Abstract
We recently demonstrated, using yeast DNA microarrays, that mRNAs of polysomes that coisolate with mitochondria code for a subset of mitochondrial proteins. The majority of these mRNAs encode proteins of prokaryotic origin. Herein, we show that a similar association occurs between polysomes and mitochondria in human cells. To determine whether mRNA transport machinery is conserved from yeast to human cells, we examined the subcellular localization of human OXA1 mRNA in yeast. Oxa1p is a key component in the biogenesis of mitochondrial inner membrane and is conserved from bacteria to eukaryotic organelles. The expression of human OXA1 cDNA partially restores the respiratory capacity of yeast oxa1– cells. In this study, we demonstrate that 1) OXA1 mRNAs are remarkably enriched in mitochondrion-bound polysomes purified from yeast and human cells; 2) the presence of the human OXA1 3′ untranslated region (UTR) is required for the function of the human Oxa1p inside yeast mitochondria; and 3) the accurate sorting of the human OXA1 mRNA to the vicinity of yeast mitochondria is due to the recognition by yeast proteins of the human 3′ UTR. Therefore, it seems that the recognition mechanism of OXA1 3′ UTR is conserved throughout evolution and is necessary for Oxa1p function.
INTRODUCTION
A key feature of eukaryotic cells is their organization into separate subcellular compartments, each containing distinct sets of proteins. The sorting of several proteins destined to organelles involves mRNA localization. This specific localization might be preferable to protein localization; indeed, one mRNA molecule can serve as template for multiple rounds of translation. Thus, localizing an mRNA rather than the protein to its site of action offers obvious advantages (Jansen, 2001; Tekotte and Davis, 2002). Mitochondrial biogenesis is a complex process that requires the concerted expression of both nuclear and mitochondrial genomes. More than 98% of mitochondrial proteins are encoded by the nucleus and synthesized in the cytoplasm. Mitochondrial sorting of mRNAs encoding mitochondrial proteins might likely represent a key step to ensure the functionality of the corresponding polypeptides inside the organelle. In this case, a cotranslational phase might assist the import of the precursors (Corral-Debrinski et al., 1999, 2000; Fünfschilling and Rospert, 1999; George et al., 2002). In the yeast Saccharomyces cerevisiae, the impairment of ATP2 mRNA targeting to the vicinity of mitochondria leads to a respiratory deficiency due to an inefficient import of the protein (Margeot et al., 2002). Therefore, mRNA sorting to the vicinity of mitochondria seems to be essential for the organelle function. Yeast DNA microarrays allowed us to demonstrate that >100 mRNAs encoding mitochondrial proteins localize to mitochondrion-bound polysomes and that their 3′ untranslated regions (UTRs) are required for their localization (Marc et al., 2002). Herein, we present evidence indicating that in higher eukaryotes mRNAs encoding mitochondrial proteins exhibit the same distribution pattern between free and mitochondrion-bound polysomes as in S. cerevisiae. We then address the question of whether a common mechanism of 3′ UTR recognition exists in yeast and human cells. To do this, we used the OXA1 gene as a model system. OXA1 encodes a membrane protein highly conserved in both prokaryotes and eukaryotes (Hermann and Neupert, 2000; Luirink et al., 2001; Saint-Georges et al., 2001). In yeast mitochondria, Oxa1p is involved in the insertion of inner membrane proteins, and a null mutation in the OXA1 gene leads to a complete respiratory deficiency. A human Oxa1p homolog has been described that partially rescues the respiratory capacity of a yeast oxa1 mutant, suggesting that the proteins play essentially the same role in both organisms (Bonnefoy et al., 1994). In this study, we first demonstrate that OXA1 mRNA is enriched in mitochondrion-bound polysomes purified from both yeast and HeLa cells. The human cDNA described in the initial report codes for a protein devoid of the first 60 amino acids, corresponding to the mitochondrial targeting sequence (mts) (Rötig et al., 1997). Therefore, we decided to construct the full-length human OXA1 cDNA and to examine the cellular distribution of the two human mRNA species in yeast cells. Both transcripts were detected almost exclusively in mitochondrion-bound polysomes, indicating that the machinery of mRNA targeting to the vicinity of mitochondria is able to recognize both human and yeast sequences. Both transcripts share the same 3′ UTR, and a human OXA1 cDNA devoid of this sequence is unable to rescue cell respiratory capacity of yeast oxa1 mutant, confirming the importance of this 3′ UTR to ensure the function of the human Oxa1p inside the organelle. Moreover, we show that the 3′ UTR of OXA1 mRNA possesses mitochondrial targeting properties, because fluorescent RNAs that encompass either the yeast or the human OXA1 3′ UTRs are found in proximity to mitochondria in living yeast cells. Hence, these data argue for the existence of a common recognition mechanism of the OXA1 3′ UTR in both yeast and human cells.
MATERIALS AND METHODS
Human Cell Culture
HeLa cells were grown in American Type Culture Collection medium until confluence (Eagle's minimum essential medium with 2 mM l-glutamine and Earle's balanced salt solution adjusted to contain 1.5 g/l sodium bicarbonate, 0.1 M nonessential amino acids, 1 mM pyruvate, and 10% fetal bovine serum). These cells have an adenocarcinoma origin and present an epithelial morphology.
Obtention of Human cDNA Clones
The truncated human OXA1 cDNA was cloned by functional complementation of an oxa1–mutation, by using a human cDNA library constructed in pFL61 (Bonnefoy et al., 1994). The yeast expression vector pFL61 contains the replication origin of the yeast 2μ plasmid, the URA3 gene, and the promoter and terminator of the phosphoglycerate kinase gene PGK1 (Minet et al., 1992) The full-length human OXA1 cDNA was constructed by overlap extension using polymerase chain reaction (PCR) (Ho et al., 1989). The first DNA fragment corresponds to the first exon of the human OXA1 gene amplified from a genomic clone (Rötig et al., 1997), and the second one to the truncated human cDNA cloned by complementation. The two fragments were combined in a fusion reaction in which the overlapping ends (63 nucleotides) annealed, and the resulting fusion was amplified by PCR, cloned in pFL61 by using the unique NotI site, and sequenced. To obtain constructions with human OXA1 cDNAs but devoid of their 3′ UTRs, 230 nucleotides long, we purified the complete cDNAs from the pFL61 plasmid digested with NotI. The inserts were then submitted to EcoR1 digestion, a unique site that is present 30 nucleotides downstream from the stop codon of the human OXA1 open reading frame (ORF). The obtained fragments were subsequently cloned in pFL61 linearized by NotI.
Yeast Strains and Culture
The S. cerevisiae strains were the oxa1– mutant NBT1 (mat alpha, oxa1::LEU2, ade2-1, ura3-1, his3-11,-15, trp1-1, leu 2-3,-112) and the corresponding wild-type [WT] OXA1 strain CW04 (mat alpha, ade2-1, ura3-1, his3-11,-15, trp1-1, leu 2-3,-112) (Bonnefoy et al., 1994). Yeast cells were transformed using a simplified lithium method (Gietz et al., 1992).
For the analysis of respiratory function, cells were grown in synthetic medium containing 2% glucose until OD600 of 2. Then, they were serially diluted (1:5) and spotted either on synthetic medium containing 2% glucose supplemented with the appropriate nutritional requirements, or on 2% glycerol medium (1% yeast extract, 2% peptone, 2% glycerol, 10% sodium phosphate buffer, pH 6.2). Plates were incubated at 28°C for either 72 or 96 h.
Biochemical Fractionation of Polysome Populations
Polysomes associated with mitochondria and free cytoplasmic polysomes of yeast cells were isolated as described previously (Margeot et al., 2002). For HeLa cells, we applied the following modifications: cells were collected by trypsination and spun down at 2500 rpm/min for 6 min at room temperature. Cells were resuspended in 20 ml of fresh medium, cycloheximide was added at 250 μg/ml, and the cells were incubated for 15 min at 37°C. After centrifugation, the cells were washed in mannitol-polysome buffer (0.6 M mannitol, 30 mM Tris-HCl, pH 7.4, 5 mM MgAc, 100 mM KCl, 200 μg/ml cycloheximide, 500 μg/ml heparin, 1 g/l bovine serum albumin, and 5 mM β-mercaptoethanol). The pellet was resuspended in 6 ml of mannitol-polysomes buffer, 100 μg/ml digitonin was added, and the cells were incubated on ice for 4 min. Cells were disrupted with 15 strokes by using a glass Teflon pestle. The homogenate was centrifuged at 2500 rpm for 7 min; the pellet was once again disrupted and centrifuged. Both homogenates were combined and centrifuged twice to eliminate unbroken cells and nuclei. The resulting supernatant was further centrifuged at 11,000 rpm for 30 min. The pellet of crude mitochondria associated with polysomes was washed twice in mannitol-polysome buffer before storage at –80°C. Free cytoplasmic polysomes were purified from the post-mitochondrial supernatant by sedimentation through a step gradient of 2 and 0.5 M sucrose. The gradients were centrifuged at 40,000 rpm for 20 h at 4°C by using the TST41 rotor. The pellet, containing free cytoplasmic polysomes, was used for RNA extraction.
RNA Extraction and Northern Blot Analyses
RNA extractions from mitochondrion-bound polysomes, and from free cytoplasmic polysomes, were performed using the RNEasy Protect MIDI kit (QIAGEN, Hilden, Germany). For Northern blots, 8 μg of RNA was separated by electrophoresis through denaturating formaldehyde-agarose gels and transferred to nylon membranes. The blots were stained with methylene blue before prehybridization and hybridization with specific probes (Sambrook et al., 1989). The PhosphorImager system and TINA software were used to compare the relative abundance of each mRNA species. Probes used were obtained by reverse transcription (RT)-PCR by using specific oligonucleotides (Table 1). For the amplifications, we used 200 ng of total RNA purified from HeLa cells and the Access RT-PCR system (Promega, Madison, WI). Labeling of DNA fragments was performed using the Nona Primer kit from Appligene (Illkirch, France).
Table 1.
Human or yeast cDNA probes
| ORF and function | 5′ Primer (5′-3′) | 3′ Primer (5′-3′) | RT-PCR product length (bp) |
|---|---|---|---|
| ACO2 | tccacgagaccaacctgaaga | ctgatggcacacgtggagct | 320 |
| Aconitase, TCA cycle | |||
| AK2 | atgctgagggccatggtggcttct | ccccggtgatgtcatctttcatgg | 363 |
| Adenylate kinase, nucleotide metabolism | |||
| ALDH2 | atggcatgaccatcgccaaggagg | ccatcaatggctgagggaggaagc | 374 |
| Aldehyde dehydrogense 2, carbohydrate metabolism | |||
| ATP5b | atggatggtacagaaggcttggtt | catttcatggtataaatcattgcc | 413 |
| β subunit of F1 ATPase | |||
| COX6c | atggctcccgaagttttgccaaaa | tgttaattgtttatttatcaagag | 354 |
| Cytochrome c oxidase subunit | |||
| COX10 | gttgctaaataaccatttgagagc | acttaaagatagagtttcaaataa | 540 |
| Heme A biosynthesis | |||
| OXA1 | gtggcttctggagagactgcagatgta | gggcacaggaaggttggccatctctct | 420 |
| Cytochrome oxidase assembly | |||
| NDUFV1 | atcgcgccagttcctcagcctcag | ctgcagattggaggcctcattgta | 579 |
| NADH ubiquinone oxidoreductase | |||
| NDUFV2 | atacaatgtataatcgaaagccag | catattttatttctctagtgacaac | 438 |
| NADH ubiquinone oxidoreductase | |||
| UQCRFS1 | gcaggccacggtgcccgccacccc | cctctccatttgaaagccatgttc | 422 |
| Rieske, Ubiquinol-cytochrome c reductase | |||
| SOD2 | ggtacgaccagcactagcagcatg | cagcataacgatcgtggttta | 707 |
| Manganese superoxide dismutase | |||
| IRP1 | tgatcgttctggctggcaaag | cgaccaagtgcacgtctccta | 391 |
| Iron regulatory protein 1 | |||
| FERRITIN H chain | ttgtgtgacttcattgagacac | aagtggatgttttggtacaact | 277 |
| MtDNA (8282-13851) | cctctagagcccactgtaaagc | ttgaggtctagggctgtta | 5569 |
| ATPase8, ATPase6, COXIII, ND3, ND4, ND4L, ND5 | |||
| COX2 | tataggctaaatcctatatat | aggtcgcctggttctaggaat | 402 |
| Yeast OXA1 | ttccgcttctacttcggaccttatcgc | attggaatttgtagcatgggtgcggcc | 420 |
| Cytchrome oxidase assembly | |||
| Yeast COX3 | atgacacatttagaaagaagtagacatcaa | tattacctgcgattaaggcatgatgactat | 250 |
| Cytochrome oxidase subunit | |||
| Yeast COX6 | tatacaacaatgttatcaagggccata | ttcatcatgtgcgtcagaatacttcct | 675 |
| Cytochrome c oxidase subunit | |||
| Yeast ACT1 | gaggttgctgctttggttatt | gtggtgaacgatagatggacc | 800 |
| Cytoskeleton protein | |||
| Yeast ATP2 | tagaagaaataaagcttaaaccaagggg | catgtccagtgggaaagcgaggcaaga | 660 |
| β subunit of F1 ATPase |
The 15 first ORFs correspond to human sequences, whereas the five others are from yeast. The oligonucleotides used for the RT-PCR analysis are indicated in columns 2 and 3 and the size of the PCR products in the fourth column.
RT-PCR Analyses
The respective amounts of human OXA1 mRNAs in yeast and human cells were measured by RT-PCR by using the Access RT-PCR system (Promega, Madison, WI). After DNase treatment of each RNA preparation, 200 ng of RNA was used for reverse transcription. The products obtained were subjected to 25 cycles of PCR with specific oligonucleotides inside each open reading frame (Table 1). Ten percent of the amplified products were run in agarose gels, and the quantities of amplified products reflecting the amount of each mRNA in each polysomal population was measured with the TINA software.
Fluorescence Microscopy
The coat protein (CP) of bacteriophage MS2 was fused to the green fluorescent protein (GFP) and expressed from the plasmid pCP-GFP, generously provided by D. Beach and K. Bloom (Beach et al., 1999). The pCP-GFP is a low copy HIS3 selectable plasmid that produced CP-GFP under the regulation of the MET25 promoter. Cells grown in the presence of methionine produced no detectable CP-GFP protein product. To induce CP-GFP production, cells were switched to a medium without methionine for 2 h. To obtain reporter RNA, we used the pIIIA/MS2–2 plasmid, which contains two tandem copies of the CP-binding site. This plasmid can express a transcript tagged by the two tandem copies of the CP-binding site, under RNase P promoter control, which maintains RNA levels throughout the cell cycle. The single SmaI site allowed us to introduce the nucleotide sequences with potential targeting properties. The oligonucleotides used to amplify the human 3′ UTR (237 nucleotides in length) were as follows: forward primer, 5′tcccccgggggacaaagtatccctggcacgacacatttg3′ and reverse primer, 5′tcccccgggggatttttgtagtacagaggtttactaact3′. To further characterize the region within the human OXA1 3′ UTR having targeting properties in vivo, we studied two fragments of 100 and 137 nucleotides, respectively. For the 100-nucleotide fragment close to the stop codon, we combined the precedent forward primer with the reverse primer 5′tcccccgggggactaggactggggcaaggacacagtg3′. For the 137-nucleotide fragment encompassing the polyadenylation signal, we used the forward primer 5′tcccccgggggaagtcctaggaactgtggcacacacagagat, combined with the reverse primer used to amplify the full-length 3′ UTR.
For the yeast 3′ UTR, the forward primer was 5′cccaagcttgggattaataacaaaaaatgaataaaggc 3′ and the reverse primer was 5′cccaagcttgggtccaaatgattatttcaagcaataaa 3′, which allows the amplification of the complete yeast OXA1 3′ UTR (158 nucleotides in length). In all cases, the binding site precedes the sequence examined. For each inserted sequence, we studied both possible orientations. CW04 cells expressing the “green” RNAs and the CP-GFP protein were observed by fluorescence microscopy with a chilled charge-coupled device camera (Hamamatsu Photonics, Hamamatsu City, Japan). For all microscopic observations, cells were harvested in early-log phase. Mitochondrial DNA and nuclear DNA were visualized using the Hoetsch reagent at 1 nM (Molecular Probes, Eugene, OR). Mitochondria were also labeled with the specific mitochondrial dye MitoTracker Red CMX Ros at 0.02 nM (Molecular Probes). We also performed the induction of the CP-GFP protein for 2 h in a medium devoid of methionine, but instead of 2% galactose, 2% glycerol was added. In this condition, several cells show mitochondria as branched tubular networks. To process the cell images, Leica 4000 software was used.
RESULTS
Detection of mRNAs Exclusively Associated with Mitochondrion-bound Polysomes Isolated from HeLa Cells
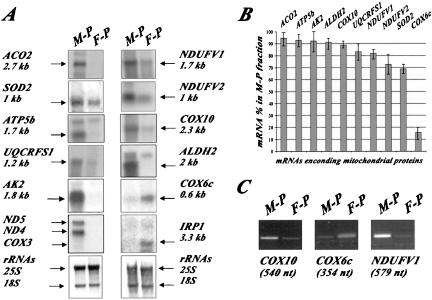
We have previously reported that >100 mRNAs encoding mitochondrial proteins localized to the vicinity of the organelle in yeast cells (Marc et al., 2002). To determine whether this process is conserved throughout evolution, free cytoplasmic and mitochondrion-bound polysomal RNAs purified from HeLa cells were analyzed by Northern blot. We used a 5.6-kb mtDNA fragment (encompassing the region from nucleotide 8282 to 13851) as a mitochondrial marker and the cDNA for IRP1, encoding the nuclear iron regulatory protein as a cytoplasmic marker. Mitochondrial preparations seemed to be devoid of IRP1 mRNA; furthermore, ND5, ND4, and COX3, mitochondrial mRNAs, were not detected in free cytoplasmic polysomes, confirming that there is little cross-contamination between the two polysomal fractions examined (Figure 1). We next investigated whether the mitochondrial fraction contained nuclear mRNAs coding for mitochondrial proteins. Ten genes covering an array of functions within the organelle were chosen for mRNA localization analysis (Table 1 and Figure 1A). Messenger RNAs encoding Aco2, Ndufv1, Ndufv2, Atp5b, Ak2, Aldh2, Sod2, Cox10, and Uqcrf1 proteins were remarkably enriched in the polysomes bound to the mitochondrial surface. Consistently, these nine transcripts were found overrepresented in polysomes bound to mitochondria in four independent experiments (Figure 1B). In contrast, the COX6c transcript mostly localized to free cytoplasmic polysomes, as we previously observed for the yeast homolog (Figure 1). RT-PCR analyses were performed with 200 ng of RNA purified from mitochondrion-bound polysomes or free cytoplasmic polysomes and specific oligonucleotides for COX10, COX6c, and NDUFV1 sequences. Figure 1C shows that the cellular distribution of COX10, NDUFV1, and COX6c mRNAs measured by Northern blot and RT-PCR are quite similar. These results provide evidence that in HeLa cells, as in the yeast S. cerevisiae, several mRNAs encoding mitochondrial proteins localized to the proximity of the organelle in vivo.
Figure 1.
Subellular distribution of mRNAs encoding mitochondrial proteins in HeLa cells. (A) Northern blots were performed with RNA prepared from free polysomes (F-P) and mitochondrion-bound polysomes (M-P) by using probes for different genes encoding mitochondrial proteins (Table 1). At the bottom, methylene blue staining of the ribosomal RNAs is shown. The exposure times of the autoradiograms were ∼14 h at –80°C by using Amersham intensifying screens for all the probes except the 5.6-kb mtDNA, which reveals the ND5, ND4, and COX3 genes, the sizes of these three transcripts are 1.9, 1.45, and 1 kb, respectively, and required an exposure time of only 1 h. (B) Quantifications of the hybridization signals were made using the PhosphorImager system and TINA software. The signal obtained for an individual transcript in the M-P fraction was normalized with the COX3 signal. Normalization for the signal obtained in the F-P fraction was performed using the IRP1 signal. For a given mRNA, addition of both signals after normalization was considered as 100%. The approximate percentage is a mean of four independent biochemical purification experiments, and the Northern blot analyses were performed twice for each polysomal preparation. (C) RT-PCR analysis were performed with 200 ng of RNA purified from M-P and F-P by using specific oligonucleotides within COX10, COX6c, and NDUFV1 ORFs (Table 1), the size of each amplified product is shown at the bottom. The four independent polysomal RNA purifications were subjected twice to RT-PCR analyses.
Both the Yeast and Human OXA1 mRNAs Localize to the Vicinity of Mitochondria
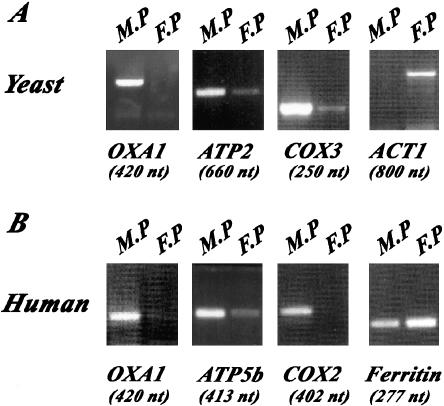
Oxa1p is a member of the conserved OxaI/Yidc/Alb protein family involved in the insertion of proteins into mitochondrial, choloroplast, and prokaryote membranes. To determine the subcellular distribution of OXA1 mRNA, we purified mitochondrion-bound polysomes and free cytoplasmic polysomes from both yeast and HeLa cells. RNA from each polysomal fraction was subjected to RT-PCR analysis. Figure 2 shows that OXA1 mRNA is preferentially found in mitochondrion-bound polysomes; indeed, very little signal was detected in free cytoplasmic polysomes. Additionally, it seems that mRNA localization is identical in yeast and HeLa cells. The distribution of OXA1 mRNA was very similar to that of COX3 and COX2 mRNAs transcribed, respectively, from yeast and human mitochondrial genomes. Furthermore, RT-PCR products for ACT1 and Ferritin confirmed that preparations of mitochondrion-bound polysomes presented little contamination with free cytoplasmic polysomes in both yeast and human cells (Figure 2). Moreover, the distribution of OXA1 mRNA in yeast is quite similar to that of ATP2 mRNA, a transcript almost exclusively found in the vicinity of mitochondria (Margeot et al., 2002). In HeLa cells, OXA1 mRNA is particularly enriched in mitochondrion-bound polysomes, as is ATP5b mRNA (Figure 1). Thus, OXA1 mRNA is mostly found at the proximity of mitochondria, and this particular subcellular distribution is conserved in yeast and human cells.
Figure 2.
OXA1 mRNA localizes to the vicinity of mitochondria in both yeast and human cells. RT-PCR analyses were performed with RNAs purified from mitochondrion-bound polysomes (M-P), free cytoplasmic polysomes (F-P) in yeast and HeLa cells. Specific oligonucleotides were chosen inside each coding region for human OXA1, ferritin, and ATP5b genes, yeast OXA1, ATP2, ACT1, and COX3 genes (Table 1) to perform RT-PCR analyses. Ten percent of the amplified products were subjected to electrophoresis; the size of each amplified product is shown at the bottom.
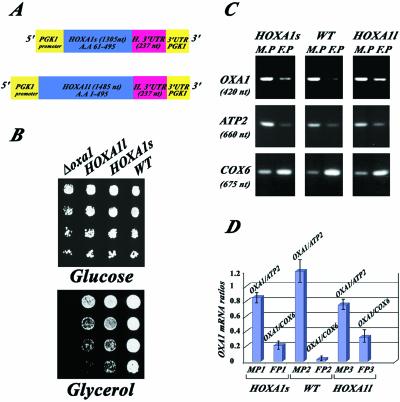
Human OXA1 mRNA Expressed in Yeast Cells Localizes to the Vicinity of Mitochondria
In S. cerevisiae, the oxa1 null mutation leads to a complete respiratory deficiency. A partial restoration of cell respiration is observed when oxa1– cells are transformed with the human homolog, suggesting that the proteins play essentially the same role in both organisms (Bonnefoy et al., 1994). To address the question whether the human OXA1 mRNA expressed in yeast cells is delivered to the proximity of mitochondria, we studied two versions of the human gene (Figure 3A). One directing the expression of the full-length protein (HOXA1l cDNA; see MATERIALS AND METHODS for construction) and the second allowing the expression of a truncated protein in which the first 60 amino acids coding the mitochondrial targeting sequence are absent (HOXA1s cDNA) (Bonnefoy et al., 1994). The expression of human Oxa1p was under the regulation of the yeast PGK1 promoter in each one of the plasmids studied. We first examined the ability of each one of these sequences to complement the respiratory defect of oxa1 yeast mutants. The oxa1– cells plated on glycerol medium are unable to grow at 28°C. Both the truncated and the complete forms of the human OxaI protein rescue the ability to grow on glycerol. Interestingly, cells expressing the truncated version of the human protein grow better than cells expressing the full-length protein (Figure 3B). To determine the subcellular localization of the corresponding transcripts, RT-PCR analyses were performed using RNAs purified from mitochondrion-bound polysomes and free cytoplasmic polysomes (Figure 3, C and D). Several mRNAs were examined by RT-PCR to confirm that little cross-contamination between the two polysomal populations exists. As expected, ATP2 mRNA is mainly detected in mitochondrion-bound polysomes, whereas COX6 transcript is predominantly found in free cytoplasmic polysomes. In wild-type cells, OXA1 mRNA is almost exclusively detected in mitochondrion-bound polysomes, as shown in Figure 2 (Figure 3, C and D, WT). In cells expressing the truncated version of the human protein, the corresponding transcript is strongly enriched in mitochondrion-bound polysomes (Figure 3, C and D, HOXA1s), very little of the transcript is detected in free cytoplamsic polysomes. The mRNA encoding the complete form of human Oxa1p is also very abundant in mitochondrion-bound polysomes, even though the mRNA level in free cytoplasmic polysomes is slightly higher than that found for the yeast mRNA and mRNA encoding the truncated form of the human protein (Figure 3, C and D, HOXA1l). Thus, both human OXA1 mRNAs are recognized and delivered to the vicinity of mitochondria in yeast cells.
Figure 3.
Human OXA1 mRNAs expressed in yeast cells are found associated with polysomes bound to the mitochondrial surface. Yeast oxa1– mutant cells (Δoxa1, strain NBT1) were transformed with plasmids directing the expression of either the full-length Oxa1 protein (HOXA1l) or the N-terminal truncated form (HOXA1s). (A) Schematic representation of the two plasmids. The expression of both human proteins is under the regulation of the yeast PGK1 promoter. Both constructions share at their 3′ extremities 237 nucleotides of the human OXA1 3′ UTR sequence and the yeast PGK1 3′ UTR. (B) The respiratory growth of the transformants and wild-type cells (WT) was examined on glycerol medium at 28°C. The image represents an incubation of 3 d. (C) RT-PCR analysis were performed with RNAs from free cytoplasmic polysomes (F-P), mitochondrion-bound polysomes (M-P) purified from WT cells and oxa1– cells expressing either the HOXA1l or the HOXA1s plasmid. Specific oligonucleotides were chosen inside the coding region for human OXA1, yeast OXA1, ATP2, and COX6 sequences (Table 1). Ten percent of each amplified product was subjected to electrophoresis. (D) Quantifications were made using the TINA software. For the M-P polysomes, the OXA1 RT-PCR product signal was normalized using the ATP2 signal. For the F-P polysomes, the OXA1's RT-PCR signal was normalized using the COX6 signal. The experiments were performed with four independent polysomal RNA purifications, and each polysomal preparation was subjected to RT-PCR analysis twice.
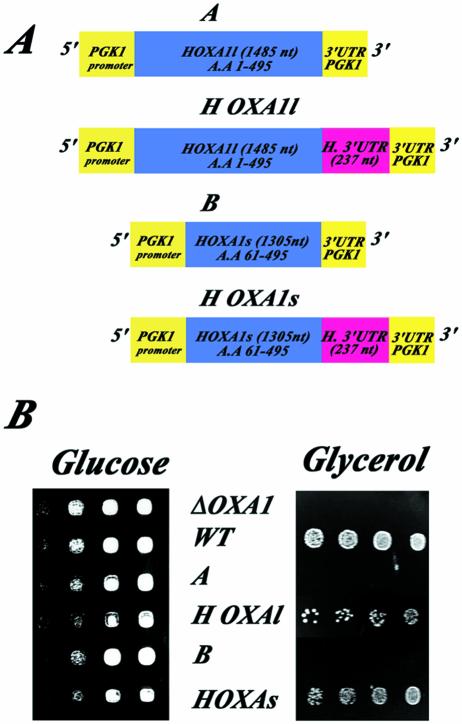
Presence of the Human OXA1 3′ UTR Is Essential to Allow the Human Protein to Rescue the Respiratory Function of Yeast oxa1 Mutant Cells
The two plasmids HOXA1l and HOXA1s direct the expression of human OxaI proteins under the regulation of the yeast PGK1 promoter. The 5′ UTR of the human OXA1 gene is absent in both constructions; however, they share a 237-nucleotide stretch at their 3′ extremities that in the human gene follows the stop codon of the OXA1 ORF (Figure 3A). To determine the role of the 237-nucleotide stretch in the ability of the human protein to rescue the respiratory capacity of oxa1 mutants, we constructed plasmids with either the full-length or the truncated form of the proteins devoid of the entire 3′ UTR (Figure 4A). In these constructions, human OXA1 coding regions are followed by the PGK1 3′ UTR. The PGK1 gene encodes a cytoplasmic protein and its 3′ UTR did not allow a reporter RNA to localize to the proximity of mitochondria in yeast living cells (Corral-Debrinski et al., 2000). Figure 4B clearly shows that the presence of the human 3′ UTR is essential for the respiratory function complementation of oxa1– cells; indeed, cells transformed with the plasmids, in which the human OXA1 3′ UTR is absent, are unable to grow on glycerol medium at 28°C. Thus, the presence of the human OXA1 3′ UTR is required to allow the human Oxa1p to play essentially the same role than its yeast counterparts in the inner mitochondrial membrane.
Figure 4.
The human OXA1 3′ UTR is essential to rescue the respiratory function of yeast oxa1 mutant cells. (A) Schematic representation of the four constructions. The complete human OXA1 3′UTR was deleted from the original constructions directing the expression of either the full-length human Oxa1 protein (HOXA1l) or the N-terminal truncated version (HOXA1s) to give HOXA1/A et HOXA1s/B. (B) Yeast oxa1-mutant cells (Δoxa1, strain NBT1) were transformed with either one of the four plasmids (A) and the respiratory capacity of the transformed cells was tested by serially diluting cells at OD600 of 2 and plating in both glucose medium or glycerol medium. Images represent an incubation of 4 d at 28°C.
3′ UTR of the Human OXA1 mRNA Is Able to Localize a Reporter RNA to the Vicinity of Mitochondria in Living Yeast Cells
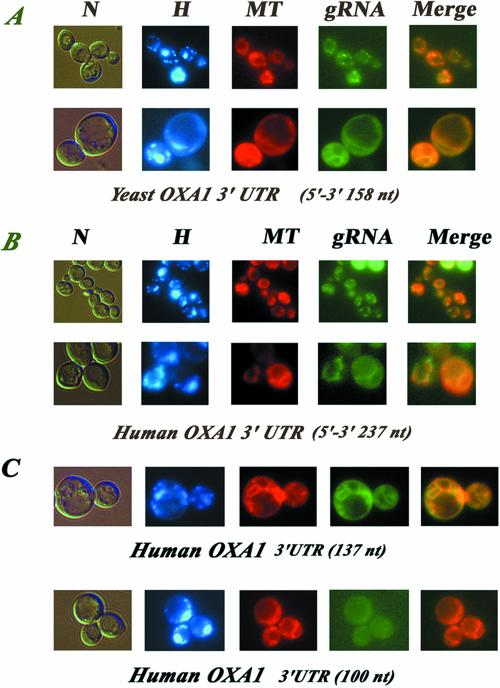
Because we demonstrated that human OXA1 transcripts localized to mitochondrion-bound polysomes and that the presence of the human OXA1 3′ UTR is necessary for the function of the human protein in yeast mitochondria, we next determined whether this sequence possesses mitochondrial targeting properties, by visualizing fluorescent RNAs in living yeast cells (Beach et al., 1999). The 237-nucleotide human sequence, as well as a stretch of 158 nucleotides downstream the stop codon of the yeast OXA1 ORF were fused to two CP (coat protein from the MS2 bacteriophage) binding sites in the pIIIA/MS2-2 plasmid, which directs the synthesis of reporter RNAs. These RNAs can be visualized by fluorescence microscopy when cells express the CP-GFP fusion protein. Cells expressing RNAs in which the 3′ UTRs is inserted in the opposite orientation, such that the noncoding sequence is transcribed, showed a low amount of fluorescence distributed throughout the cytoplasm (our unpublished data). Figure 5A clearly shows that the reporter RNA containing the yeast OXA1 3′ UTR, in the coding orientation, produced a punctuate distribution of fluorescent speckles or reticulum structures representing mitochondria as confirmed by staining with the MitoTracker probe. The overlay of gRNA and MitoTracker labeling (Figure 5A, merge) indicates that the hybrid RNA mostly colocalized with the mitochondria. The human OXA1 3′ UTR is also able to target a reporter RNA to close contact to mitochondria (Figure 5B). When cells were grown on galactose, mitochondria look like small, discrete fluorescent spots within the cytoplasm. A 2-h incubation in 2% glycerol allows visualization of mitochondria as more elaborated networks (Figure 5, A and B, merge). Reporter RNAs containing either the yeast or the human sequence were often visualized in proximity to mitochondria under both conditions examined (Figure 5A and B, merge). Hence, both yeast and human 3′ UTR sequences are recognized by yeast proteins that allow the targeting of reporter RNAs to the proximity of mitochondria in vivo.
Figure 5.
Imaging fluorescent RNAs in living yeast cells. Coexpression of CP-GFP plasmid and reporter RNAs, leads a GFP-labeled RNA, which was visualized using fluorescence microscopy techniques. Cells were grown in 2% galactose medium or incubated in a 2% glycerol medium to detect cells in which mitochondria consist of a branched tubular network and visualized at early log phase. gRNA indicates the green RNA labeling, H the Hoechst staining, N the cells photographed with Nomarski optics, and MT cells labeled with the MitoTracker dye. The merge at the right of the figure represents the superposition of both green RNA fluorescence and mitochondrial labeling with MitoTracker. (A) Reporter RNA with the yeast OXA1 3′ UTR of 158 nucleotides in length cloned in the coding orientation. Top line, CP-GFP expression was induced for 2 h in 2% galactose medium devoid of methionine Bottom line, CP-GFP expression was induced for2hin2% glycerol medium devoid of methionine. (B) Reporter RNA with the human 3′ UTR of 237 nucleotides in length cloned in the coding orientation. Top line, CP-GFP expression was induced for 2 h in 2% galactose medium devoid of methionine. Bottom line, CP-GFP expression was induced for2hin2% glycerol medium devoid of methionine. (C) Reporter RNAs encompassing two subfragments of 100 and 137 nucleotides of the human OXA1 3′ UTR amplified with specific oligonucleotides, and cloned in the unique SmaI site of the pIIIA/MS2–2 plasmid (see MATERIALS AND METHODS). The insert orientations inside the plasmid were checked by PCR by using an internal pIIIA/MS2-2 oligonucleotide. The CP-GFP expression was induced for 2 h in 2% glycerol medium devoid of methionine.
To map more precisely cis-acting elements within the 237 nucleotides of human OXA1 3′ UTR, the subcellular distribution of two deletion RNAs was analyzed. The first one contains the 100 nucleotides immediately downstream of the stop codon, and the other contains the 137 nucleotides close to the putative polyadenylation signal at the end of the human cDNA sequence. When either one of these sequences was placed in the opposite orientation, such that the noncoding sequence was transcribed, both reporter RNAs were diffuse in their distribution throughout the cytoplasm (our unpublished data). The hybrid RNA containing the 100 nucleotides downstream the stop codon did not localize to the vicinity of mitochondria; indeed, a diffuse and weak fluorescent staining in the cytoplasm was consistently observed (Figure 5C). In contrast, the hybrid RNA containing the last 137 nucleotides of the 3′ UTR close to the polyadenylation signal was as efficient as the complete 3′ UTR (Figure 5B). Indeed, we were able to visualize the reporter RNA in branched and elaborated cytoplasmic networks also stained by MitoTracker, which represented mitochondrial structures (Figure 5C, MT, gRNA, and merge). These results indicate that the mechanism by which the human Oxa1p rescues respiratory function of yeast oxa1– cells implies that the corresponding mRNA localizes to the proximity of the organelle in vivo. To achieve this specific localization the human 3′ UTR is required and is probably recognized by yeast proteins allowing its accurate targeting.
DISCUSSION
Subcellular RNA localization is used by eukaryotic cells to achieve high local concentrations of protein products. More than 90 localized mRNAs are known so far. Mounting evidence shows that mRNA localization is not restricted to cell fate determination; it is also required for the efficient import of proteins destined to the nucleus and organelles in somatic cells (Jansen, 2001). A classic example has been found in oligodendrocytes, which are involved in myelination of neurons. In these cells, the mRNA encoding myelin basic protein is localized to the peripheral processes as granules containing multiple mRNA molecules and some components of the translation machinery (Bassell et al., 1999; Tekotte and Davis, 2002). Another example well studied in yeast is the regulation of mating-type switching, which requires the concentration of Ash1p within the daughter nucleus. This asymmetric distribution is achieved by the localization of ASH1 mRNA to the bud tip (Chartrand et al., 1999; Jansen, 2001).
We recently found that yeast mRNAs transcribed in the nucleus and coding for mitochondrial proteins are asymmetrically distributed in the cytoplasm (Corral-Debrinski et al., 2000; Marc et al., 2002). To determine whether this mRNA sorting process is conserved in human cells, we purified polysomes bound to mitochondria from HeLa cells. We clearly demonstrated that, as in yeast, several mRNAs encoding mitochondrial proteins are sorted to the vicinity of mitochondria. Transcripts encoding Aco2, Aldh2, Ak2, Atp5b, Cox10, Uqcrf1, Ndufv1, and Ndufv2 are almost exclusively found in polysomes specifically associated with the mitochondrial surface. In contrast, COX6c mRNA was mostly found in the free cytoplasmic polysomes. Interestingly, the Aldh2 and Ak2 precursors have been shown to use a cotranslational pathway of import (Nobumoto et al., 1998; Ni et al., 1999). Both mRNAs seemed remarkably enriched in polysomes bound to the mitochondrial surface in HeLa cells, confirming the link between mRNA delivery to mitochondrial surface and cotranslational import. Furthermore, 8 of 10 transcripts examined possess yeast counterparts, and their sorting in yeast is similar to that observed in human cells, showing the conservation of this process throughout evolution. In yeast, no simple explanation can be found to account of the specific mitochondrial targeting of mRNAs, except that they code for proteins of procaryotic origin (Marc et al., 2002). Although, we have only analyzed a limited set of human mRNAs, this correlation could also hold true in human cells because Cox6c, a complex IV subunit is not conserved in bacteria and all the other proteins have prokaryotic homologs.
The YidC/Oxa1p/Alb3 protein family is required for the proper biogenesis of a subset of bacterial, mitochondrial, and thylakoid membrane proteins (Luirink et al., 2001). In this study, we showed that two forms of the human OXA1 mRNA, one encoding a truncated version of the protein, devoid of the first 60 amino acids, and the other the full-length protein are mainly targeted to the proximity of yeast mitochondria. Surprisingly, cells expressing the truncated mRNA grow even more rapidly on respiratory medium than those expressing the full-length RNA and the relative amount of RNA bound to mitochondria also seems higher. The two forms of human OXA1 mRNA have no human 5′ UTR sequence but share a 237-nucleotide sequence at their 3′ extremities. We demonstrated in this study, that the deletion of this 3′ UTR sequence abolishes the respiratory growth of the oxa1 mutant cells. Thus this 3′ UTR human sequence seems essential for the mitochondrial function of the human OxaI proteins, whereas the 5′ UTR and the beginning of the ORF are not. Furthermore, the 237 nucleotides and even a subfragment corresponding to the last 137 nucleotides of this 3′ UTR are able to deliver a reporter RNA to the vicinity of mitochondria in living yeast cells, showing that yeast proteins involved in mRNA sorting to the vicinity of mitochondria are able to recognize the human OXA1 3′ UTR. Therefore, as in the case of the ATP2 mRNA (Margeot et al., 2002), there is a direct correlation between human protein function inside the organelle and human OXA1 mRNA localization to the vicinity of yeast mitochondria.
Interestingly, yeast proteins involved in mRNA sorting to the vicinity of mitochondria are able to recognize the human OXA1 3′ UTR. It has already been shown that stem-loop structures are required for the localization of ASH1 mRNA in yeast (Chartrand et al., 1999; Gonzalez et al., 1999). In an attempt to define a conserved structural motif in the 3′ UTR involved in the specific targeting of OXA1 mRNA to the vicinity of mitochondria, we folded stretches of 100 nucleotides within the putative 3′ UTRs (Zuker, 1989) of five OXA1 orthologs, presenting similar functions: Homo sapiens, S. cerevisiae, the two genes of Schizosaccharomyces pombe (Bonnefoy et al., 2000), Neurospora crassa (Nargang et al., 2002), and Arabidopsis thaliana (Hamel et al., 1997). Even though a perfectly conserved structure cannot be defined, motifs encompassing stem-loop structures formed by stems and asymmetrical bulges were observed for all the sequences folded (our unpublished data). This conserved stem loop structure might be recognized by transacting factors involved in mRNA transport to the vicinity of mitochondria.
In conclusion, in human cells, mRNAs encoding a subset of mitochondrial proteins are sorted to the vicinity of mitochondria, as we first demonstrated for the yeast S. cerevisiae. Because motifs within the 3′ UTR of the human OXA1 RNA are functional in yeast cells, we can conclude that the recognition mechanism is also conserved in both cells. The conservation of this process throughout evolution suggests that it is essential for the organelle biogenesis in higher eukaryotes. Therefore, mutations affecting this machinery are likely responsible for some mitochondrial disorders.
Acknowledgments
We thank Michèle Kermorgant for technical assistance. We thank Drs. Elvira Carvajal, Thierry Delaveau and Olga Groudinsky for helpful discussions and critical reading of the manuscript and Dr. Yu-Chun Lone for kindly providing HeLa cells. This study was supported by funds from the Centre National de la Recherche Scientifique, the Ecole Normale Supérieure, and the Association Française contre les Myopathies (grants MNM 2000 to M.C.-D. and G.D.). A.M. is the recipient of a Centre National de la Recherche Scientifique/Bourse de l'Ingénieur fellowship.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–02–0074. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-02-0074.
References
- Bassell, G.J., Oleynikov, Y., and Singer, R.H. (1999). The travels of mRNAs through all cells large and small. FASEB J. 13, 447–454. [DOI] [PubMed] [Google Scholar]
- Beach, D.L., Salmon, E.D., and Bloom, K. (1999). Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9, 569–578. [DOI] [PubMed] [Google Scholar]
- Bonnefoy, N., Kermorgant, M., Groudinsky, O., and Dujardin, G. (2000). The respiratory gene OXA1 has two fission yeast orthologs which together encode a function essential for cellular viability. Mol. Microbiol. 35, 1135–1145. [DOI] [PubMed] [Google Scholar]
- Bonnefoy, N., Kermorgant, M., Groudinsky, O., Minet, M., Slonimski, P.P., and Dujardin, G. (1994). Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1– mutation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91, 11978–11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand, P., Meng, X.-H., Singer, R.H., and Long, R.M. (1999). Structural elements required for the localization of Ash1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 9, 333–336. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski, M., Belgareh, N., Blugeon, C., Claros, M.G., Doye, V., and Jacq, C. (1999). Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol. Microbiol. 31, 1499–1511. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski, M., Blugeon, C., and Jacq, C. (2000). In yeast, the 3′ Untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol. Cell. Biol. 20, 7881–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fünfschilling, U., and Rospert, S. (1999). Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 10, 3289–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, R., Walsh, P., Beddoe, T., and Lithgow, T. (2002). The nascent polypeptide-associated complex (NAC) promotes interaction of ribosome with the mitochondrial surface in vivo. FEBS Lett. 516, 213–216. [DOI] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, I., Buonomo, S.B.C., Nasmyth, K., and Ahsen, U.V. (1999) ASH1 mRNA. localization in yeasts involves multiple secondary elements and Ash1 protein translation. Curr. Biol. 9, 337–340. [DOI] [PubMed] [Google Scholar]
- Hamel, P., Sakamoto, W., Wintz, H., and Dujardin, G. (1997). Functional complementation of an oxa1– yeast mutation identifies an Arabidopsis thaliana cDNA involved in the assembly of respiratory complexes. Plant J. 12, 1319–1327. [DOI] [PubMed] [Google Scholar]
- Hermann, J.M., and Neupert, W. (2000). Protein transport into mitochondria. Curr. Opin. Microbiol. 3, 210–214. [DOI] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pearse, L.R. (1989). Site-directed mutageneses by overlap extension using the polymerase chain reaction. Gene 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Jansen, R.-P. (2001). mRNA localization: message on the move. Nature reviews. Mol. Cell. Biol. 2, 247–256. [DOI] [PubMed] [Google Scholar]
- Luirink, J., Samuelsson, T., and Gier, J.-W.D. (2001) YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501, 1–5. [DOI] [PubMed] [Google Scholar]
- Marc, P., Margeot, A., Devaux, F., Blugeon, C., Corral-Debrinski, M., and Jacq, C. (2002). Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 3, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeot, A., Blugeon, C., Sylvestre, J., Jacq, C., and Corral-Debrinski, M. (2002). In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 21, 6893–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.-E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Nargang, F.E., Preuss, M., Neupert, W., and Herrmann, J.M. (2002). The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J. Biol. Chem. 277, 12846–12853. [DOI] [PubMed] [Google Scholar]
- Ni, L., Heard, T.S., and Weiner, H. (1999). In vitro mitochondrial import: a comparison of leader sequence charge and structural relationships with the in vitro model resulting in evidence for co-translational import. J. Biol. Chem. 274, 12685–12691. [DOI] [PubMed] [Google Scholar]
- Nobumoto, M., Yamada, M., Song, S., Inouye, S., and Nakasawa, A. (1998). Mechanism of mitochondrial import of adenylate kinase enzymes. J. Biochem. 123, 128–135. [DOI] [PubMed] [Google Scholar]
- Rötig, A., Parfait, B., Heidet, L., Dujardin, G., Rustin, P., and Munnich, A. (1997). Sequence and structure of the human OXA1L and its upstream elements. Biochem. Biophys. Acta 1361, 6–10. [DOI] [PubMed] [Google Scholar]
- Saint-Georges, Y., Hamel, P., Lemaire, C., and Dujardin, G. (2001). Role of positively charged transmembrane segments in the insertion and assembly of mitochondrial inner-membrane proteins. Proc. Natl. Acad. Sci. USA 98, 13814–13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, K.J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Tekotte, H., and Davis, I. (2002). Intracellular mRNA localization: motors move messages. Trends Genet. 18, 636–642. [DOI] [PubMed] [Google Scholar]
- Zuker, M. (1989). On finding all suboptimal foldings of an RNA molecule. Science 244, 48–52. [DOI] [PubMed] [Google Scholar]