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- 5.Swanson H I, Bradfield C A. Pharmacogenetics. 1993;3:213–230. doi: 10.1097/00008571-199310000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. Biochem Biophys Res Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 7.Burbach K M, Poland A, Bradfield C A. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffman E C, Reyes H, Chu F F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 9.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 10.Nambu J R, Lewis J O, Wharton K A, Jr, Crews S T. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 11.Coumailleau P, Poellinger L, Gustafsson J A, Whitelaw M L. J Biol Chem. 1995;270:25291–25300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- 12.Dolwick K M, Swanson H I, Bradfield C A. Proc Natl Acad Sci USA. 1993;90:8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukunaga B N, Probst M R, Reisz-Porszasz S, Hankinson O. J Biol Chem. 1995;270:29270–29278. doi: 10.1074/jbc.270.49.29270. [DOI] [PubMed] [Google Scholar]
- 14.Denissenko M F, Pao A, Tang M, Pfeifer G P. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 15.Thorgeirsson S S, Nebert D W. Adv Cancer Res. 1977;25:149–193. doi: 10.1016/s0065-230x(08)60634-x. [DOI] [PubMed] [Google Scholar]
- 16.Pirkle J L, Wolfe W M, Patterson D G J, Needham L L, Michalek J E, Miner J C, Peterson M R, Phillips D L. J Toxicol Environ Health. 1989;27:165–171. doi: 10.1080/15287398909531288. [DOI] [PubMed] [Google Scholar]
- 17.Abbott B D, Birnbaum L S, Perdew G H. Dev Dyn. 1995;204:133–143. doi: 10.1002/aja.1002040204. [DOI] [PubMed] [Google Scholar]
- 18.Abbott B D, Perdew G H, Birnbaum L S. Toxicol Appl Pharmacol. 1994;126:16–25. doi: 10.1006/taap.1994.1085. [DOI] [PubMed] [Google Scholar]
- 19.Nebert D W. Biochem Pharmacol. 1994;47:25–37. doi: 10.1016/0006-2952(94)90434-0. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt J V, Su G H, Reddy J K, Simon M C, Bradfield C A. Proc Natl Acad Sci USA. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Salguero P, Pineau T, Hilbert D M, McPhail T, Lee S S, Kimura S, Nebert D W, Rudikoff S, Ward J M, Gonzalez F J. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- 22.Okkema P G, Fire A. Development (Cambridge, UK) 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 23.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X Y, Hirsh D. Proc Natl Acad Sci USA. 1989;86:8640–8644. doi: 10.1073/pnas.86.22.8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waterston R, Sulston J. Proc Natl Acad Sci USA. 1995;92:10836–10840. doi: 10.1073/pnas.92.24.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 28.Bartel P L, Chien C, Sternglanz R, Fields S. In: Cellular Interactions in Development: A Practical Approach. Hartley D A, editor. Oxford: IRL Press; 1993. pp. 153–179. [Google Scholar]
- 29.Carver L A, Jackiw V, Bradfield C A. J Biol Chem. 1994;269:30109–30112. [PubMed] [Google Scholar]
- 30.Bacsi S G, Hankinson O. J Biol Chem. 1996;271:8843–8850. doi: 10.1074/jbc.271.15.8843. [DOI] [PubMed] [Google Scholar]
- 31.Fukunaga B N, Hankinson O. J Biol Chem. 1996;271:3743–3749. doi: 10.1074/jbc.271.7.3743. [DOI] [PubMed] [Google Scholar]
- 32.Hahn M E, Poland A, Glover E, Stegeman J J. Arch Biochem Biophys. 1994;310:218–228. doi: 10.1006/abbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 33.Lusska A, Shen E, Whitlock J P. J Biol Chem. 1993;268:6575–6580. [PubMed] [Google Scholar]
- 34.Bacsi S G, Reisz-Porszasz S, Hankinson O. Mol Pharmacol. 1995;47:432–438. [PubMed] [Google Scholar]
- 35.Sogawa K, Nakano R, Kobayashi A, Kikuchi Y, Ohe N, Matsushita N, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1995;92:1936–1940. doi: 10.1073/pnas.92.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swanson H I, Chan W K, Bradfield C A. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 37.Hogenesch J B, Chan W K, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Tukey R H. J Biol Chem. 1996;271:26261–26266. doi: 10.1074/jbc.271.42.26261. [DOI] [PubMed] [Google Scholar]
- 39.Carrier F, Owens R A, Nebert D W, Puga A. Mol Cell Biol. 1992;12:1856–1863. doi: 10.1128/mcb.12.4.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berghard A, Gradin K, Pongratz I, Whitelaw M, Poellinger L. Mol Cell Biol. 1993;13:677–689. doi: 10.1128/mcb.13.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zelzer E, Wappner P, Shilo B-Z. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan W K, Chu R, Jain S, Reddy J K, Bradfield C A. J Biol Chem. 1994;269:26464–26471. [PubMed] [Google Scholar]
- 43.Hahn M E, Karchner S I, Shapiro M A, Perera S A. Proc Natl Acad Sci USA. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duncan D., Burgess E. & Duncan, I. (1998) Genes Dev., in press. [DOI] [PMC free article] [PubMed]