Abstract
Zinc is an essential mineral for a wide variety of physiological and biochemical processes. To understand zinc transport in cereals, we identified putative zinc transporters in gene databases. Three full-length cDNAs were identified and characterized from rice (Oryza sativa). Two of the cDNAs partially complemented a yeast (Saccharomyces cerevisiae) mutant deficient in zinc uptake at low concentrations. The two transporters showed many similarities in function but differed in ionic selectivity and pH optimum of activity. Expression patterns also differed between the two genes. One gene was broadly expressed under all conditions, and the other gene was mainly induced by zinc deficiency to higher levels in roots than in leaves. Although the timing of expression differed between the two genes, localization of expression overlapped in roots. Comparisons of the protein sequences, ionic selectivity, and gene expression patterns of the two transporters suggest that they may play different roles in the physiology of the whole plant.
An appreciation for the importance of zinc in molecular biology, structural biology, and nutritional sciences has been rapidly growing over the past ten years (Berg and Shi, 1996). Zinc plays many essential unique biological roles in part because it possesses unique chemical characteristics. It is well known that a vast array of proteins use zinc for stabilizing their structures in a functional form (Christianson, 1991). In many cases, zinc interacts with cysteines and histidines in proteins. The most conspicuous examples of such interactions are the zinc finger transcription factors, which require the binding of zinc for activation of transcription (Alberts et al., 1998). Zinc is well suited for its role in protein structure because it is a borderline acid and therefore can interact strongly with ligands, it is not redox active, and it is relatively labile and therefore undergoes ligand exchange reactions rapidly.
Studies of zinc uptake in biology are critical because zinc is essential for all organisms, and human zinc deficiency ranks third in importance after iron and vitamin A deficiency (Hambidge, 2000). Food zinc content is very important because the supplementation of minerals is often difficult to achieve, particularly in developing countries. Therefore, it has been suggested that increased levels of zinc in staple foods may play a role in reducing zinc deficiency (Ruel and Bouis, 1998; Graham et al., 1999; Welch and Graham, 1999). Because zinc plays multiple roles in plant biochemical and physiological processes, even slight deficiencies will cause a decrease in growth, yield, and zinc content of edible parts. Therefore, it is essential to understand the molecular details of how plants take up, translocate, and store zinc.
Zinc is taken up from soils by root membrane transport mechanisms. The selectivity of these transporters determines whether other divalent cations are imported at the same time as zinc. Recent advances have revealed that plant genomes contain several gene families involved in the transport of divalent micronutrients (Maser et al., 2001). Some of these transporters have broad substrate specificity (Korshunova et al., 1999), but the range of specificities in plant zinc transporters is unknown because their functional characterization is lagging behind the gene discovery. There appears to be at least one family of transporters in plants that should be relatively specific for iron and zinc uptake; however, the precise metal ions that these transporters take up may vary according to the composition of metal ions in the environment (Eide et al., 1996; Grotz et al., 1998; Pence et al., 2000; Eckhardt et al., 2001; Vert et al., 2001; Moreau et al., 2002). Although the molecular identity of specific zinc transporters has become evident in plants, we know little about how the structures of these proteins interact to create differences in functional characteristics, such as ionic selectivity. Ionic selectivity is particularly important for plant zinc transporters in root cells. In soils that contain contaminants such as cadmium, zinc transport mechanisms may allow for cadmium entry into whole plants (Holmgren et al., 1993; Zhao et al., 2002). Cadmium, a toxic heavy metal, can be potentially lethal to plants, and entry into the food chain may result in human toxicity (Lasat et al., 2000; Pence et al., 2000).
The ZIP family of zinc and iron transporters is found in plants, bacteria, fungi, and humans (Gaither and Eide, 2001a). The function of several members of the ZIP family in Escherichia coli (Grass et al., 2002), yeast (Saccharomyces cerevisiae; Zhao and Eide, 1996a, 1996b), plants (Eide et al., 1996; Grotz et al., 1998; Guerinot, 2000; Pence et al., 2000; Eckhardt et al., 2001; Vert et al., 2001; Moreau et al., 2002), and humans (Gaither and Eide, 2000; Gaither and Eide, 2001b) has been studied. In plants, ZIP transporters have been described mainly from dicots, including pea (Pisum sativa), Arabidopsis, and Thalaspi caerulescens. For the most part, the expression of genes encoding the plant ZIP transporters appears to be induced by zinc or iron deprivation (Eide et al., 1996; Grotz et al., 1998). Some of the ZIP family members, such as IRT genes, are constitutively expressed, but expression levels increase with iron deprivation (Eckhardt et al., 2001; Vert et al., 2001).
In this study, we present data showing novel differential selectivities and differential expression of two zinc transporters from rice (Oryza sativa). Our studies focused on rice because it is the model cereal system, and a wealth of full-length cDNAs are available from expressed sequence tag (EST) projects. Our interest in cereals was also based on their importance as world food crops and the differences in micronutrient uptake mechanisms that may exist between certain monocot species and dicot species (Welch, 1995; von Wiren et al., 1996).
RESULTS
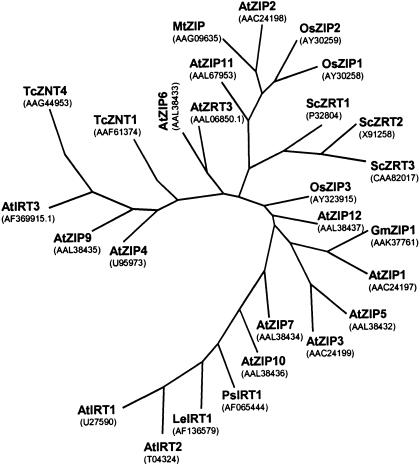
Phylogenetic analysis showed that OsZIP1 and Os-ZIP3 differ significantly in their amino acid sequence and in their relationship to other members of the ZIP family (Fig. 1). Twenty-nine zinc transporter proteins were aligned from Arabidopsis, tomato, T. caerulescens, rice, soybean, and yeast. To determine the relationships in the primary amino acid sequences between the different transporters, Pileup (GCG, Madison, WI) was used to generate an initial alignment. To create 100 different possible alignments, SeqBoot was used. Eprotpars was then used to create 100 phylogenetic trees from the alignments; Econsensus brought these trees into the most statistically likely model for a single unrooted phylogenetic tree. From the consensus tree, it appears that OsZIP1 and OsZIP2 are closely related to AtZIP2 and MtZIP. However, it appears that OsZIP3 is not closely related to any of the other previously characterized zinc transporters with the exception, perhaps, of GmZIP1.
Figure 1.
Unrooted tree that highlights the relationship among the zinc transporter proteins in different plant species. At, Arabidopsis; Gm, soybean (Glycine max); Tc, T. caerulescens; Os, rice; Sc, S. cerevisiae; Mt, Medicago truncatula; Ps, pea; Le, tomato (Lycopersicon esculentum). GenBank identification numbers are provided next to each protein.
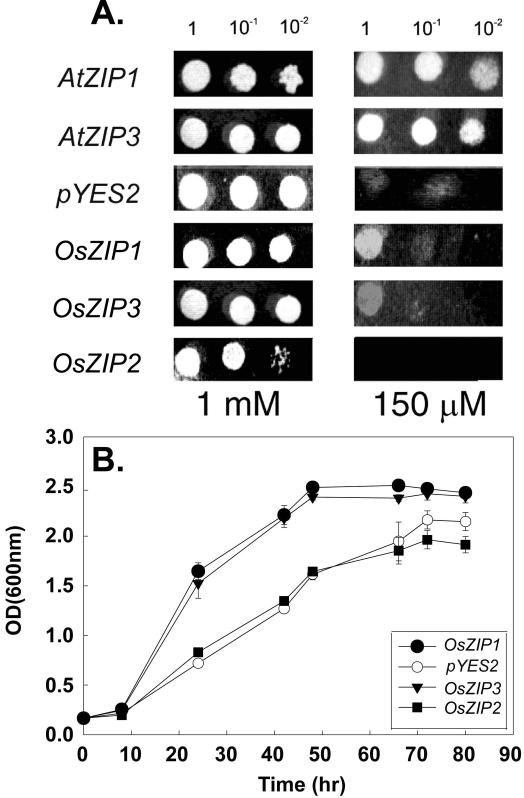
Complementation tests were performed using the ZHY3 mutant (Zhao and Eide, 1996a). As a positive control, cells expressing AtZIP1 and AtZIP3 were dropped onto solid medium containing 1 mm ZnCl2 and 150 μm ZnCl2 (Fig. 2A). These strains grew under both conditions. Cells expressing the rice cDNA clones OsZIP1 and OsZIP3 grew well on 1 mm ZnCl2 and weakly at 150 μm ZnCl2. The cells containing the empty vector pYES2 or expressing the cDNA OsZIP2 grew only on medium containing 1 mm ZnCl2. To confirm the weak complementation of ZHY3 by Os-ZIP1 and OsZIP3, cells were grown in liquid medium containing 1 mm ZnCl2. Quantitative analysis of growth clearly showed that the cells expressing Os-ZIP1 and OsZIP3 grew at a faster rate than cells expressing OsZIP2 or containing the empty vector pYES2 (Fig. 2B). The weak complementation and the faster growth rates suggested the possibility that Os-ZIP1 and OsZIP3 encoded functional zinc transporters. OsZIP1 and OsZIP3 also weakly complemented the yeast mutant defective in manganese uptake (smf1; Supek et al., 1996), but not the yeast mutant fet3fet4 defective in iron uptake (Dix et al., 1994).
Figure 2.
Complementation of the ZHY3 yeast mutant by rice and Arabidopsis cDNAs. a, Serial dilutions of yeast cells were dropped onto a low-zinc medium (LZM) supplemented with 1 mm or 150 μm ZnCl2. Yeast cells expressing the Arabidopsis cDNAs (AtZIP1 and AtZIP3) complement the ZHY3 mutant at 150 μm ZnCl2. ZHY3 cells transformed with empty vector (pYES2) and the rice cDNA OsZIP2 do not grow on medium containing 150 μm ZnCl2. The cells expressing rice cDNAs OsZIP1 and OsZIP3 grow with 150 μm ZnCl2 when cells are plated as undiluted. b, Quantitative analysis of the ZHY3 cells expressing rice cDNAs and containing an empty vector grown in medium containing 1 mm ZnCl2. Data points are the mean and se of three separate experiments, with a total of nine replicates. Error bars smaller than symbols are not visible.
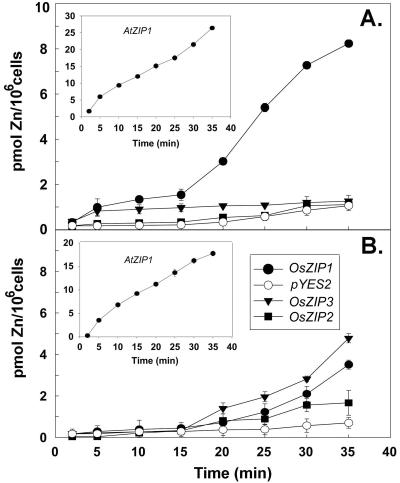
To directly measure the uptake rates of zinc into the ZHY3 cells, we used 65Zn as a tracer for uptake. At pH 4.7, we found that the cells expressing OsZIP1 had significantly higher uptake rates than the other strains tested (Fig. 3A). As a control at that pH, we tested the ZHY3 cells expressing AtZIP1 and found uptake rates similar to those that had been measured previously (Grotz et al., 1998). To determine whether a more neutral pH would alter uptake rates, we tested uptake at pH 6.0. Under these conditions, we observed a reduced uptake rate in the control cells expressing AtZIP1 (Fig. 3B, inset) and the strain expressing OsZIP1. In contrast, we found significantly higher uptake rates in the cells expressing OsZIP3; such rates were not observed at pH 4.7.
Figure 3.
Mean zinc uptake rates (±se) over time of ZHY3 cells expressing rice cDNAs and containing empty vector were measured with 6.8 μm Zn2+ labeled with 65Zn2+ as a tracer. a, Uptake measured at pH 4.7; b, Uptake measured at pH 6.0. The inset shows the Zn2+ uptake rate of ZHY3 cells expressing AtZIP1 for reference to previously published reports (Grotz et al., 1998). Data points represent three separate experiments, with nine replicates.
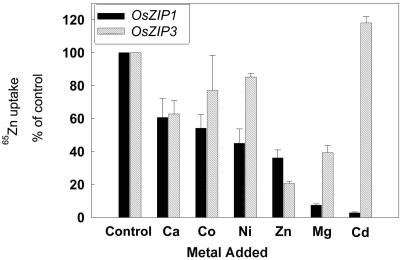
The effect of competing divalent cations on the uptake of zinc in strains expressing OsZIP1 or Os-ZIP3 was determined by measuring zinc uptake in the presence of a 10-fold excess of several divalent cations (Fig. 4). These experiments were completed at pH 4.7 for OsZIP1 and pH 6.0 for OsZIP3. In general, the uptake of zinc in cells expressing OsZIP3 was less inhibited by competing divalent cations than by cells expressing OsZIP1. In the case of a 10-fold increase in zinc, we found that zinc uptake was reduced more in cells expressing OsZIP3. Overall, it appeared that the OsZIP3 transporter was more selective for zinc uptake than for other divalent cations. Particularly striking was the effect of added cadmium. The zinc uptake in cells expressing OsZIP1 was potently inhibited by cadmium, whereas there was virtually no effect in the cells expressing OsZIP3.
Figure 4.
Competition for Zn2+ uptake by 10-fold excess of divalent cations. Zinc concentration was 6.8 μm Zn2+ labeled with 65Zn2+ as a tracer. Mean zinc uptake (±se; n = 9) was measured in ZHY3 cells expressing OsZIP1 and OsZIP3. Divalent cations were added as chloride salts.
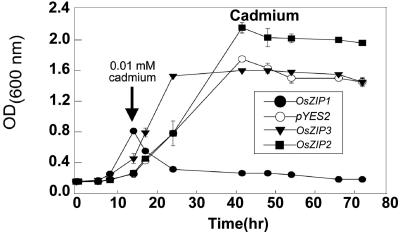
To further demonstrate the effects of cadmium on the different yeast strains, we conducted a quantitative growth study. Because we found that even low concentrations of cadmium inhibited the growth of ZHY3 expressing OsZIP1, we grew the cells to approximately mid log phase and then added cadmium (Fig. 5). Several concentrations of cadmium were tested, but only the lowest concentration of cadmium tested is shown in Figure 5. The concentration of 10 μm Cd2+ only reduced the growth of the strain expressing OsZIP1; higher concentrations of Cd2+ also slightly reduced the growth of the other strains. These quantitative growth experiments strongly suggest that cadmium was more inhibitory to the yeast cells expressing OsZIP1 than the cells expressing OsZIP3, OsZIP2, or the cells containing the empty vector.
Figure 5.
Effects of CdCl2 on the growth of ZHY3 cells expressing rice cDNAs and containing empty vector. Cells were inoculated into YNB and were allowed to grow to mid log phase, at which time 10 μm CdCl2 was added to the medium. Each point represents the mean of nine replicate samples (±se).
To determine whether the effects of cadmium were due to a blockade of zinc uptake or due to cadmium uptake and the ensuing toxicity, we used inductively coupled plasma to analyze cells that had been grown in the presence of cadmium for internal content. Those analyses confirmed that the cells expressing OsZIP1 took up three to four (se = 5%) times more cadmium than the cells containing the empty pYES2 vector or cells expressing OsZIP3.
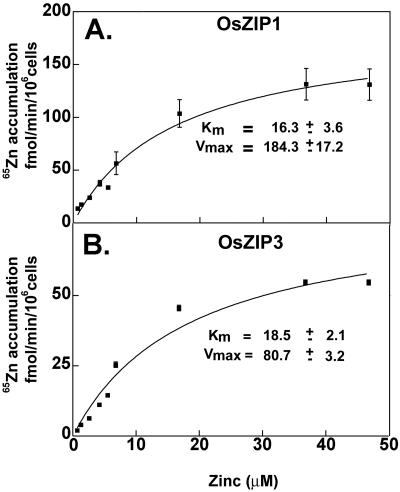
To study the kinetics of zinc uptake, we measured uptake rate as a function of external zinc concentration. The affinity for zinc for both transporters was similar: between 16 and 18 μm (Fig. 6). Interestingly, the Vmax for uptake was twice as high for the cells expressing OsZIP1 than for the cells expressing Os-ZIP3. The higher Vmax for OsZIP1 (Fig. 6A) was consistent with higher uptake rates measured as a function of time (Fig. 3).
Figure 6.
Kinetics of zinc uptake in yeast cells expressing rice cDNAs OsZIP1 (a) and OsZIP3 (b). Calculated affinity for zinc and maximal uptake rates are shown (±se). Each data point represents the mean and se of four separate experiments, with a total of 12 replicate samples. Error bars smaller than symbols are not visible.
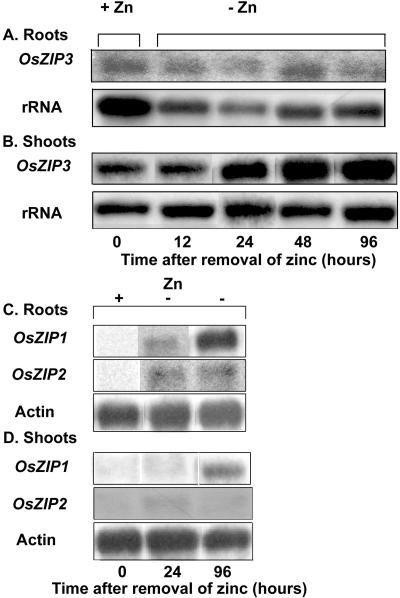
In addition to differences in ionic selectivity, clear differences emerged in the expression of genes encoding these transporters. We found differences in the level of gene expression and in how gene expression was controlled. In the case of OsZIP3, the transcript could be detected in total RNA present in roots (Fig. 7A) and shoots (Fig. 7B) of plants that were grown in zinc-replete conditions. Only a slight induction in gene expression was observed in shoots after 96 h of zinc deprivation. In contrast, the OsZIP1 transcript was of lower abundance and could only be detected in poly(A)+ mRNA. Expression of OsZIP1 was only induced in roots and in shoots 24 to 96 h after zinc deprivation (Fig. 7, C and D). After the plants were deprived of zinc, the OsZIP2 transcript was visible in the poly(A)+ mRNA fraction of roots and, to a lesser extent, in shoots. Although the genes were differentially expressed, the transcripts localize to the same cells in the root (Fig. 8, A, B, E, and F).
Figure 7.
Northern blots showing the expression of rice cDNAs in control rice plants and in rice plants that were deprived of zinc for the indicated amounts of time. The OsZIP3 transcript was detectable in total RNA (a and b), whereas OsZIP1 transcript was only detectable in poly(A)+ RNA (c and d).
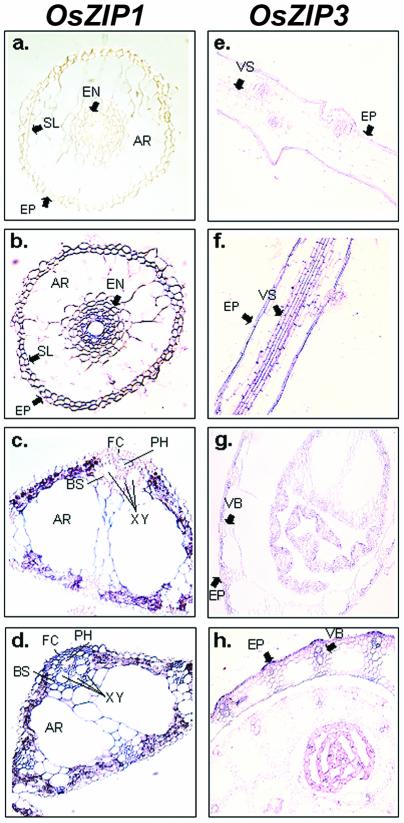
Figure 8.
Detection of OsZIP1 and OsZIP3 mRNA by in situ hybridization in zinc deprived rice. Sections were hybridized with OsZIP1 (a-d) and OsZIP3 (e-h). OsZIP1 sense probe (a) and antisense probe (b) hybridized with root cross section. OsZIP1 sense probe (c) and antisense probe (d) hybridized with leaf cross section. OsZIP3 sense probe (e) and antisense probe (f) hybridized with longitudinal section. OsZIP3 sense probe (g) and antisense probe (h) hybridized with stem cross section. AR, Aerenchyma; BS, bundle sheath; EN, endodermis; EP, epidermis; FC, fiber cells; PH, phloem; SL, sclerenchyma; VB, vascular bundle; VS, vascular system..
Only OsZIP1 could be detected in leaves, and those transcripts were mainly found in the cells that made up the vascular bundles (Fig. 8, C and D). In the stem, OsZIP3 was detected in the vascular bundles and epidermal cells (Fig. 8, G and H).
DISCUSSION
Several genes encoding zinc transporters have now been cloned, and the encoded proteins have been functionally characterized (Gaither and Eide, 2001a). Except for OsIRT1, most of these transporters come from dicot plants (Bughio et al., 2002). The study of micronutrient uptake of iron and zinc in monocots is particularly important because the Graminaceous monocot species have different strategies for iron uptake (Mori, 1999) and perhaps for zinc uptake (von Wiren et al., 1996). In addition, the monocot cereals rice, wheat (Triticum aestivum), and corn (Zea mays) are the major world food crops. It is for these reasons that we embarked on a study to characterize zinc transporters from rice.
Protein Structure
In searching gene databases, we identified at least four ESTs for what appeared to be rice zinc transporters or clones belonging to the ZIP family, which also include iron transporters. Comparison of the protein sequences of 29 ZIP transporters from a range of plant species and yeast suggested that there are distinct structural differences between the two rice transporters that we were able to functionally characterize in a yeast mutant. These two clones were named OsZIP1 and OsZIP3 based on similarity to other ZIP family members. These proteins also contain conserved sequences that identify them as belonging to the ZIP family (Marchler-Bauer et al., 2002; for more details on conserved sequences in the ZIP family, see http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).
We found that these structural differences appear to result in interesting functional differences. However, although there are many obvious differences in amino acid sequence between OsZIP1 and OsZIP3, our understanding of how zinc transporter structure determines function is limited (Moreau et al., 2002). Therefore, it is not possible to draw any conclusions about how structure determines function based on these differences in primary sequence.
General Functional Characteristics
The cDNAs OsZIP1 and OsZIP3 partially complemented the ZHY3 yeast mutant (Zhao and Eide, 1996a) defect in growth on low-zinc medium, which suggests that they may be zinc transporters. Uptake experiments provided direct evidence that these cDNAs encode transporters that are capable of transporting zinc. The uptake rates of OsZIP1 and OsZIP3 were lower than those of AtZIP1. This may account for the partial complementation of the yeast mutant. Zinc uptake activity was clearly evident in the cells expressing OsZIP1 at pH 4.7 and in cells expressing OsZIP3 at pH 6.0. We were not able to detect zinc uptake in the cells expressing OsZIP2. Northern-blot analysis of yeast cells expressing these three cDNAs indicated that transcript from OsZIP2 was barely detectable (data not shown), and, therefore, that the lack of complementation may have been due to lack of gene expression. Zinc uptake activity in Arabidopsis ZIP transporters has also been found to be pH dependent. Most of the AtZIP transporters have higher zinc uptake rates at more acidic pH (4.0). However, the pH optimum of AtZIP2 from Arabidopsis (Grotz et al., 1998) was 6.0, similar to the pH optimum measured for OsZIP3. Although OsZIP1 shows a high degree of homology to the AtZIP2 protein from Arabidopsis (Grotz et al., 1998), it has a different pH optimum for zinc transport activity. The dependence of uptake activity on pH has also been observed with many other ion transporters, such as the mannitol transporter in celery (Apium graveolens; Noiraud et al., 2001) and the human zinc transporter hZIP2 (Gaither and Eide, 2000).
The lag time for zinc uptake observed in the cells expressing the rice transporters contrasted the absence of lag in cells expressing AtZIP1. This lag could not be explained by changes in the pH of the medium by the yeast cells. The lag could have been due to some posttranslational modification in response to zinc, which caused an up-regulation of the transporter.
Ionic Selectivity
The ionic selectivity of ion transporters is a very important characteristic for plants because it impinges on the plant's tolerance and uptake of toxic minerals. Zinc binds to many different types of proteins (Christianson, 1991; Alberts et al., 1998); the different amino acid motifs that may function as zinc-binding sites have been identified mainly in soluble proteins (Alberts et al., 1998). In a majority of cases, His and Cys are key residues in binding zinc. Very little is known about how zinc is bound to membrane transport proteins, transported across the membrane, and released to the cytoplasm during the transport process. Studies on how zinc and cadmium bind to transport mechanisms in plants will be important for increasing crop yields on zinc-deficient soils, boosting the micronutrient density of foods (Kochian and Garvin, 1999; Schachtman and Barker, 1999; Welch and Graham, 1999; Guerinot and Salt, 2001), and creating crop plants that are better able to filter out toxic ions from contaminated soils.
Recent progress has identified several different classes of zinc transporter molecules, but the structures that determine the metal-binding characteristics of these proteins have not yet been elucidated (Gaither and Eide, 2001a). In Arabidopsis, there are 15 members in the ZIP family, several of which have been characterized at the molecular and functional level (Maser et al., 2001). Only small differences between zinc and cadmium transport have been demonstrated thus far among the ZIP transporters. For example, zinc transport through AtZIP2 appears to be almost fully blocked by cadmium, whereas AtZIP1 and AtZIP3 are partially blocked by cadmium (Grotz et al., 1998), as is the case for GmZIP1 (Moreau et al., 2002). These small differences in selectivity contrast the large differences in selectivity that we found between two zinc transporters in rice: OsZIP1 and OsZIP3. OsZIP1-mediated zinc uptake was strongly inhibited by cadmium, magnesium, and zinc, whereas OsZIP3-mediated zinc uptake was strongly inhibited by zinc and magnesium and, to a lesser extent, by calcium. The addition of NaCl reduced zinc uptake rates in both yeast strains. In the case of the cells expressing OsZIP3, as little as 100 μm NaCl reduced uptake rates by 50% (data not shown). In tests done on solid medium, we found that iron, manganese, and copper had no effect on the growth of the yeast strains used (data not shown). Therefore, these ions were not tested in the competition studies. Remarkably, cadmium had no effect on the zinc uptake activity mediated by OsZIP3. These results are supported by our growth studies, which show that cells expressing OsZIP3 take up less cadmium. These cells' growth was also less inhibited by cadmium. A difference in cadmium permeability, shown by growth tests on solid medium, was also found between AtIRT1 and AtIRT2 (Vert et al., 2001). In a recent study, single amino acid substitutions altered the ionic selectivity of the IRT1 transporter (Rogers et al., 2000). The change in one amino acid reduced the cadmium permeability and maintained zinc uptake but abolished iron and manganese uptake. The identification of this single amino acid substitution D136A provides the first localization of a putative cadmium-binding site in the ZIP proteins, of which IRT1 is a member. Although the characterized ZIP transporters differ in selectivity, the rice OsZIP3 is to our knowledge, the first zinc transporter to be found that is highly selective for zinc over toxic ions such as cadmium.
Gene Expression
Gene expression patterns for members of the ZIP family have been widely reported. In a recent paper, it was shown that the transcript levels of IRT1 do not necessarily correlate with the levels of protein produced (Connolly et al., 2002). Many of the ZIP family genes are induced upon starvation of zinc or iron. Similar to our findings for the rice genes OsZIP3 and OsZIP1, some of these transcripts were found in roots and leaves (Pence et al., 2000). However, the expression of many ZIP family members is mainly restricted to roots (Eide et al., 1996; Grotz et al., 1998; Eckhardt et al., 2001; Vert et al., 2001, 2002; Bughio et al., 2002) and nodules (Moreau et al., 2002). It is also interesting to note that the expression of OsZIP1 was not detectable under control conditions and was primarily localized to the roots. In contrast, OsZIP3 was expressed under control conditions, but gene expression was slightly up-regulated when zinc was removed from the medium. OsZIP3 was also expressed in roots and leaves, but expression was much higher in leaves. These results suggest that OsZIP1 is primarily involved in metal uptake when rice is deprived of zinc or other metals, and that OsZIP3 is constitutively active and involved in overall cell zinc homeostasis, particularly in leaves.
Transcripts from both genes were localized to the epidermis and stele of roots of zinc-deprived plants. These results suggest that the transporters play similar roles but are expressed under different conditions. In leaves, OsZIP1 transcripts were localized to the cells containing the vascular tissue. This suggests a role for this transporter in zinc absorption or transfer from the vascular tissue.
Resources generated from EST projects allowed us to identify several putative zinc transporters from an agronomically important crop. Using a yeast mutant, we have provided a thorough functional analysis of two of these transporters. One transporter, OsZIP1, shows a broad substrate specificity for divalent cations and has inducible gene expression. The other transporter, OsZIP3, appears to be more selective for zinc, is not permeable to toxic cadmium, and has more constitutive expression. The link between differences in ionic selectivity and gene expression patterns provide important suggestions for analyzing the physiological function of these two zinc transporters in the future.
MATERIALS AND METHODS
Yeast Strains and cDNA Information
Saccharomyces cerevisiae strain ZHY3 (Zhao and Eide, 1996a) was used to study the function of rice zinc transporters. The yeast strain ZHY3 was grown on LZM (Zhao and Eide, 1996a) or yeast nitrogen base (YNB) medium supplemented with 1 mm ZnCl2 and 2% (w/v) Gal. Three cDNA clones were identified by searching GenBank with amino acid sequences from Arabidopsis ZIP transporters. The cDNA clones were obtained from the Japanese Rice Genome Research Program. Clones were sequenced and appeared to be full length.
Complementation
For the complementation experiments, OsZIP1 (AY302058), OsZIP2 (AY302059), and OsZIP3 (AY323915) cDNAs from rice (Oryza sativa) were directionally cloned into the KpnI/NotI sites of the yeast expression vector pYES2. These constructs were introduced into the yeast strain ZHY3, and the transformants were selected on LZM medium supplemented with 1 mm ZnSO4 and lacking uracil. The cDNAs AtZIP1 and AtZIP3 in pFL61 (Minet et al., 1992) were transformed into the yeast strain ZHY3 and used as positive controls, whereas ZHY3 containing the empty plasmid pYES2 was used as a negative control. All of the yeast strains were grown overnight in LZM with 1 mm ZnSO4. The OD(600) of the cultures was adjusted to 0.1, and 5 μL of 10× serial dilutions were spotted onto LZM plates containing 150 μm or 1 mm ZnSO4 to test for complementation.
Growth Experiment
Yeast strain ZHY3 expressing the cDNAs OsZIP1, OsZIP2, and OsZIP3 and containing the empty plasmid pYES2 was used in the growth experiments. A 5-mL aliquot of YNB supplemented with 1 mm ZnCl2 was inoculated with a single colony of a yeast strain grown on plates containing the same medium and was then grown overnight at 30°C in an orbital shaker. All of the yeast strains were inoculated in 5 mL each of YNB medium supplemented with indicated concentrations of ZnCl2 (1 mm) at an optical density of 0.08 from overnight cultures. To demonstrate the effect of cadmium, cadmium chloride (0.01 mm) was added to the cultures at mid log phase of growth. The optical density of the cultures was sampled periodically, with three replicates for each data point. Experiments were replicated with the same results several times; only the final experiment is shown.
Uptake Experiments
Zinc uptake assays were performed as described by Eide and Guarente (1992) except that 65ZnCl2 (NEN Life Science Products, Boston) and LZMEDTA (LZM without EDTA) were substituted for 59FeCl3 and low-iron medium-EDTA (low-iron medium minus EDTA). The yeast strain ZHY3 with the empty pYES2 vector and strains expressing the cDNAs OsZIP1, OsZIP2, and OsZIP3 were grown to mid log phase in LZM supplemented with 1 mm ZnCl2 for uptake experiments. Exponentially growing cells were harvested, and pellets were washed twice in ice-cold assay buffer LZMEDTA and resuspended in 0.01 of the original volume culture in assay buffer. Cell suspensions were kept on ice before use. Uptake solutions were prepared by diluting ZnCl2 to the specified concentrations in the uptake buffer (LZM-EDTA). To measure uptake, 50 μL of cell suspension was added to 450 μL of uptake buffer containing the isotope. Cell suspensions were transferred to a heating block at 30°C. Tubes containing the cells were vortexed every two to 3 h. The samples were vortexed, and yeast cells were collected on a nylon membrane (0.45 μm) using vacuum. The membranes were then washed with 10 mL of ice-cold 1 mm EDTA, 20 mm Tri-sodium citrate, 1 mm KH2PO4, 1 mm CaCl2, 5 mm MgSO4, and 1 mm NaCl. Cell-associated 65Zn was measured with a scintillation counter (LS 380; Beckman Instruments, Fullerton, CA). All of the experiments were repeated three times with fresh cultures on separate days. In the results, means represent the average of three separate experiments, with nine replicates for each data point.
The effect of pH (4.7 and 6.0) on zinc uptake by the yeast strain ZHY3 expressing the rice cDNAs was studied by adjusting the pH of the uptake assay buffer by the addition of 0.1 m HCl or 0.1 m NaOH to the medium. The concentration of the zinc used in these uptake experiments was 6.8 μm.
For substrate specificity studies, the stock solutions of the competing metal ions were added to a final concentration of 68 μm in the tubes containing the uptake assay buffer with the cell suspensions. The concentration of zinc chloride was 6.8 μm. Zinc uptake rates were calculated in picomoles per 106 cells per minute and were expressed as a percentage of control: control was the zinc uptake rate with no added competing metal ion.
For zinc concentration-dependence studies, zinc uptake rates of the yeast cells expressing rice cDNAs were measured over a range of zinc concentrations (1-46.8 μm). Points were fitted to the Michaelis-Menten equation using Prism software (GraphPad Software, San Diego), and kinetic values Vmax and Km were derived.
The effect of sodium chloride on zinc uptake of ZHY3 cells expressing the OsZIP1 and OsZIP3 cDNAs was studied by preparing the uptake buffer (LZM-EDTA) without sodium and adjusting the pH of the medium to 4.7 for the yeast strain OsZIP1 and to 6.0 for the yeast strain OsZIP3. Cells were harvested and processed according to the protocol outlined, and sodium chloride was added to a final concentration of 0.1, 1, or 10 mm in medium containing 6.8 μm ZnCl2 in the uptake buffer.
Growth of Rice Plants and RNA Extraction
Surface sterilized seeds of rice (cv Jarrah) were germinated on filter paper discs (Whatman, Clifton, NJ) in petri plates in the darkness. Seedlings with emerging plumules and radicles were transferred to a hydroponic growth medium in 50-liter plastic tanks. The nutrient solution contained 1.0 mm CaCl2, 0.51 mm K2SO4, 1.4 mm NH4NO3, 0.32 mm NaH2PO4.2H20, 1.64 MgSO4, 9.5 μm MnCl2.4H20, 19 μm H3BO3, 0.4 μm CuSO4, 0.07 μm NH4Mo7O24, and 35 μm FeEDTA. Zinc was added to a final concentration of 12 μm as required. The growth medium in all tanks was changed every 5 d. Plants were grown with a light intensity of 150 μmol m-2 s-1, in a 16:8-h light:dark photoperiod, and at a temperature of 28°C. After 3 weeks of growth in hydroponic tanks, plants were deprived of zinc for 0, 12, 24, 48, and 96 h and were then harvested. For zinc deprivation, deionized water and nutrient solution that did not contain zinc were used. Shoots and roots were harvested and frozen in liquid nitrogen and were used for RNA extraction.
Northern Blots
Total RNA was extracted from roots and shoots of rice plants using the RNeasy plant mini kit From Qiagen (Valencia, CA) per the manufacturer's protocol. Poly(A)+ RNA was extracted from roots and shoots of rice plants using PolyATract mRNA Isolation System I (Promega, Madison, WI) per the manufacturer's protocol.
Four micrograms of total or poly(A)+ RNA was loaded onto a 1.2% (w/v) denaturing agarose gel. After running the gel, RNA samples were transferred onto a Hybond N+ membrane (Amersham, Buckinghamshire, UK) for at least 16 h. The membrane was then briefly rinsed in 2× SSC, and RNA was fixed by UV cross-linking using a UV Stratalinker (model no. 1800; Stratagene, La Jolla, CA). The purified fragments OsZIP1, OsZIP2, and OsZIP3 were used as templates for synthesizing the probes with the Giga-prime DNA labeling kit (Geneworks, Adelaide, South Australia, Australia) according to the manufacturer's instructions. The membranes containing RNA samples were hybridized in modified Denhardt's buffer (50× Denhardt's reagent, 25% [w/v] Dextran sulfate, 20× SSPE, 10% [w/v] SDS, deionized formamide, and 5 mg mL-1 salmon sperm DNA) at 42°C after the addition of the denatured probes for at least 16 to 18 h. Membranes were washed twice in 2× SSC (150 mm NaCl and 15 mm trisodium citrate, pH 7.0) and 0.1% (w/v) SDS for 10 min at 65°C, and once in 0.1× SSC and 0.1% (w/v) SDS for 15 min at 65°C; they were then exposed to film (Biomax; Eastman-Kodak, Rochester, NY) with intensifying screen at -80°C. Autoradiographs were developed after 5 d (total RNA) or after 30 min [poly(A)+].
In Situ Localization
Rice cv Taipei 309 was grown in hydroponic culture as described above for 26 d with 16 h of light per day (200 μmol m-2 s-1) at 22°C. Plants were transferred into hydroponic medium without zinc for 96 h and were then harvested. Tissue from these plants was fixed in 4% (w/v) paraformaldehyde fixative for 36 h, and was then dehydrated in an ethanol series. After dehydration, tissue was infused with paraplast and then sectioned to 8 μm and mounted on slides. Probes from the 3′ end of each of the cDNAs were labeled using the DIG system and protocol (Roche, Mannheim, Germany). After hydrolysis of the labeled probes and further treatment of tissue, the slides were hybridized overnight at 55°C and washed. The tissue was then incubated with anti-DIG AP conjugate (Roche) for 2 h at room temperature, and the antibody was detected with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate after an overnight incubation. Slides were air dried, and results were visualized using a microscope (Eclipse 800; Nikon, Melville, NY) and were documented with a Retiga Q-imaging CCD camera (Burnaby, Canada).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Mark Thomas (Commonwealth Scientific and Industrial Research Organization Plant Industry Horticulture Research Unit, Glen Osmond, South Australia, Australia) for his support during the project. We also thank Mark Running for help with the in situ hybridizations, and Cathy Kromer and Janet Oriatti for their assistance in manuscript preparation (Danforth Plant Science Center, St. Louis).
This work was supported by the Cooperative Research Centre for Molecular Plant Breeding (Adelaide, Australia) and by the Korea Science and Engineering Foundation (postdoctoral fellowship to R.S.).
References
- Alberts IL, Nadassy K, Wodak SJ (1998) Analysis of zinc binding sites in protein crystal structures. Protein Sci 7: 1700-1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Shi Y (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science 271: 1081-1085 [DOI] [PubMed] [Google Scholar]
- Bughio N, Yamaguchi H, Nishizawa NK, Nakanishi H, Mori S (2002) Cloning an iron-regulated metal transporter from rice. J Exp Bot 53: 1677-1682 [DOI] [PubMed] [Google Scholar]
- Christianson DW (1991) Structural biology of zinc. Adv Protein Chem 42: 281-355 [DOI] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347-1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ (1994) The Fet4 gene encodes the low affinity Fe2+ transport protein of Saccharomyces cerevisiae. J Biol Chem 269: 26092-26099 [PubMed] [Google Scholar]
- Eckhardt U, Mas Marques A, Buckhout TJ (2001) Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol Biol 45: 437-448 [DOI] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot M (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624-5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Guarente L (1992) Increased dosage of a transcriptional activator gene enhances iron-limited growth of Saccharomyces cerevisiae. J Gen Microbiol 138: 347-354 [DOI] [PubMed] [Google Scholar]
- Gaither LA, Eide DJ (2000) Functional expression of the human hZIP2 zinc transporter. J Biol Chem 275: 5560-5564 [DOI] [PubMed] [Google Scholar]
- Gaither LA, Eide DJ (2001a) Eukaryotic zinc transporters and their regulation. Biometals 14: 251-270 [DOI] [PubMed] [Google Scholar]
- Gaither LA, Eide DJ (2001b) The human ZIP1 transporter mediates zinc uptake in human K562 erythroleukemia cells. J Biol Chem 276: 22258-22264 [DOI] [PubMed] [Google Scholar]
- Graham R, Senadhira D, Beebe S, Iglesias C, Monasterio I (1999) Breeding for micronutrient density in edible portions of staple food crops: conventional approaches. Field Crops Res 60: 57-80 [Google Scholar]
- Grass G, Wong MD, Rosen BP, Smith RL, Rensing C (2002) ZupT is a Zn(II) uptake system in Escherichia coli. J Bacteriol 184: 864-866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci USA 95: 7220-7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465: 190-198 [DOI] [PubMed] [Google Scholar]
- Guerinot ML, Salt DE (2001) Fortified foods and phytoremediation: two sides of the same coin. Plant Physiol 125: 164-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambidge M (2000) Human zinc deficiency. J Nutr 130: 1344S-1349S [DOI] [PubMed] [Google Scholar]
- Holmgren GGS, Meyer MW, Chaney RL, Daniels RB (1993) Cadmium, lead, zinc, copper, and nickel in agricultural soils of the United States of America. J Environ Qual 22: 335-348 [Google Scholar]
- Kochian LV, Garvin DF (1999) Agricultural approaches to improving phytonutrient content in plants: an overview. Nutr Rev 57: S13-S18 [DOI] [PubMed] [Google Scholar]
- Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37-44 [DOI] [PubMed] [Google Scholar]
- Lasat MM, Pence NS, Garvin DF, Ebbs SD, Kochian LV (2000) Molecular physiology of zinc transport in the Zn hyperaccumulator Thlaspi caerulescens. J Exp Bot 51: 71-79 [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH (2002) CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res 30: 281-283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646-1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour M, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2: 417-422 [DOI] [PubMed] [Google Scholar]
- Moreau S, Thomson RM, Kaiser BN, Trevaskis B, Guerinot ML, Udvardi MK, Puppo A, Day DA (2002) GmZIP1 encodes a symbiosis-specific zinc transporter in soybean. J Biol Chem 277: 4738-4746 [DOI] [PubMed] [Google Scholar]
- Mori S (1999) Iron acquisition by plants. Curr Opin Plant Biol 2: 250-253 [DOI] [PubMed] [Google Scholar]
- Noiraud N, Maurousset L, Lemoine R (2001) Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13: 695-705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence NS, Larsen PB, Ebbs SD, Letham DLD, Lasat MM, Garvin DF, Eide D, Kochian LV (2000) The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc Natl Acad Sci USA 97: 4956-4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Eide D, Guerinot ML (2000) Altered selectivity in an Arabidopsis metal transporter. Proc Natl Acad Sci USA 97: 12356-12360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel MT, Bouis HE (1998) Plant breeding: a long-term strategy for the control of zinc deficiency in vulnerable populations. Am J Clin Nutr Suppl 68: 488S-494S [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Barker SJ (1999) Molecular approaches for increasing the micronutrient density in edible portions of food crops. Field Crops Res 60: 81-92 [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N (1996) A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc Natl Acad Sci USA 93: 5105-5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Briat JF, Curie C (2001) Arabidopsis IRT2 gene encodes a root-periphery iron transporter. Plant J 26: 181-189 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223-1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wiren N, Marschner H, Romheld V (1996) Roots of iron-efficient maize also absorb phytosiderophore-chelated zinc. Plant Physiol 111: 1119-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RM (1995) Micronutrient nutrition of plants. Crit Rev Plant Sci 14: 49-82 [Google Scholar]
- Welch RM, Graham RD (1999) A new paradigm for world agriculture: meeting human needs: productive, sustainable, nutritious. Field Crops Res 60: 1-10 [Google Scholar]
- Zhao FJ, Hamon RE, Lombi E, McLaughlin MJ, McGrath SP (2002) Characteristics of cadmium uptake in two contrasting ecotypes of the hyper-accumulator Thlaspi caerulescens. J Exp Bot 53: 535-543 [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide D (1996a) The ZRT2 gene encodes the low-affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem 271: 23203-23210 [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide DJ (1996b) The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA 93: 2454-2458 [DOI] [PMC free article] [PubMed] [Google Scholar]