Abstract
The DNA damage/replication checkpoints act by sensing the presence of damaged DNA or stalled replication forks and initiate signaling pathways that arrest cell cycle progression. Here we report the cloning and characterization of Xenopus orthologues of the RFCand PCNA-related checkpoint proteins. XRad17 shares regions of homology with the five subunits of Replication factor C. XRad9, XRad1, and XHus1 (components of the 9-1-1 complex) all show homology to the DNA polymerase processivity factor PCNA. We demonstrate that these proteins associate with chromatin and are phosphorylated when replication is inhibited by aphidicolin. Phosphorylation of X9-1-1 is caffeine sensitive, but the chromatin association of XRad17 and the X9-1-1 complex after replication block is unaffected by caffeine. This suggests that the X9-1-1 complex can associate with chromatin independently of XAtm/XAtr activity. We further demonstrate that XRad17 is essential for the chromatin binding and checkpoint-dependent phosphorylation of X9-1-1 and for the activation of XChk1 when the replication checkpoint is induced by aphidicolin. XRad17 is not, however, required for the activation of XCds1 in response to dsDNA ends.
INTRODUCTION
During each cell cycle, cells must ensure that DNA replication is completed accurately and that DNA damage is repaired before the onset of nuclear division. Failure to do this will lead to genomic instability that can contribute to the development of cancer in humans. To help maintain genome stability, eukaryotic cells have evolved a complex network of surveillance mechanisms termed checkpoints (Weinert and Hartwell, 1988; Elledge, 1996). These checkpoint pathways detect DNA lesions and convey a signal that halts cell cycle progression and facilitates DNA repair.
In Schizosaccharomyces pombe, a group of 6 proteins (Rad1, Rad3, Rad9, Rad17, Rad26, and Hus1) were found to be essential for the checkpoint response to both DNA damage and blocked DNA replication (Al-Khodairy and Carr, 1992; Enoch et al., 1992). In addition, two protein kinases, Chk1 and Cds1, mediate the DNA damage and DNA replication checkpoint pathways, respectively (Murakami and Okayama, 1995; Walworth and Bernards, 1996; Lindsay et al., 1998; Martinho et al., 1998). These downstream kinases mediate cell cycle arrest by both the positive and negative regulation of proteins that modulate Cdc2-cyclinB kinase activity (Sanchez et al., 1997; Furnari et al., 1999; O'Connell et al., 2000). Components of the checkpoint pathways have been highly conserved through evolution. However, there are significant differences in the organization of the checkpoint pathways in higher eukaryotes compared with the yeasts. In S. pombe, the DNA damage and replication checkpoints are both dependent on Rad3. Rad3 is a member of the phosphatidylinositol-3 (PI-3) protein kinase family and shows significant homology to the PI-3–like protein kinases ATR and ATM of higher eukaryotes (Bentley et al., 1996; Cimprich et al., 1996). In mammalian cells ATR is required for Chk1 activation in response to UV and blocks to replication (Liu et al., 2000; Zhao and Piwnica-Worms, 2002) and overexpression of a kinase-dead ATR mutant renders cells sensitive to DNA damaging agents and replication inhibitors (Cliby et al., 1998). The closely related ATM kinase, which is encoded by the gene mutated in the cancer prone syndrome ataxia-telangectasia, is required for the activation of Chk2 (Cds1) predominantly in response to ionizing radiation (Zhou and Elledge, 2000).
Observations using Xenopus extracts show that XChk1 is phosphorylated and activated in response to aphidicolin or to the addition of UVor MMS-treated pronuclei. This activation is dependent on the initiation of DNA replication (Lupardus et al., 2002; Stokes et al., 2002) and the PI-3–like protein kinase XAtr (Guo et al., 2000; Hekmat-Nejad et al., 2000). On the other hand, XCds1 is phosphorylated by the presence of double-strand DNA ends (Guo and Dunphy, 2000), though it is unknown whether this is dependent on XAtm. Therefore, in Xenopus and other higher eukaryotes it appears that there is a greater distinction between the DNA damage checkpoint and the DNA replication checkpoint at the level of the ATM and ATR kinases and that the roles of Chk1 and Cds1 in higher eukaryotes appear to have been interchanged. For review see Melo and Toczyski (2002).
In S. pombe the checkpoint proteins Rad17, Rad9, Rad1, and Hus1 are essential for both the DNA damage and DNA replication checkpoints. Homologues of these proteins have been identified in humans (Lieberman et al., 1996; Bluyssen et al., 1998; Freire et al., 1998; Parker et al., 1998a, 1998b), demonstrating their conservation through evolution. Rad17 contains regions homologous to all the five subunits that form Replication Factor C (RFC) and has been shown to interact with the four small RFC subunits to form an alternative RFC-like complex (Shimomura et al., 1998; Shimada et al., 1999; Green et al., 2000; Lindsey-Boltz et al., 2001). Bioinformatic analysis of Rad9, Rad1, and Hus1 shows that all three proteins share structural similarity to PCNA (Caspari et al., 2000; Venclovas and Thelen, 2000), and studies in yeast and human systems have demonstrated that Rad9, Rad1, and Hus1 can be detected as a hetero-trimeric complex that is thought to be analogous to the PCNA homo-trimer (Volkmer and Karnitz, 1999; Caspari et al., 2000).
Molecular modeling predicts that Rad9, Rad1, and Hus1 will form a PCNA-like ring structure, whereas Rad17, in conjunction with the four small RFC subunits, will form a complex with structure typical of the RFC clamp-loader (Venclovas and Thelen, 2000). Using electron microscopy, the 9-1-1 proteins have been shown to form a trimeric ring structure similar to PCNA, whereas Rad17 and the four small RFC subunits form a pentameric complex structurally similar to the RFC clamp-loader (Griffith et al., 2002). Together these observations suggest that Rad17, in conjunction with the small subunits of RFC, form a complex required for loading other checkpoint proteins and (or) DNA repair proteins at sites of DNA damage or stalled replication. Indeed several studies suggest that chromatin association of 9-1-1 after DNA damage is dependent on Rad17 function (Melo et al., 2001; Zou et al., 2002). It remains unclear whether the loading of the 9-1-1 complex onto DNA by Rad17 is part of the mechanism that initially senses the presence of damaged DNA and stalled replication forks or whether this occurs downstream of the initial detection of the lesion. In S. cerevisiae and human cells, 9-1-1 subunits can associate with damaged DNA independently of the PI-3–like protein kinases (Kondo et al., 2001; Melo et al., 2001; Zou et al., 2002).
To investigate the role of Rad17 in checkpoint signaling, we cloned and characterized the Xenopus homologues of Rad17 (XRad17), Rad9 (XRad9), Rad1 (XRad1), and Hus1 (XHus1). Using a Xenopus cell-free extract system we find that these proteins become associated with chromatin and become phosphorylated when replication is blocked by aphidicolin. Ablation of XRad17 from egg extracts prevents the aphidicolin-dependent phosphorylation and chromatin association of the X9-1-1 complex and prevents phosphorylation of XChk1. Inhibition of ATR function with caffeine also prevents phosphorylation but does not prevent chromatin association. Thus XRad17 acts in the XAtr/XChk1 pathway and, as seen for DNA damage in yeast and human cells, can associate with DNA when replication is stalled in a manner independent of PI-3–like kinase activity.
MATERIALS AND METHODS
Cloning the Xenopus Homologue of Rad17
Using degenerate primers (ytxgcxgtxcayaaraaraarat, catdatxgtxggxgcxacxacxggrttraa, where x = Inosine) corresponding to the regions of conserved homology between the S. pombe and human protein sequences, a 500-bp fragment of XRad17 was generated using Xenopus oocyte cDNA as template. This fragment was used to screen a Xenopus kidney cell Lambda Zap cDNA library (Stratagene, La Jolla, CA) yielding a 1.5-kb clone that lacked the complete 5′ sequence. To obtain the 5′ end of the gene, RACE PCR (Clonetech, Palo Alto, CA) was carried out on cDNA generated from mRNA isolated from Xenopus tissue culture cells. The resulting 300-bp overlapping RACE PCR product was used to screen the same cDNA library as before yielding a 2.2-kb clone. This clone containing the complete XRad17 ORF. Xenopus checkpoint homologues of Cds1 and Chk1 were cloned by a similar procedure. During preparation of this work a sequence for XRad17 was published and is essentially identical to our independently isolated clone (Lee et al., 2003). Xenopus homologues of XRad9, XRad1, and XHus1 were identified by translated blast searches (tBLASTn) using the respective human amino acid sequences. Xenopus ESTs of potential clones were obtained from the UK HGMP resource center and the clones were characterized. Because many of these clones came from post-midblastula transition stages of development, we designed PCR primers and used egg cDNA as a template. In each case we obtained a sequence identical to that found in the EST clones. IMAGE clones obtained from UK HGMP Resource Center were as follows: XRad1: 3557566, 4435085 XHus1:4678866, 4679890. XRad9 was cloned from egg cDNA by PCR using sequence data from ESTs GenBank accession numbers: BJ099796, BJ070583, BJ095923, and BJ087612.
Generation of Antibodies
Full-length polypeptides of XRad9, XRad1, and XHus1 and a 320-amino acid C-terminal fragment of XRad17 were expressed in E. coli (BL21DE5) and purified on Nickel NTA agarose (Qiagen, Crawley, UK) under denaturing conditions according to the manufacturer's instructions. Purified protein was used for the production of polyclonal antisera (Eurogentec Ltd, Herstal, Belgium). Antibodies were affinity-purified on antigen coupled to amino-link plus resin (Perbio, Chester, UK). Antibodies to the downstream kinases XCds1 and XChk1 were raised against an N-terminal 25.3-kDa fragment and a C-terminal 26.5-kDa, fragment respectively. Antibodies to Phospho (S345) human Chk1 and phospho (S645) human Rad17 were purchased from Cell Signaling Technologies (Beverly, MA).
Preparation of Xenopus Extracts and DNA Templates
Low-speed “cycling” extracts and demembranated sperm heads were prepared essentially as previously described (Murray, 1991). Briefly, eggs were dejellied and activated with calcium ionophore in 25% Barth X for 2 min. After removal of the ionophore by several changes of 25% Barth X, the eggs were incubated for 20 min, washed five times in ice-cold XB buffer (100 mM KCl, 50 mM sucrose, 0.1 mM CaCl2, 1 mM MgCl2, 10 mM potassium HEPES, pH 7.7), and centrifuged at 10,000 × g (HB-4 rotor) for 10 min at 4°C. The cytoplasmic fraction was removed and treated with 50 μg/ml cytochalasin B and 10 μg/ml aprotinin. Extract was centrifuged as above to clarify the extract and stored on ice before use.
Depletion of XRad17 from Xenopus Extracts
Xenopus low-speed extract was incubated with 20% vol/vol of protein A sepharose (Pharmacia, Bucks, UK) cross-linked to affinity-purified XRad17 antibodies for 40 min at 4°C with occasional resuspension of the beads. Two rounds of depletion were carried out. Mock-depleted extracts were treated identically except protein A beads were coated and cross-linked to nonspecific rabbit IgGs (Sigma, Poole, UK). Antibodies were cross-linked with dimethyl pimilimidate using the method of (Harlow and Lane, 1988).
Isolation of Nuclei and Chromatin from Extracts
Typically, 50 μl of extract containing nuclei (3 × 103/μl) was diluted with 200 μl of XB buffer. Diluted extract was overlaid on 800 μl of XB containing 750 mM sucrose and centrifuged at 5000 × g for 10 min. The supernatant was removed and the pellet was resuspended in SDS-PAGE sample buffer.
To isolate chromatin, the nuclear pellet was disrupted with a fine pipette tip using 200 μl of XB with 0.6% Triton X-100. This was overlaid on 800 μl 750 mM sucrose cushion and centrifuged at 10,000 × g for 5 min. The supernatant was removed, and the chromatin pellet was resuspended in SDS-PAGE sample buffer. For phosphatase treatment the chromatin pellet was resuspended in phosphatase buffer (5 mM MgCl2, 50 mM Tris, pH 8.0) with 20 Ui calf intestinal phosphatase (Roche) and incubated at 37°C for 30 min.
Replication Assays
Replication assays were carried out as previously described (Hutchison et al., 1988).
Expression and Purification of Recombinant Geminin
The plasmid encoding GST-geminin was a generous gift from Dr. Matthew Michael. The recombinant protein was expressed and purified as previously described (Stokes et al., 2002).
SDS-PAGE and Western Blotting
Samples were run on 10% acrylamide gels using Protogel 30:0.8 acrylamide:bis acrylamide (National Diagnostics, Hull, UK). Protein was transferred to nitrocellulose (Nitrobind, Osmonics, Minnetonka, MN) at 100V for 1 h using Towbin transfer buffer (48 mM Tris-base, 39 mM glycine, 20% methanol). Protein transfer was visualized with Ponceau S before membranes were blocked with Blotto (PBS containing 5% nonfat milk powder and 0.5% Tween 20). Membranes were incubated with antibodies at various dilutions (1/1000–1/5000) overnight at 4°C. After washing with Blotto, membranes were incubated with a peroxidase-conjugated anti-rabbit secondary antibody (Dako, Cambs, UK) at a dilution of 1/5000 in Blotto. After extensive washing with PBS, proteins were detected using chemiluminescence. Quantitation of Western blots was carried out using the Uvitec system.
Site-directed Mutagenesis and In Vitro Transcription
Mutations were introduced into cDNAs using the Quick-Change site-directed mutagenesis kit (Stratagene). Mutation was confirmed by sequencing. The complete ORF of each positive clone was sequenced to ensure no additional mutations had been generated by the procedure. The predicted ORF of each cDNA was cloned into the expression vector pEPEXHA. This vector places a single HA tag at the N-terminus of any expressed protein. It is a modified form of the plasmid pEPEX (Gautier et al., 1991). Each mRNA was produced using the “Message Machine” in vitro transcription kit (Ambion, Austin, TX). mRNAs were translated in rabbit reticulocyte lysate (Promega, Madison, WI).
RESULTS
Isolation of Xenopus Checkpoint Gene Orthologues
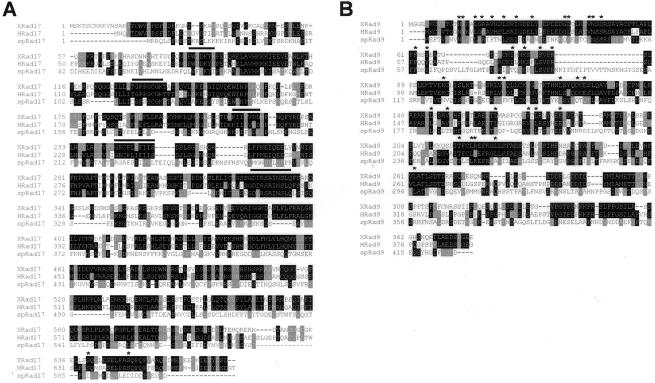
To investigate activation of the DNA replication checkpoint in Xenopus we cloned the Xenopus orthologues of Rad17, Rad9, Rad1, and Hus1. This was performed by a combination of degenerate PCR, database searches, and RACE PCR (see MATERIALS AND METHODS). The predicted amino acid sequences of all four proteins show a high degree of identity when compared with the human (65–70%) and the S. pombe (25–28%) sequences. While this manuscript was being prepared, identical sequences for XRad1 and XHus1 and XRad17 were published (Lupardus et al., 2002; You et al., 2002, Lee et al., 2003). Sequences for XRad17, XRad9, XHus1, and XRad1 have been deposited in databases (GenBank accession numbers AY253230, AY253231, AY253232, and AY253233, respectively). Figure 1, A and B, shows sequence alignments for the Xenopus homologues of XRad17 and XRad9. In Figure 1A, regions of homology between XRad17 homologues and the five subunits of RFC are indicated below solid lines. It is interesting to note that these domains coincide with regions where there is a high degree of similarity between the human, Xenopus, and S. pombe sequences. In Figure 1B conserved residues common to XRad9, XRad1, XHus1, and PCNA are indicated with asterisks. The PCNA-like domain in XRad9 is at the N-terminus of the protein, as in the yeast and human homologues (Caspari et al., 2000; Venclovas and Thelen, 2000).
Figure 1.
Rad17 contains regions of homology to RFC subunits and XRad9 is related to PCNA. (A) Multiple sequence alignment of XRad17 with human and S. pombe orthologues. Regions showing extensive homology to RFC subunits are indicated as solid black line. The SQ residues conserved between the human and Xenopus sequences are marked with asterisks. (B) Amino acid sequence alignment of XRad9 with the human and S. pombe orthologues. Residues that are conserved between XRad9, XRad1, XHus1, and PCNA are indicated with asterisks. Alignment was carried out using Clustal-W (Thompson et al., 1994), and the alignments subsequently were processed using BOXSHADE (V3.21). Identical residues are shaded black, and similar residues are shaded in gray.
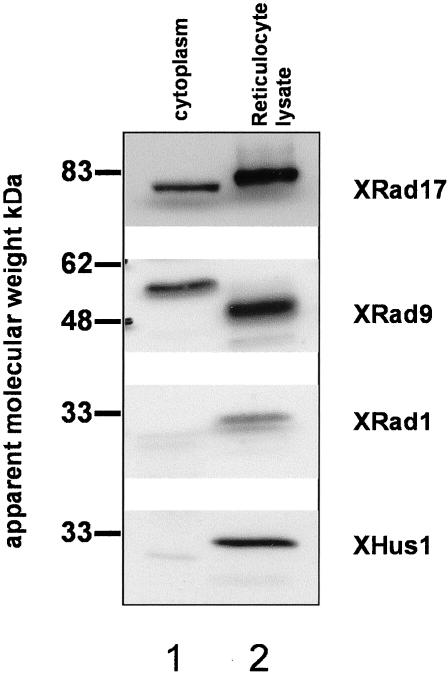
Polyclonal antibodies were raised against a 320-amino acid C-terminal fragment of XRad17 and the full-length XRad9, XRad1, and XHus1 proteins expressed in E. coli. Each of the antibodies was affinity-purified against its respective antigen. All four antibodies specifically detect the relevant protein expressed in reticulocyte lysate (Figure 2) and show no cross-reaction with other proteins expressed in this way (unpublished data). In Xenopus extract XRad17 antibodies detect a single band of ∼78 kDa, the predicted molecular weight of XRad17 (Figure 2). Antibodies to XRad9 also detect a single band in Xenopus extract, although this band, at ∼55 kDa, is significantly larger than the predicted molecular weight of ∼43 kDa (Figure 2). In vitro–translated XRad9 runs at 43 kDa as predicted. However, incubation of recombinant XRad9 in Xenopus extract converts it to the 55-kDa form (unpublished data). Antibodies to XRad1 and XHus1 also detect bands of the expected molecular weight of 32 kDa in Xenopus extract (Figure 2). Using the appropriate recombinant protein as a standard, we estimate that all four of the endogenous proteins are found at concentrations in the range 3–5 ng/μl of extract.
Figure 2.
Characterization of antibodies raised to Xenopus checkpoint proteins. Samples, 2-μl, of Xenopus low-speed extract (lane 1) were immunoblotted with antibodies raised to XRad17, XRad9, XRad1, and XHus1. To confirm specificity of the antibodies, they were also tested against 2 μl of rabbit reticulocyte lysate (Promega T7, TnT) expressing the appropriate cDNA (lane 2). The vector used to express the cDNAs places a single HA epitope at the N-terminus of the protein. This is consistent with the slightly higher apparent molecular weight of the in vitro–translated proteins.
Chromatin Binding of XRad17 Is Stimulated when Replication Is Inhibited by Aphidicolin
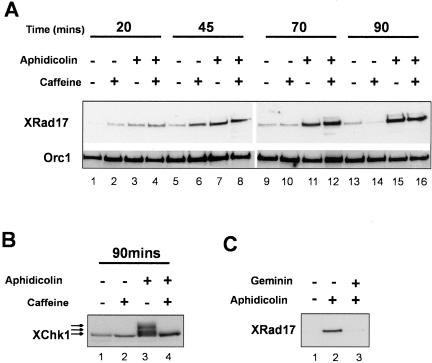
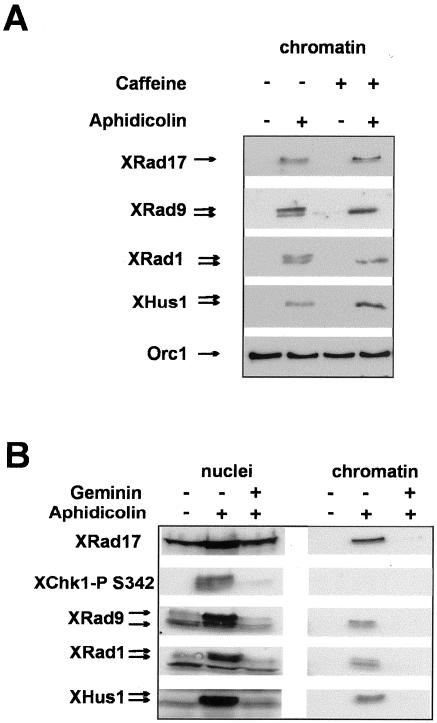
Rad17 homologues in other systems have been shown to be constitutively associated with chromatin throughout the cell cycle. In S. pombe and human cells, Rad17 chromatin binding increased when cells are treated with DNA damaging agents or replication inhibitors (Griffiths et al., 1995; Kai et al., 2001; Zou et al., 2002). We examined the chromatin association of XRad17 in low-speed Xenopus egg extracts. Very little XRad17 associated with chromatin in untreated extracts (Figure 3A, lanes 1, 5, 9, and 13). However, when DNA replication was inhibited with aphidicolin (an inhibitor of DNA polymerase α), XRad17 chromatin binding was significantly increased within 45 min and continued to increase over the 90-min duration of the incubation (Figure 3A, lanes 7, 11, and 14). The activation of the replication checkpoint under these conditions is indicated by phosphorylation of the checkpoint kinase XChk1 (Figure 3B, lane 3). The addition of 5 mM caffeine was sufficient to prevent the phosphorylation of XChk1 in response to aphidicolin (Figure 3B, lane 4) but did not reduce XRad17 chromatin binding. Because caffeine is a potent inhibitor of the ATM and ATR kinases, both in vitro and in vivo (Blasina et al., 1999; Hall-Jackson et al., 1999; Sarkaria et al., 1999), this result suggests that XRad17 chromatin binding is independent of the ATR or ATM kinase activities. The addition of caffeine to aphidicholin-treated extract may in fact enhance XRad17 chromatin binding (Figure 3A, compare lanes 3 and 4, 7 and 8, 11 and 12, 15 and 16), though the extent of this increase varied between experiments. The inclusion of caffeine in extracts incubated with MMS-treated sperm pronuclei also resulted in an increase in XRad17 chromatin binding (Stokes et al., 2002). More recently Lee et al. (2003) also demonstrated that caffeine increased the aphidicolin-dependent chromatin binding of XRad17. In addition they showed that depletion of XAtr also led to increased chromatin binding of checkpoint proteins in extracts treated with aphidicolin (Lee et al., 2003). Together these data suggest that ATR/ATM kinases may play some role in the subsequent release of checkpoint complexes from chromatin.
Figure 3.
Chromatin binding of XRad17 is stimulated when replication is inhibited with aphidicolin. (A) Aliquots, 50 μl, of extract containing nuclei (3 × 103/μl) and cycloheximide, 50 μg/ml, were treated with caffeine (5 mM), aphidicolin (100 μg/ml), or both drugs together. At the time points indicated chromatin was isolated and associated protein analyzed by SDS-PAGE and immunoblotting using the XRad17 antibody. Western blots were subsequently stripped and reprobed with an antibody to ORC1 to provide a loading control. Caffeine prevents aphidicholin-dependent phosphorylation of XChk1. (B) A duplicate set of samples was taken at 90 min, and the nuclear fraction was isolated. This nuclear fraction was analyzed for Western blotting using an antibody raised to Xenopus Chk1 (XChk1). The appearance of the phosphorylated forms of XChk1 (lane 3) shows that aphidicolin has activated the replication checkpoint. Lane 4 shows the effect of caffeine in abrogating the activation of XChk1 despite the presence of aphidicolin. (C) XRad17 chromatin binding in response to aphidicolin requires the initiation of replication. Extract, 50 μl, containing nuclei (3 × 103/μl) and cycloheximide (50 μg/ml) was incubated at 21°C for 90 min in the presence of aphidicolin (100 μg/ml) or aphidicolin with 250 nM recombinant geminin. Chromatin was purified and subjected to Western blot analysis with XRad17 antibody.
Recent studies have shown that the chromatin association of checkpoint proteins can be enhanced by incubation of MMS-treated or UV-treated sperm pronuclei in egg extract. Under these conditions, chromatin binding was found to be dependent on the initiation of DNA replication as it was abrogated by the addition of the replication inhibitors geminin or p27kip (Lupardus et al. 2002, Stokes et al., 2002). Geminin blocks DNA replication before prereplication complex formation and p27kip inhibits the cdk2/cyclinE kinase activity that is necessary for the initiation step. Both these restriction points are upstream of the aphidicolin-sensitive polα step and thus prevent activation of the replication checkpoint (Michael et al., 2000). Consistent with this, we find that the XRad17 chromatin binding induced by aphidicolin can be prevented if replication is inhibited upstream of initiation by the preaddition of geminin to extracts (Figure 3C).
Activation of the Replication Checkpoint Leads to Phosphorylation of XRad9, XRad1, and XHus1
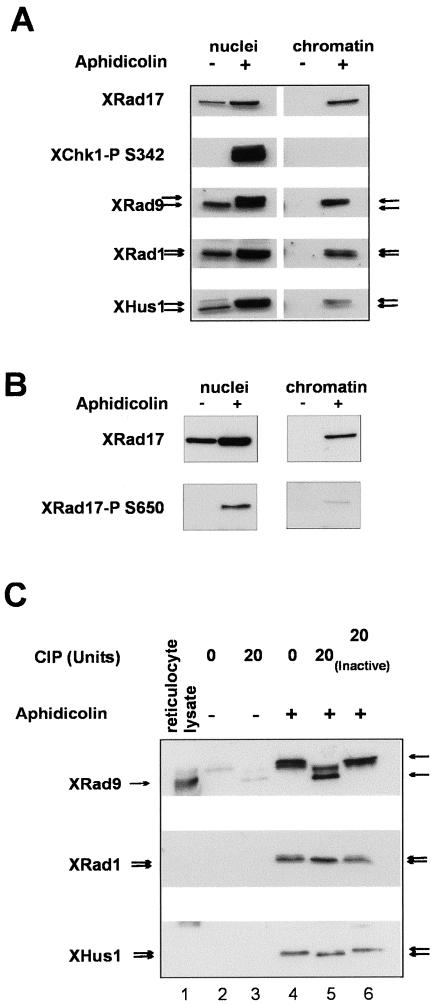
Having established that the chromatin association of XRad17 was stimulated when replication was blocked by aphidicolin, we wanted to determine if the distribution of XRad9, XRad1, and XHus1 proteins in the nuclear and chromatin fractions was similarly affected. The concentration of XRad17, XRad9, XRad1, and Hus1 found in nuclei isolated from aphidicolin-treated extract showed a significant increase (∼5-fold by densitometer analysis) when compared with nuclei from untreated extract (Figure 4A, left panel). To demonstrate the aphidicholin-dependent activation of the replication checkpoint in these experiments, we analyzed the phosphorylation status of XChk1. XChk1 is phosphorylated on Ser 342 in an ATR-dependent manner, and this phosphorylation is a reliable marker for replication checkpoint activation in Xenopus extracts (Guo et al., 2000). We thus used a phosphospecific human Chk1 antibody that has been shown to specifically recognize the equivalent phosphorylated residue on XChk1 (Figure 4A, left panel; Shimuta et al., 2002). XRad17 contains two conserved SQ motifs in the c-terminus of the protein (see Figure 1A). In the human protein these sites are phosphorylated in an ATR/ATM-dependent manner in response to a variety of DNA damaging agents or to DNA replication inhibitors (Bao et al., 2001; Post et al., 2001). To investigate XRad17 phosphorylation during activation of the DNA replication checkpoint, we used a phosphospecific antibody directed at Ser645 of the human Rad17 protein. This antibody only detected XRad17 in the nuclear and chromatin fractions from aphidicolin-treated extracts (Figure 4B, left and right panels). We conclude that XRad17 becomes phosphorylated on Ser650 after activation of the replication checkpoint in Xenopus.
Figure 4.
XRad9, XRad1, and XHus1 associate with chromatin and undergo posttranslational modification when replication is blocked with aphidicolin. (A) Extract, 50 μl, with nuclei (3 × 103/μl) and cycloheximide (50 μg/ml) were incubated in the absence or presence of 100 μg/ml aphidicolin. After 90 min duplicate samples were then diluted in XB buffer and centrifuged through a 750 mM sucrose cushion to isolate the nuclear fraction. The chromatin fraction was obtained by disruption of this nuclear fraction in XB buffer containing 0.6% Triton X-100, followed by centrifugation through an identical sucrose cushion. Proteins were resolved by SDS-PAGE on 10% gels. After transfer to nitrocellulose, proteins were detected with polyclonal antibodies to XRad9, XRad1, XHus1, and human phospho-specific(S345) Chk1 (Cell Signaling Technologies). (B) XRad17 becomes phosphorylated on Ser650 when replication is blocked with aphidicolin. Samples treated as described above, and Western blots were probed with antibodies to XRad17 or human antiphospho (S645) Rad17 (Cell Signaling Technologies). (C) Phosphatase treatment of chromatin from extract incubated with aphidicolin shows that modification of XRad9, XRad1, and XHus1 is due to phosphorylation. Chromatin was isolated from extracts incubated in the absence (lanes 2 and 3) and presence of 100 μg/ml aphidicolin (lanes 4–6). The chromatin pellets were resuspended in phosphatase buffer (5 mM MgCl2, 50 mM Tris, pH 8.0) and incubated at 37°C for 30 min with either 0 or 20 Ui of CIP or 20 Ui of CIP that had previously been inactivated by heating at 80°C for 20 min). After CIP incubation the reactions were stopped with sample buffer and subjected to SDS-PAGE. The proteins were then immunoblotted with XRad9, XRad1, or XHus1 antibodies. Lanes 1 contains 2 μl of rabbit reticulocyte lysate reaction expressing XRad9 from the pepexHA vector.
In addition to XChk1, we observed that XRad9, XRad1, and XHus1 all underwent posttranslational modification after aphidicolin treatment as judged by the appearance of slower mobility forms on SDS-PAGE gels (Figure 4A, left panel). When we analyzed the chromatin fraction, we found that XRad9, XRad1, and Hus1 showed similar chromatin association profiles as we had previously seen forXRad17. All three proteins associated with chromatin after replication was blocked with aphidicolin (Figure 4A, right panel). We also found that it was predominantly the modified form of XRad9 and XHus1 that was recovered in the chromatin fraction, whereas both forms of XRad1 were detected (Figure 4, A right panel).
To demonstrate that these modified forms were produced by phosphorylation events, we treated the chromatin fraction isolated from aphidicolin-treated extracts with calf intestinal phosphatase (CIP). In each case, CIP treatment resulted in loss of the aphidicolin-dependent slower mobility form (Figure 4B). In the case of XRad9, CIP treatment resulted in a more extensive decrease in apparent molecular weight. The CIP-treated XRad9 migrated at a similar size to the recombinant protein (43 kDa), indicating that XRad9 undergoes checkpoint-independent phosphorylation in Xenopus extracts. This is consistent with our observation that the recombinant protein migrates at 56 kDa after incubation in Xenopus extract.
We were interested to determine the significance of the phosphorylation events that occur when the checkpoint is activated by aphidicolin. Caffeine has been shown to abrogate the replication checkpoint by inhibiting the kinase activity of the PI-3–like kinases. We analyzed the chromatin binding and phosphorylation of XRad9, XRad1, and XHus1 in the presence of aphidicolin and 5 mM caffeine (Figure 5A) and found that the presence of caffeine did not significantly affect the aphidicolin-induced chromatin binding of the three checkpoint proteins, but did inhibit their aphidicolin-induced phosphorylation. These observations suggest that the phosphorylation of XRad9, XRad1, and XHus1, which is induced by aphidicolin, is dependent on a PI-3–like kinase activity. These data also show that although it is predominantly the phosphorylated form of the X9-1-1 proteins that become associated with chromatin, such phosphorylation is not a prerequisite for chromatin binding. This indicates that the checkpoint-dependent chromatin binding of these proteins is independent of PI-3–like kinase activity. This is consistent with a recent report where depletion of XAtr from a soluble, nucleus-free replication system did not prevent the aphidicolin-dependent chromatin association of XHus1 (You et al., 2002).
Figure 5.
Checkpoint-dependent phosphorylation of XRad9, XRad1, and XHus1 is sensitive to caffeine. (A) Extract with nuclei (3 × 103/μl) and cycloheximide, 50 μg/ml, was incubated with 100 μg/ml aphidicolin, 5 mM caffeine, or both drugs together for 90 min at 21°C. Equal volumes of either DMSO or H2O were added to ensure that the volume of additions to each sample was the same. The chromatin fraction was isolated as before, and the pellet was resuspended in SDS-PAGE sample buffer. Proteins found in the pellet were visualized by immunoblotting using the antibodies shown. The Western blots were stripped and reprobed with an antibody to Orc1 to act as a loading control. Replication is required for chromatin binding and phosphorylation of XRad9, XRad1, and XHus1. (B) Extract with nuclei (3 × 103/μl) and cycloheximide, 50 μg/ml, was incubated with 100 μg/ml aphidicolin or with 100 μg/ml aphidicolin with 250 nM geminin for 90 min at 21°C. Duplicate samples were then diluted in XB buffer and centrifuged through a 750 mM sucrose cushion to isolate the nuclear fraction. The chromatin fraction was obtained by disruption of this nuclear fraction in XB buffer containing 0.6% Triton X-100, followed by centrifugation through an identical sucrose cushion. Samples were run on 10% SDS-PAGE gels and immunoblotted with the antibodies indicated.
Replication Is Required for XRad17, XRad9, XHus1, and XRad1 Chromatin Binding and the Phosphorylation of XRad9, XRad1, and XHus1
We have previously shown that the checkpoint-induced chromatin association of XRad17 is dependent on the initiation of DNA replication (Stokes et al., 2002). To investigate the relationship between DNA replication and the nuclear distribution of XRad9, XRad1, and XHus1 proteins, we analyzed the nuclear and chromatin fractions of extracts treated with aphidicolin or with aphidicolin plus 250 nM geminin. We find that, as with XRad17 (see Figure 4C), the chromatin association of XRad9, XRad1, and XHus1 induced by aphidicolin is prevented if the initiation of DNA replication is inhibited by geminin (Figure 5B). This is consistent with the findings of You et al. (2002) and Lupardus et al. (2002), who have also shown replication-dependent chromatin binding for XHus1 and XRad1, respectively. In addition, the nuclear population of XRad9, XRad1, and XHus1 did not become phosphorylated and the nuclear protein levels did not accumulate to those observed in extracts treated with aphidicolin alone (Figure 5B). It is unclear whether the increase in nuclear protein is due to an increase in nuclear import or a decrease in nuclear export, though we believe that the loading of X9-1-1 onto chromatin is a contributory factor. We conclude from these observations that the chromatin association and phosphorylation of the X9-1-1 proteins requires DNA replication arrest subsequent to the initiation step blocked by geminin.
XRad17 Is Required for the Activation of XChk1 in Response to Aphidicolin
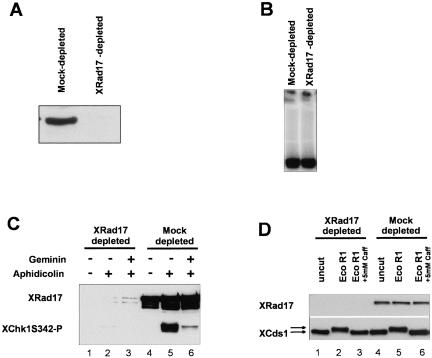
In S. pombe, Rad17 and the 9-1-1 proteins are required for both the DNA replication and the DNA damage checkpoint response. However, in S. cerevisiae, Rad17 and the 9-1-1 homologues are not required for a DNA replication checkpoint response. To analyze the relationship between XRad17, XRad9, XRad1, and XHus1 and the activation of the DNA replication checkpoint in Xenopus extracts, we established conditions where XRad17 could be efficiently depleted (Figure 6A). Because activation of the replication checkpoint in Xenopus is dependent on the initiation of DNA replication, we established that the level of replication seen in XRad17-depleted extract was the same as that observed in extracts after mock depletion (Figure 6B). In the absence of XRad17, extracts challenged with aphidicolin did not activate the replication checkpoint, as judged by significantly reduced levels of XChk1 S342 phosphorylation (Figure 6C, lane 2). On average (3 independent experiments) we estimate that XChk1 S342 phosphorylation was reduced by >80% compared with mock-depleted extracts (Figure 6C, lane 5). This significant reduction in the level of XChk1 phosphorylation in XRad17-depleted extract demonstrates that XRad17 is an important component of the pathway leading to XChk1 phosphorylation when DNA replication is inhibited with aphidicolin.
Figure 6.
Immunodepletion of XRad17 prevents the activation of XChk1 when replication is blocked with aphidicolin. (A) Western blot showing levels of XRad17 protein remaining in mockand XRad17-depleted extract. Extract was incubated with 1/20th volume sepharose beads cross-linked with either XRad17 antibody (1 mg/ml) or nonspecific rabbit IgG (Mock-depleted) for 40 min at 4°C with gentle mixing. Extract was recovered from beads by gentle centrifugation. A second round of depletion was performed. (B) XRad17 is not required for DNA replication in Xenopus extracts. Twenty microliters of mockor XRad17-depleted extract was incubated with sperm pronuclei (3 × 103/μl), cycloheximide (50 μg/ml), and 1 μCi [α-32P]dCTP at 21°C. Samples were run on 0.8% agarose gels, dried down, and exposed to x-ray film. (C) XRad17 is required for XChk1 phosphorylation on S342 when DNA replication is inhibited with aphidicolin. Aliquots, 50 μl, of Xrad17or mock-depleted extract with sperm pronuclei (3 × 103/μl) and cycloheximide (50 μg/ml) were incubated with either aphidicolin (100 μg/ml) or an equal volume of DMSO. After incubating reactions at 21°C for 2 h nuclei were isolated and processed for immunoblotting using XRad17 antibody and phospho-specific (S342) Chk1 antibody. (D) XRad17 is not required of XCds1 phosphorylation in response to double-strand DNA ends. Fifty-microliter volumes of mockand XRad17-depleted extract were incubated with uncut plasmid DNA (12 ng/μl), plasmid DNA (12 ng/μl) cut with EcoRI, or with cut plasmid and 5 mM caffeine for 60 min at 21°C. Samples were taken for Western blotting using antibodies to XRad17 and XCds1.
XRad17 Is Not Required for XCds1 Phosphorylation in Response to DNA Double-strand breaks
In S. pombe Rad17 is essential for activation of both Chk1 and Cds1 in response to DNA damage and blocks to replication, respectively. However, in higher eukaryotes it is Chk1 that is predominantly activated in response to UV and other agents that inhibit replication. The Cds1 homologue (Chk2) responds more strongly to ionizing radiation where the major lesion is thought to be double-strand DNA breaks. In Xenopus, when plasmid DNA cut with restriction enzymes to generate double-strand ends is incubated in egg extract, XCds1 is phosphorylated and becomes active as a kinase (Guo and Dunphy, 2000). We incubated XRad17-depleted and mock-depleted extract with either circular plasmid DNA (12 ng/μl), plasmid DNA cut with restriction endonuclease EcoRI, or cut plasmid and 5 mM caffeine and analyzed the phosphorylation of XCds1 by Western blotting (Figure 6D). XCds1 became phosphorylated (as judged by retardation of the protein on SDS-PAGE gels) in response to cut plasmid in both XRad17and mock-depleted extracts (Figure 6D, lanes 2 and 5). XCds1 did not become phosphorylated in response to an equal concentration of uncut plasmid. The inclusion of 5 mM caffeine prevented XCds1 phosphorylation in response to plasmid DNA cut with EcoRI (Figure 6D, lanes 3 and 6) as expected (Guo and Dunphy., 2000). As a further control, we analyzed the phosphorylation status of XMre11. XMre11 has been shown to undergo caffeine-sensitive phosphorylation when plasmid DNA cut with restriction endonucleases is incubated in Xenopus extracts (Costanzo et al., 2001). Consistent with this, we observed phosphorylation of XMre11 only when cut plasmid, but not uncut plasmid or cut plasmid with 5 mM caffeine, was incubated with Xenopus extract. XMre11 still underwent phosphorylation in XRad17-depleted extract incubated with cut plasmid DNA (our unpublished results).
Together these observations suggest that XRad17 function is not normally required for the phosphorylation of XCds1 or XMre11 in response to double-strand DNA breaks. The phosphorylation of XCds1 also occurs in egg extract treated with geminin (our unpublished results), indicating that phosphorylation of XCds1 in response to DNA ends is not dependent on the initiation of DNA replication. This observation further demonstrates the distinction between the pathways responsive to stalled replication and damaged DNA in Xenopus.
Addition of XRad17 Protein to Depleted Extract Restores Checkpoint Signaling
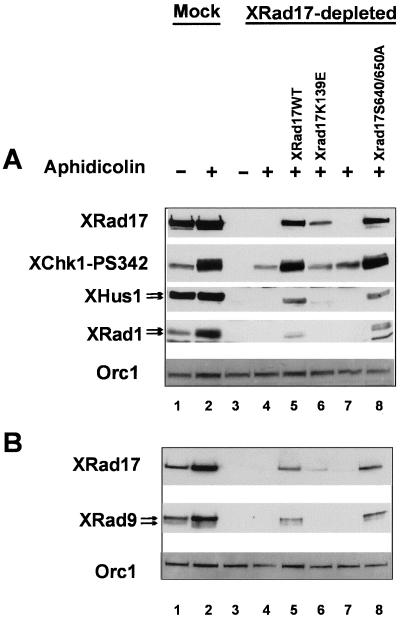
To formally demonstrate that loss of XChk1 phosphorylation in XRad17-depleted aphidicolin arrested extracts is due to the specific depletion of XRad17, we incubated XRad17and mock-depleted extract with aphidicolin and supplemented the reactions with reticulocyte lysate in which various mRNAs had been translated. The efficiency of the depletion can be seen by comparing the level of XRad17 protein found in the nuclei of mock-depleted extract (Figure 7A, lanes 1 and 2) with XRad17-depleted extract (Figure 7A, lanes 3 and 4). In extract where XRad17 had been removed, the addition of aphidicolin did not induce the phosphorylation of XChk1 on Ser342 above the background levels seen in mock-depleted extract (compare Figure 7A, lanes 3 and 4 with lanes 1 and 2). The addition of reticulocyte lysate containing XRad17 protein to XRad17-depleted extract was able to rescue the phosphorylation of XChk1 on Ser342 to a significant level (densitometer analysis estimates 75–80% of mock depleted levels Figure 7A, lane 6). In addition, XRad1 and XHus1 did not accumulate in the nuclear fraction in XRad17-depleted extract in either the presence or absence of aphidicolin, but the aphidicolin-dependent nuclear accumulation and phosphorylation of XRad1 and XHus1 was restored by addition of in vitro–translated XRad17 (Figure 7A, lane 6). When we looked at chromatin binding under these conditions, we found that the exogenous XRad17 protein was able to rescue the chromatin binding and phosphorylation of XRad9 (Figure 7B, lane 6). Similar rescue for XRad1 and XHus1 chromatin binding was also observed (unpublished data).
Figure 7.
Readdition of recombinant XRad17 to depleted extracts can rescue aphidicolin-dependent XChk1 phosphorylation as well as nuclear localization of the X9-1-1 complex. (A) Aliquots, 50 μl (all containing nuclei, 3 × 103/μl, and cycloheximide, 100 μg/ml), of XRad17-depleted (lanes 3–8) or mock-depleted extract (lanes 1 and 2) treated with aphidicolin was supplemented with 1/10th volume of rabbit reticulocyte lysate that had translated mRNA to wild-type XRad17 (lane 5), XRad17K139E (lane 6), XRad17S640A, S650A (lane 8), or mRNA to a control protein (luciferase, lanes 1–4 and 7). Therefore, in all conditions, the concentration of endogenous reticulocyte lysate proteins is the same. After incubating for 2 h at 21°C, samples were processed to obtain the nuclear fraction. Samples were subjected to SDS-PAGE and Western blotting with the antibodies indicated. Readdition of recombinant XRad17 to depleted extracts can rescue the aphidicholin-dependent chromatin binding and phosphorylation of the X9-1-1 complex. (B) Duplicate samples of those described above were processed to obtain a chromatin sample. This chromatin-enriched fraction was solubilized in sample buffer and subjected to SDS-PAGE and Western blotting. The appropriate portion of the membranes for both nuclear and chromatin samples were subsequently stripped with 200 mM glycine, pH 2.3, and reprobed with an antibody to Orc1 to act as a loading control.
By comparison of Western blots of XRad17 expressed in reticulocyte lysate and equal volumes of Xenopus extract, we estimate that the XRad17 protein concentration in reticulocyte lysate is approximately threefold higher than that of the endogenous XRad17 protein found in low-speed Xenopus extracts (see Figure 2). Therefore, when adding one tenth volume of XRad17 reticulocyte lysate to XRad17-depleted extract we restored XRad17 protein to ∼20–30% of its original levels. Because XRad17 is concentrated in the nucleus, we actually restored the XRad17 protein levels to ∼50% of the nuclear concentration of endogenous XRad17 protein found in mock-depleted extract (compare Figure 7A, lanes 1 and 2 with lane 5).
The addition of XRad17 protein mutated in its ATP binding motif (XRad17K139E), whose ability to bind chromatin is severely impaired (Figure 7B lane 6), or reticulocyte lysate that had translated a nonspecific mRNA (luciferase) was unable to rescue any of the checkpoint-dependent events (Figure 7, A and B, lanes 6 and 7), respectively. This demonstrates that rescue is dependent on the readdition of functional XRad17 and not an endogenous reticulocyte protein.
After XRad17 depletion there was no significant decrease in the cytoplasmic protein levels of XRad9, XRad1, and XHus1 (unpublished data). This indicates that the X9-1-1 complex is not codepleted under these conditions. This is consistent with the fact that readdition of XRad17 alone is sufficient to restore checkpoint signaling in XRad17-depleted extracts. Interestingly, a mutant form of XRad17, where the serine residues of two conserved SQ sites (S640 and S650) had been mutated to alanine was also able to rescue checkpoint dependent chromatin binding and phosphorylation of the X9-1-1 complex as well as phosphorylation of XChk1 (Figure 7, A and B, lane 8). We have demonstrated that XRad17 is phosphorylated on Ser650 is response to aphidicolin (Figure 4A). Although we cannot rule out that XRad17 is not phosphorylated on alternative residues, this observation suggests that phosphorylation on these conserved SQ residues is not required for the nuclear localization, chromatin binding, and phosphorylation of XRad9, XRad1, and XHus1, or the activation of XChk1 when the replication checkpoint is induced by aphidicolin.
DISCUSSION
In this study we report the identification of the Xenopus homologues of the Rad17, Rad9, Rad1, and Hus1 checkpoint proteins. These proteins become associated with chromatin and the X-9-1-1 complex subunits are phosphorylated when replication is blocked by aphidicolin. Treatment of extracts with caffeine abrogates the aphidicolin-induced phosphorylation of XRad9, XRad1, and XHus1 but does not impair their chromatin association. This suggests that the 9-1-1 proteins can associate with chromatin independently of a PI-3–like protein kinase activity. Furthermore, we show in depletion/rescue experiments that XRad17 plays an essential role in the nuclear localization, chromatin binding, and phosphorylation of XRad9, XRad1, and XHus1. XRad17 is required for the phosphorylation of the checkpoint kinase XChk1 when replication is blocked by aphidicholin (replication checkpoint) but not for XCds1 activation in response to dsDNA ends.
XRad9, XRad1, and XHus1 Proteins Associate with Chromatin when the Replication Checkpoint Is Activated
In human cells RFCand PCNA-like checkpoint proteins have been found constitutively associated with chromatin. The level of chromatin-associated protein increases after DNA damage or when problems are encountered during replication (Burtelow et al., 2000; Roos-Mattjus et al., 2002; Zou et al., 2002). In Xenopus, we and others have demonstrated that many of the proteins involved in DNA checkpoint responses can only be detected on chromatin when replication is blocked with drugs such as aphidicolin or when replication is initiated on a damaged DNA template (Lupardus et al., 2002; Stokes et al., 2002; You et al., 2002). The difference in the apparent chromatin association of these proteins in Xenopus and mammalian systems may be an artifact of the chromatin isolation procedures used in the two systems or it may reflect differences in the way checkpoint signaling is organized in embryonic and somatic cell cycles. However the mechanisms that lead to checkpoint-dependent chromatin loading are likely to be conserved.
Phosphorylation of XRad9, XRad1, and XHus1 Is Not Required for Chromatin Binding
In these experiments we have demonstrated that the caffeine-sensitive phosphorylation of the X9-1-1 components induced by checkpoint activation is not required for chromatin binding. It is possible that chromatin-bound protein is more likely to become phosphorylated when the checkpoint is activated, because of proximity to activated ATR. This is supported by a recent study where the human equivalent of the XRad17K138E mutant was shown to be unable to bind chromatin, and, as a result showed significantly reduced levels of phosphorylation after treatment of cells with DNA-damaging agents (Zou et al., 2002).
Our observations indicate that XRad17 and all the X9-1-1 proteins can associate with chromatin without XAtr activity when replication is perturbed. This is consistent with observations in yeast (Kondo et al., 2001; Melo et al., 2001), and humans (Zou et al., 2002), where independent loading of ATR and PCNA-like checkpoint proteins at sites of DNA damage has been demonstrated. Thus, our data point to conservation of the mechanism where distinct checkpoint complexes associate independently at sites of stalled replication or at sites of DNA damage. It is clear, however, that both complexes then play an essential role in the activation of the checkpoint (see below).
XRad17 Is Required for the Activation of XChk1
In Xenopus it has been shown that the activation of XChk1 and XCds1 occurs in response to replication arrest or the presence of double-strand DNA ends, respectively (Kumagai et al., 1998; Guo and Dunphy, 2000). The distinction between these two pathways is also observed at the level of the XAtr and XAtm kinases, where XAtr is required for XChk1 activation in a replication-dependent manner but not for the XCds1 activation in response to dsDNA ends (Guo and Dunphy, 2000). Depletion of XRad17 reduced XChk1 phosphorylation levels on average by 80%. The remaining XChk1 phosphorylation may be due to residual XRad17 left in the extract, though XRad17 protein levels were reduced below the limit of detection by Western blotting. Alternatively the removal of XRad17 may reveal an additional minor pathway that can contribute to XChk1 phosphorylation during checkpoint activation. In S. cerevisiae an alternative RFC-like protein, Chl12 (Ctf18), has been identified that also forms a complex with the small subunits of RFC and has been shown to act redundantly with RAD24 (homologue of XRad17) in the replication checkpoint (Naiki et al., 2001). In S. pombe, Rad17 mutants are defective in the replication checkpoint; however, they do show a low level of Cds1 kinase activity compared with Rad3-(ATR homologue) and Rad26-(ATRIP) deleted strains (Lindsay et al., 1998). Therefore, it is possible that the residual phosphorylation of XChk1 observed in XRad17-depleted extract when replication is blocked with aphidicholin may be due to the presence of a Xenopus homologue of Chl12. However, we have shown that XRad17 is an important component of the checkpoint pathway that activates XChk1 when replication is stalled. In contrast to S. pombe, XRad17 does not appear to be required for the phosphorylation of XCds1 in response to dsDNA ends.
Nuclear Localization of XRad9, XRad1, and XHus1 Is Lost when XRad17 Depleted Extract Is Treated with Aphidicolin
One important observation arising from this work is that extracts depleted for XRad17, show severely reduced levels of XRad9, XRad1, and XHus1 protein in nuclei even before checkpoint activation. This is consistent with observations in fission yeast where the nuclear localization of Rad9 and Hus1 was lost in a Rad17 deletion strain (Caspari et al., 2000). A physical interaction between S. pombe Rad17 and Rad1 has been shown by two-hybrid analysis and in vitro using XRad17-coated sepharose beads and overexpressed Rad1 (Caspari et al. 2000). An interaction between Rad17 and Rad1 was not, however, detected in soluble cell extracts without overexpressing one of the proteins, suggesting the interaction may be transient or unstable (Caspari et al., 2000).
In human cells, interaction between the Rad17/RFC and 9-1-1 complexes has been observed after coexpression in a baculovirus system (Rauen et al., 2000; Lindsey-Boltz et al., 2001). Depletion of XRad17 from Xenopus extract under the conditions described did not significantly alter the total protein levels of XRad9, XRad1, or XHus1, suggesting that XRad17 does not interact with X9-1-1 in the cytoplasm. This is supported by the observation that the nuclear accumulation of XRad9, XRad1, and XHus1 can be rescued by the addition of XRad17 protein on its own. It is possible, however, that any interaction between XRad17/RFC and X9-1-1 is disrupted during immunodepletion of the XRad17. Depletion of XHus1 from extracts also abrogates XChk1 phosphorylation in response to aphidicolin, but in this case readdition of XHus1 protein alone is insufficient to rescue the checkpoint (You et al., 2002). Presumably this is due to codepletion of XRad9 and XRad1 and possibly other proteins that are also critical for checkpoint function. The interaction between XRad17 and the X9-1-1 complex may occur in the nucleus, and this interaction may be necessary to maintain the nuclear localization of X9-1-1 and for subsequent loading onto chromatin when the checkpoint is activated. In human cells a reduction in hRad17 by siRNA treatment reduced the loading of hRad9 onto chromatin without apparently reducing the level of hRad9 protein in the nucleus (Zou et al., 2002). However, in these experiments there was still residual hRad17 detectable after siRNA expression. Further experiments will be required to resolve this issue. As yet, we do not know whether the dependency of nuclear localization for the X9-1-1 proteins on XRad17 acts at the level of nuclear import or export. The homology between XRad17 and RFC makes it likely that XRad17 is directly responsible for loading the X9-1-1 (PCNA-like) complex onto chromatin at sites of stalled replication.
A major advantage of the Xenopus cell-free system over other higher eukaryote systems is the ability to rapidly and selectively remove specific proteins or protein complexes. This presents the opportunity to dismantle cellular pathways and then to reconstitute them using purified proteins or mutant constructs. We have used immunodepletion and add-back procedures to dissect the XRad17-dependent steps in the activation of the replication checkpoint. We found that readdition of XRad17 was able to rescue the chromatin loading and phosphorylation of the X9-1-1 proteins as well as the phosphorylation of XChk1. A mutant XRad17 protein in which an essential lysine residue in the Walker A box was mutated to glutamic acid was unable to rescue the checkpoint. The equivalent mutation in S. pombe (K118E), encodes a protein that cannot bind ATP, does not interact with the Rfc2 subunit, and fails to associate with chromatin (Kai et al., 2001). Our observations show that the Xenopus Rad17 protein containing this mutation does not accumulate in the nucleus when the replication checkpoint is activated. Therefore we suggest that the failure to rescue the checkpoint defect of XRad17-depleted extracts is caused predominantly by an inability to bind chromatin.
Phosphorylation of Conserved SQ Motifs in the C Termini of XRad17 Is Not Required for the Checkpoint Response to Aphidicolin
The XRad17 protein contains two conserved SQ motifs (S640, S650). When these sites are mutated to alanine, the mutant XRad17 protein is still able to load X9-1-1 onto chromatin, restore the phosphorylation of X9-1-1, and rescue the activation of XChk1 when added to XRad17-depleted extracts. In human cells these Rad17 SQ sites have been shown to be targets of ATM and ATR kinases in response to a number of DNA-damaging agents (Bao et al., 2001; Post et al., 2001). Overexpression of nonphosphorable Rad17 SQ mutants suggest that phosphorylation of these sites is required for checkpoint responses to ionizing radiation (Bao et al., 2001; Post et al., 2001). However, in both these studies the phosphorylation of Chk1 or the chromatin association of checkpoint proteins was not investigated. It would seem, however, that in spite of XRad17 becoming phosphorylated on Ser650 (see Figure 4A) and presumably Ser640 in response to aphidicolin, phosphorylation of these sites is not required for the Xenopus replication checkpoint responses involving XChk1 and the X9-1-1 complex that we have analyzed.
The functional consequence of X9-1-1 checkpoint-dependent phosphorylation has yet to be determined. It will be interesting to ascertain whether X9-1-1 proteins that lack SQ motifs are proficient in rescuing XChk1 phosphorylation when the endogenous protein has been immunodepleted. Alternatively, these phosphorylation events may enable the X9-1-1 proteins to fulfill some other aspect of the checkpoint response. It has been shown in yeast that even a short period of replication arrest in strains deleted of checkpoint genes leads to rapid loss in viability despite the presence of a mitotic arrest (Enoch et al., 1992). This demonstrates that the checkpoint plays a role in the recovery from stalled replication, possibly by helping reinitiation in addition to halting cell cycle progression. In human cells Rad9 has been shown to interact with c-Abl tyrosine kinase and may be involved in DNA damage–induced apoptosis (Yoshida et al., 2002). In C. elegans, homologues to Rad1 and Hus1 have been implicated in the activation of apoptotic pathways during development and after DNA damage (Ahmed et al., 2001; Hofmann et al., 2002). Therefore, it is tempting to speculate that in higher eukaryotes, the recruitment and phosphorylation of a complex analogous to PCNA may allow the recruitment of additional factors to the sites of replication arrest. These factors may be necessary to stabilize the replication apparatus or may have other specialized functions. The development of an in vitro system will allow these hypotheses to be tested experimentally and provide fresh insight into checkpoint responses and the maintenance of genome integrity in higher eukaryotes.
Acknowledgments
We thank Dr. Matthew Michael for the GST-Geminin construct and for interesting discussion. Antibodies to Orc1 were a generous gift of Dr. Julian Blow. We also thank Dr. Elaine Taylor for critical reading of the manuscript. This work was supported by MRC Grant G9901480 and CRUK Grants SP2396/0201 and SP2396/0101. R.J. is supported by an MRC studentship.
Submitted January February 22, 2003; Revised March April May 10, 2003; Accepted May June 20, 2003
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–03–0138. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-03-0138.
References
- Ahmed, S., Alpi, A., Hengartner, M.O., and Gartner, A. (2001). C. elegans RAD-5/CLK-2 defines a new DNA damage checkpoint protein. Curr. Biol. 11, 1934–1944. [DOI] [PubMed] [Google Scholar]
- Al-Khodairy, F., and Carr, A.M. (1992). DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11, 1343–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, S., Tibbetts, R.S., Brumbaugh, K.M., Fang, Y., Richardson, D.A., Ali, A., Chen, S.M., Abraham, R.T., and Wang, X.F. (2001). ATR/ATM-mediated phosphorylation of human Rad17 is required for genotoxic stress responses. Nature 411, 969–674. [DOI] [PubMed] [Google Scholar]
- Bentley, N.J., Holtzman, D.A., Flaggs, G., Keegan, K.S., DeMaggio, A., Ford, J.C., Hoekstra, M., and Carr, A.M. (1996). The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 15, 6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Blasina, A., Price, B., Turenne, G., and McGowan, C. (1999). Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 9, 1135–1138. [DOI] [PubMed] [Google Scholar]
- Bluyssen, H.A., van Os, R.I., Naus, N.C., Jaspers, I., Hoeijmakers, J., and de Klein, A. (1998). A human and mouse homolog of the Schizosaccharomyces pombe rad1+ cell cycle checkpoint control gene. Genomics 1, 331–337. [DOI] [PubMed] [Google Scholar]
- Burtelow, M.A., Kaufmann, S.H., and Karnitz, L.M. (2000). Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J. Biol. Chem. 34, 26343–26348. [DOI] [PubMed] [Google Scholar]
- Caspari, T., Dahlen, M., Kanter-Smoler, G., Lindsay, H.D., Hofmann, K., Papadimitriou, K., Sunnerhagen, P., and Carr, A.M. (2000). Characterization of Schizosaccharomyces pombe Hus 1, a PCNA-related protein that associates with Rad1 and Rad9. Mol. Cell. Biol. 20, 1254–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich, K.A., Shin, T.B., Keith, C.T., and Schreiber, S.L. (1996). cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. USA, 93, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby, W.A., Roberts, C.J., Cimprich, K.A., Stringer, C.M., Lamb, J.R., Schreiber, S.L., and Friend, S.H. (1998). Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo, V., Robertson, K., Bibikova, M., Kim, E., Grieco, D., Gottesman, M., Carroll, D., and Gautier, J. (2001). Mre11 protein complex prevents double-strand break accumulation during chromosomal DNA replication. Mol. Cell 8, 137–147. [DOI] [PubMed] [Google Scholar]
- Elledge, S.J. (1996). Cell cycle checkpoints: preventing an identity crisis. Science 274, 1664–1672. [DOI] [PubMed] [Google Scholar]
- Enoch, T., Carr, A.M., and Nurse, P. (1992). Fission yeast genes involved in coupling mitosis to completion of DNA replication. Genes Dev. 6, 2035–2046. [DOI] [PubMed] [Google Scholar]
- Freire, R., Murguia, J.R., Tarsounas, M., Lowndes, N.F., Moens, P.B., and Jackson, S.P. (1998). Human and mouse homologs of Schizosaccharomyces pombe rad1(+) and Saccharomyces cerevisiae RAD 17, linkage to checkpoint control and mammalian meiosis. Genes Dev. 12, 2560–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari, B., Blasina, A., Boddy, M.N., McGowan, C.H., and Russell, P. (1999). Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10, 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, J., Solomon, M.J., Booher, R.N.J.F., Bazan, J.F., and Kirschner, M.W. (1991). cdc25 is a specific tyrosine phosphatase that directly activates p34cdc2. Cell 67, 197–211. [DOI] [PubMed] [Google Scholar]
- Green, C.M., Erdjument-Bromage, H., Tempst, P., and Lowndes, N.F. (2000). A novel Rad24 checkpoint protein complex closely related to replication factor C. Curr. Biol. 10, 39–42. [DOI] [PubMed] [Google Scholar]
- Griffith, J., Lindsey-Boltz, L.A., and Sancar, A. (2002). Structures of the human Rad17-replication factor C and checkpoint Rad 9-1-1 complexes visualized by glycerol spray/low voltage microscopy. J. Biol. Chem. 277, 15233–15236. [DOI] [PubMed] [Google Scholar]
- Griffiths, D.J.F., Barbet, N.C., McCready, S., Lehmann, A.R., and Carr, A.M. (1995). Fission yeast rad17: a homologue of budding yeast RAD24 that shares regions of sequence similarity with DNA polymerase accessory proteins. EMBO J. 14, 5812–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., and Dunphy, W.G. (2000). Response of Xenopus Cds1 in cell-free extracts to DNA templates with double-stranded ends. Mol. Biol. Cell 11, 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z., Kumagai, A., Wang, S.X., and Dunphy, W.G. (2000). Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 21, 2745–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Jackson, C., Cross, D., Morrice, N., and Smythe, C. (1999). ATR is a caffeine-sensitive, DNA-activated protein kinase with a substrate specificity distinct from DNA-PK. Oncogene 48, 6707–13. [DOI] [PubMed] [Google Scholar]
- Harlow, E. and Lane, D. (1988). Antibodies. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Hekmat-Nejad, M., You, Z., Yee, M.C., Newport, J.W., and Cimprich, K.A. (2000). Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 10, 1565–73. [DOI] [PubMed] [Google Scholar]
- Hofmann, E.R., Milstein, S., Boulton, S.J., Ye, M., Hofmann, J.J., Stergiou, L., Gartner, A., Vidal, M., and Hengartner, M.O. (2002). Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12, 1908–1918. [DOI] [PubMed] [Google Scholar]
- Hutchison, C.J., Cox, R., and Ford, C.C. (1988). The control of DNA replication in a cell free extract that recapitulates a basic cell cycle in vitro. Development 103, 553–566. [DOI] [PubMed] [Google Scholar]
- Kai, M., Tanaka, H., and Wang, T. (2001). Fission yeast Rad17 associates with chromatin in response to aberrant genomic structures. Mol. Cell. Biol. 21, 3289–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, T., Wakayama, T., Naiki, T., Matsumoto, K., and Sugimoto, K. (2001). Recruitment of mec1 and ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294, 867–870. [DOI] [PubMed] [Google Scholar]
- Kumagai, A., Guo, Z., Emami, K.H., Wang, S.X., and Dunphy, W.G. (1998). The Xenopus Chk1 protein kinase mediates a caffeine-sensitive pathway of checkpoint control in cell-free extracts. J. Cell. Biol. 142, 1559–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Kumagai, A., and Dunphy, W.G. (2003). Claspin, a Chk1-regulatory protein monitors DNA replication on chromatin independently of RPA, and Rad17. Mol. Cell 11, 329–340. [DOI] [PubMed] [Google Scholar]
- Lieberman, H.B., Hopkins, K.M., Nass, M., Demetrick, D., and Davey, S. (1996). A human homologue of the Schizosaccharomyces pombe rad9+ checkpoint control gene. Proc. Natl. Acad. Sci. USA 93, 13890–13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, H.D., Griffiths, D.J.F., Edwards, R.J., Christensen, P.U., Murray, J.M., Osman, F., Walworth, N., and Carr, A.M. (1998). S-phase specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12, 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey-Boltz, L.A., Bermudez, V.P., Hurwitz, J., and Sancar, A. (2001). Purification and characterization of human DNA damage checkpoint Rad complexes. Proc. Natl. Acad. Sci. USA 98, 11236–11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. et al. (2000). Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 14, 1448–1459. [PMC free article] [PubMed] [Google Scholar]
- Lupardus, P.J., Byun, T., Yee, M.C., Hekmat-Nejad, M., and Cimprich, K.A. (2002). A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 16, 2327–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinho, R.G., Lindsay, H.D., Flaggs, G., DeMaggio, A., Hoekstra, M., Carr, A.M., and Bentley, N.J. (1998). Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17, 7239–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, J., and Toczyski, D. (2002). A unified view of the DNA-damage checkpoint. Curr. Biol. 14, 237–245. [DOI] [PubMed] [Google Scholar]
- Melo, J.A., Cohen, J., and Toczyski, D.P. (2001). Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15, 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael, W.M., Ott, R., Fanning, E., and Newport, J. (2000). Activation of the DNA replication checkpoint through RNA synthesis by primase. Science 289, 2133–2137. [DOI] [PubMed] [Google Scholar]
- Murakami, H., and Okayama, H. (1995). A kinase from fission yeast responsible for blocking mitosis in S phase. Nature 374, 817–819. [DOI] [PubMed] [Google Scholar]
- Murray, A. (1991). Cell cycle extracts. Methods Cell Biol. 36, 581–605. [PubMed] [Google Scholar]
- Naiki, T., Kondo, T., Nakada, D., Matsumoto, K., and Sugimoto, K. (2001). Chl12 (Ctf18) forms a novel replication factor C-related complex and functions redundantly with Rad24 in the DNA replication checkpoint pathway. Mol. Cell. Biol. 21, 5838–5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, M.J., Walworth, N.C., and Carr, A.M. (2000). The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10, 296–303. [DOI] [PubMed] [Google Scholar]
- Parker, A.E., van de Weyer, I., Laus, M.C., Oostveen, I., Yon, J., Verhasselt, P., and Luyten, W.H. (1998a). A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J. Biol. Chem. 273, 18332–18339. [DOI] [PubMed] [Google Scholar]
- Parker, A.E., Van de Weyer, I., Laus, M.C., Verhasselt, P., and Luyten, W.H. (1998b). Identification of a human homologue of the Schizosaccharomyces pombe rad17+ checkpoint gene. J. Biol. Chem. 273, 18340–18346. [DOI] [PubMed] [Google Scholar]
- Post, S., Weng, Y., Cimprich, K., Chen, L., Xu, Y., and Lee, E. (2001). Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G(1)/S checkpoint activation in response to DNA damage. Proc. Natl. Acad. Sci. USA 98, 13102–13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen, M., Burtelow, M.A., Dufault, V.M., and Karnitz, L.M. (2000). The human checkpoint protein hRad17 interacts with the PCNA-like proteins hRad1, hHus1, and hRad9. J. Biol. Chem. 275, 29767–29771. [DOI] [PubMed] [Google Scholar]
- Roos-Mattjus, P., Vroman, B., Burtelow, M., Rauen, M., Eapen, A., and Karnitz, L. (2002). Genotoxin-induced Rad9-Hus1-Rad1 (9-1-1) chromatin association is an early checkpoint signaling event. J. Biol. Chem. 46, 43809–43812. [DOI] [PubMed] [Google Scholar]
- Sanchez, Y., Wong, C., Thoma, R.S., Richman, R., Wu, Z., Piwnica-Worms, H., and Elledge, S.J. (1997). Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- Sarkaria, J.N., Busby, E.C., Tibbetts, R.S., Roos, P., Taya, Y., Karnitz, L.M., and Abraham, R.T. (1999). Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 59, 4375–4382. [PubMed] [Google Scholar]
- Shimada, M., Okuzaki, D., Tanaka, S., Tougan, T., Tamai, K.K., Shimoda, C., and Nojima, H. (1999). Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol. Biol. Cell 10, 3991–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura, T., Ando, S., Matsumoto, K., and Sugimoto, K. (1998). Functional and physical interaction between Rad24 and Rfc5 in the yeast checkpoint pathways. Mol. Cell. Biol. 18, 5485–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimuta, K., Nakajo, N., Uto, K., Hayano, Y., Okazaki, K., and Sagata, N. (2002). Chk1 is activated transiently and targets Cdc25A for degradation at the Xenopus midblastula transition. EMBO J. 21, 3694–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes, M.P., Van Hatten, R., Lindsay, H.D., and Michael, W.M. (2002). DNA replication is required for the checkpoint response to damaged DNA in Xenopus egg extracts. J. Cell Biol. 158, 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venclovas, C., and Thelen, M.P. (2000). Structure-based predictions of rad1, rad9, hus1 and rad17 participation in sliding clamp and clamp-loading complexes. Nucleic Acids Res. 28, 2481–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmer, E., and Karnitz, L.M. (1999). Human homologs of Schizosaccharomyces pombe Rad1, Hus1, and Rad9 form a DNA damage-responsive protein complex. J. Biol. Chem. 274, 567–570. [DOI] [PubMed] [Google Scholar]
- Walworth, N., and Bernards, R. (1996). rad-dependent responses of the chk1-encoded protein kinase at the DNA damage checkpoint. Science 271, 353–356. [DOI] [PubMed] [Google Scholar]
- Weinert, T.A., and Hartwell, L.H. (1988). The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science 241, 317–322. [DOI] [PubMed] [Google Scholar]
- Yoshida, K., Komatsu, K., Wang, H.G., and Kufe, D. (2002). c-Abl tyrosine kinase regulates the human Rad9 checkpoint protein in response to DNA damage. Mol. Cell. Biol. 22, 3292–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Z., Kong, L., and Newport, J. (2002). The role of single-stranded DNA and polymerase alpha in establishing the ATR, Hus1 DNA replication checkpoint. J. Biol. Chem. 30, 27088–27093. [DOI] [PubMed] [Google Scholar]
- Zhao, H., and Piwnica-Worms, H. (2002). ATR-Mediated Checkpoint Pathways Regulate Phosphorylation and Activation of Human Chk1. Mol. Biol. Cell 21, 4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, B.B., and Elledge, S.J. (2000). The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- Zou, L., Cortez, D., and Elledge, S.J. (2002). Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 16, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]