Abstract
FsPP2C1 was previously isolated from beech (Fagus sylvatica) seeds as a functional protein phosphatase type-2C (PP2C) with all the conserved features of these enzymes and high homology to ABI1, ABI2, and PP2CA, PP2Cs identified as negative regulators of ABA signaling. The expression of FsPP2C1 was induced upon abscisic acid (ABA) treatment and was also up-regulated during early weeks of stratification. Furthermore, this gene was specifically expressed in ABA-treated seeds and was hardly detectable in vegetative tissues. In this report, to provide genetic evidence on FsPP2C1 function in seed dormancy and germination, we used an overexpression approach in Arabidopsis because transgenic work is not feasible in beech. Constitutive expression of FsPP2C1 under the cauliflower mosaic virus 35S promoter confers ABA insensitivity in Arabidopsis seeds and, consequently, a reduced degree of seed dormancy. Additionally, transgenic 35S:FsPP2C1 plants are able to germinate under unfavorable conditions, as inhibitory concentrations of mannitol, NaCl, or paclobutrazol. In vegetative tissues, Arabidopsis FsPP2C1 transgenic plants show ABA-resistant early root growth and diminished induction of the ABA-response genes RAB18 and KIN2, but no effect on stomatal closure regulation. Seed and vegetative phenotypes of Arabidopsis 35S:FsPP2C1 plants suggest that FsPP2C1 negatively regulates ABA signaling. The ABA inducibility of FsPP2C1 expression, together with the transcript accumulation mainly in seeds, suggest that it could play an important role modulating ABA signaling in beechnuts through a negative feedback loop. Finally, we suggest that negative regulation of ABA signaling by FsPP2C1 is a factor contributing to promote the transition from seed dormancy to germination during early weeks of stratification.
The phytohormone abscisic acid (ABA) plays important regulatory roles in many plant stress and developmental responses throughout the plant life cycle, particularly in the ability to sense and respond to various unfavorable environmental conditions, including drought, salt, and cold stresses during vegetative growth (Marcotte at al., 1992; Koornneef et al., 1998; Leung and Giraudat, 1998). In seeds, ABA is involved in the acquisition of nutritive reserves, desiccation tolerance, maturation, development, and maintenance of dormancy and germination (Marcotte at al., 1992; Rock and Quatrano, 1995; Koornneef et al., 1998).
Genetic analysis has identified the crucial role of ABA in seed dormancy, as well as the requirement for gibberellins (GAs) in germination (Koornneef and Karssen, 1994), mainly using Arabidopsis because of its excellent suitability for genetic and molecular studies (Koornneef et al., 1984), and because its germination responses are similar to those of many species used in seed physiology research (revised in Koornneef et al., 2002). However, beech (Fagus sylvatica) seeds represent a suitable model to study seed dormancy of woody plants exhibiting a specially deep degree of dormancy maintained by ABA and overcome by stratification or gibberellic acid treatment (Nicolás et al., 1996, 1997, 1998).
Most of the physiological responses regulated by ABA include changes in gene expression, and many genes and proteins have been identified as involved in ABA signaling, although the signal transduction cascades are not yet clearly established (Leung and Giraudat, 1998). However, substantial progress has been made in the characterization of several ABA signaling molecules, including second messengers such as cADPR and Ca2+ (Wu et al., 1997; Pandey et al., 2000). In particular, ABA signaling appears to involve RNA-binding proteins HYL1, ABH1, or SAD1 (Lu and Fedoroff, 2000; Hugouvieux et al., 2001; Xiong et al., 2001), and a complex network of positive and negative regulators, including kinases, phosphatases, and transcriptional regulators (for review, see Finkelstein et al., 2002; Abe et al., 2003).
A role for protein phosphorylation/dephosphorylation in these ABA-mediated processes has been assessed involving several specific protein kinases and phosphatases (Leung and Giraudat, 1998; Finkelstein et al., 2002). For instance, the protein kinase PKABA1, which is induced by ABA and suppresses GA-inducible gene expression in barley (Hordeum vulgare) aleurone layers (Gómez-Cadenas et al., 1999, 2001), the guard cell-specific protein kinase AAPK essential for ABA-induced stomatal closing (Li et al., 2000), or OST1 as a key element mediating stomatal regulation in response to drought (Mustilli et al., 2002). In addition, genetic evidences have shown the involvement of three Arabidopsis Ser/Thr protein phosphatases 2C (ABI1, ABI2, and AtPP2CA) as negative regulators of the ABA signal transduction cascade (Gosti et al., 1999; Merlot et al., 2001; Tahtiharju and Palva, 2001). Whereas ABI1 and ABI2 are key regulators of ABA signaling in seeds and in vegetative tissues (Leung et al., 1994, 1997; Meyer et al., 1994; Rodríguez et al., 1998), AtPP2CA does not appear to regulate ABA signaling in dehydration responses (Tahtiharju and Palva, 2001). A pharmacological approach has been very useful in the identification of other protein phosphatases that may function in ABA signaling. For instance, cyclosporin A, an inhibitor of Ser/Thr protein phosphatases of the type 2B (PP2B), reduces the ABA response in pea (Pisum sativum) epidermal peels (Hey et al., 1997). The inhibitor of Ser/Thr protein phosphatases of the type 1 (PP1) and the type 2A (PP2A), okadaic acid, reduces ABA-induced stomatal closure in Arabidopsis (Pei et al., 1997), but in contrast, it enhances ABA-induced stomatal closure in fava bean (Vicia faba; Schmidt et al., 1995) and activates ABA-responsive promoters in tomato (Lycopersicon esculentum) hypocotyl cells (Wu et al., 1997). The only disruption mutant found is in a regulatory subunit of PP2A (RCN1) and confers ABA insensitivity to Arabidopsis (Kwak et al., 2002). Finally, the stomatal closure induced by ABA in Commelina communis is prevented by phenylarsine oxide, a specific inhibitor of protein Tyr phosphatases (MacRobbie, 2002).
Most knowledge of the genes and pathways involved in ABA signaling has been mainly based on loss-of-function experiments. However, gain-of-function mutants (or transgenic plants) are more significant from a biotechnological viewpoint because the character of interest can be easily transferred to crop plants by transformation (Wilkinson et al., 1997).
We previously reported the cloning of FsPP2C1, a functional PP2C from beechnuts, and showed that FsPP2C1 is up-regulated upon addition of ABA to seeds and also during early weeks of stratification (Lorenzo et al., 2001). Therefore, as tools to transform beech are not available, we took advantage of the constitutive expression of FsPP2C1 in Arabidopsis to investigate the role of this protein in the regulation of ABA responses in dormancy and germination. Here, we show that gain-of-function of FsPP2C1 is sufficient to confer ABA insensitivity in seed dormancy and germination under unfavorable conditions. Furthermore, FsPP2C1 transgenic plants show ABA-resistant early root growth and diminished induction of the ABA-response genes RAB18 and KIN2, but no effect on the stomatal closure regulation was observed. Taken together, these results are consistent with a role of FsPP2C1 as a negative regulator of ABA signaling in beech seeds.
RESULTS
FsPP2C1 Is a Plant PP2C
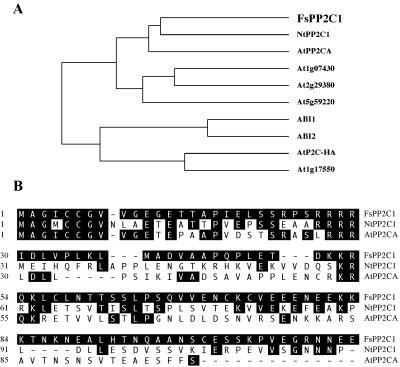
We have previously reported the isolation and characterization of FsPP2C1 as a functional plant PP2C, with all the conserved features of the catalytic domain of these proteins (Lorenzo et al., 2001). The phylogenetic relationship between FsPP2C1 and the closely related plant PP2Cs is shown in Figure 1A. The cluster contains PP2Cs that have been physiologically characterized and proposed to participate in ABA signaling, in particular, Arabidopsis ABI1, ABI2 (Merlot et al., 2001), and PP2CA (Sheen, 1998; Tahtiharju and Palva, 2001). Additionally, two subgroups can be clearly distinguished, one including FsPP2C1 and several PP2CA-like proteins, and the other one including ABI1 and ABI2.
Figure 1.
A, Phylogenetic tree of the plant PP2Cs reported to the updated databases with high similarity to beech FsPP2C1 (AJ277743). Arabidopsis ABI1 (X77116), ABI2 (Y08965), AtP2C-HA (AJ003119), AtPP2CA (D38109), AtPP2Cs (At1g17550, At1g07430, At2g29380, and At5g59220), and tobacco (Nicotiana tabacum) NtPP2C1 (Q9FEW0). Accession numbers are inside brackets. B, Alignment of the FsPP2C1 N-terminal extension with AtPP2CA from Arabidopsis and NtPP2C1 from tobacco. Positions with identical amino acids residues are highlighted in black.
Whereas the catalytic domain among these PP2Cs shows high identity, the N-terminal extension of FsPP2C1 is only similar to that of AtPP2CA (Kuromori and Yamamoto, 1994) and NtPP2C1, showing some stretches of sequence identity (about 35%; Fig. 1B) and suggesting a functional similarity. AtPP2CA is a negative regulator of ABA responses during cold acclimation (Tahtiharju and Palva, 2001). NtPP2C1 is up-regulated in response to drought stress, but its role in ABA signaling has not been investigated (Vranova et al., 2000).
Generation and Characterization of 35S:FsPP2C1 Transgenic Lines
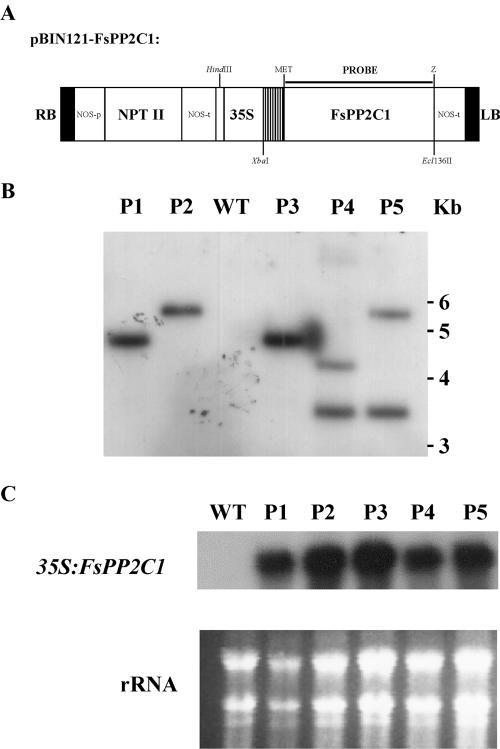
Previously, we showed that FsPP2C1 was specifically expressed in ABA-treated dormant seeds and this expression negatively correlated with germination (Lorenzo et al., 2001). To gain genetic evidence on the role of FsPP2C1 in seed dormancy and germination, we used an overexpression approach in Arabidopsis because transgenic work is not feasible in beechnut. Transgenic plants were created as described in “Materials and Methods” (Fig. 2A). T1 and T2 kanamycin-resistant lines were recovered and five T3 homozygous lines were finally selected. Southern-blot analysis displayed double and single insertions of the 35S:FsPP2C1 transgene (Fig. 2B). Expression levels of the 35S:FsPP2C1 transgene in the five different lines analyzed by northern blot are shown in Figure 2C. High expression was observed in all the transgenic lines, whereas no expression was detected in Arabidopsis wild-type plants, as expected. Out of the five T3 homozygous lines obtained, the three lines with a single insertion were selected for further analysis.
Figure 2.
Generation and molecular analysis of 35S:FsPP2C1 transgenic lines. A, Construct used for plant transformation. RB, Right T-DNA border; LB, left T-DNA border; 35S, cauliflower mosaic virus 35S promoter; NPTII, neomycin phosphotransferase II; NOS-p, NOS promoter; NOS-t, NOS terminator. B, Southern-blot analysis of transgenic lines overexpressing FsPP2C1. Wild type (WT), Col-0 background; P1 to P5, transgenic lines harboring a 35S:FsPP2C1 transgene. Genomic DNA was digested with HindIII, blotted onto a nylon membrane, and hybridized with the FsPP2C1 probe depicted in A. DNA ladder was used as molecular size markers. C, RNA-blot analysis of transgenic lines overexpressing FsPP2C1. Total RNA (10 μg/line) from wild-type (Col-0) plants and P1 to P5 transgenic plants was isolated and hybridized with FsPP2C1. Bottom, ethidium bromide-stained gel showing rRNAs.
Constitutive Expression of FsPP2C1 in Arabidopsis Confers ABA Insensitivity in Seeds
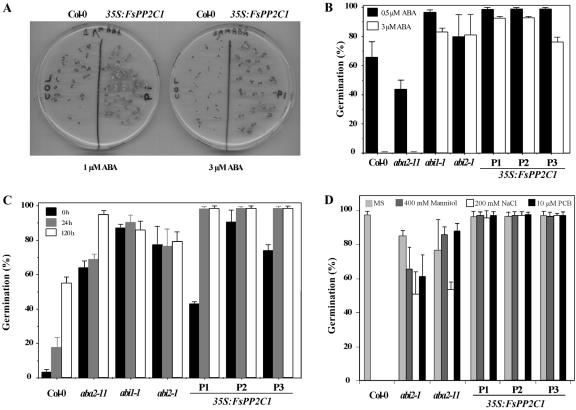
The specific induction of FsPP2C1 expression by ABA in beech seeds (Lorenzo et al., 2001) as well as the sequence homology of FsPP2C1 to AtPP2CA, ABI1, and ABI2, prompted us to test whether constitutive expression of FsPP2C1 in Arabidopsis would affect ABA sensitivity in seeds. Seed germination in media supplemented with ABA of FsPP2C1-overexpressing transgenic plants is shown in Figure 3A. After 10 d poststratification, radicle emergence and development of green and expanded cotyledons was partially inhibited in wild-type seeds at 1 μm ABA and completely under 3 μm ABA. In contrast, 35S:FsPP2C1 seeds were able to germinate and grow under these conditions, and, therefore, they showed reduced sensitivity to ABA. To further substantiate this result, we compared the germination of ABA-deficient (aba2-11), ABA-insensitive mutants (abi1 and abi2), and wild-type seeds with that of 35S: FsPP2C1 transgenics in media supplemented with ABA (Fig. 3B). The addition of 0.5 μm ABA decreased the percentages of germination in wild type and aba2-11 to 65% and 45%, respectively, whereas no germination at all was observed under 3 μm ABA. Clearly, under a concentration fully inhibitory for the wild-type (3 μm ABA), FsPP2C1-overexpressing lines reached over 80% germination, which is very similar to the one observed for the ABA-insensitive abi1-1 and abi2-1 mutants.
Figure 3.
A, ABA germination assay of 35S:FsPP2C1 seeds. Picture showing the differences in germination of FsPP2C1-overexpressing seeds and wild-type (Col-0) plants after 10 d in 1 and 3 μm ABA. B, Percentage of seeds that germinated and developed green cotyledons in the presence of 0.5 μm ABA (black bar) and 3 μm ABA (white bar). Seeds were scored 7 d after sowing. C, Dormancy assay of FsPP2C1-overexpressing seeds. Germination percentage was determined at 5 d after 0 h (black bar), 24 h (gray bar), and 120 h (white bar) of stratification at 4°C. Three independent FsPP2C1 transgenic lines in wild-type (Col-0) backgrounds were used, as well as the indicated ABA mutants and wild-type plants. D, Stress germination assays. Germination rating is represented as the percentage of seeds that germinated and developed green cotyledons in the presence of Murashige and Skoog medium, 400 mm mannitol, 200 mm NaCl, or 10 μm PCB. Three independent FsPP2C1 transgenic lines in wild-type (Col-0) backgrounds were used (P1, P2, and P3), as well as the indicated mutants and wild-type plants. Seeds were scored 7 d after sowing. Error bars represent ± sd of three independent experiments with about 100 seeds plated per data point and carried out with similar results.
Mature Arabidopsis seeds exhibit primary dormancy when freshly released from the mother plant, which means that seeds are unable to germinate under the appropriate environmental conditions without the help of dormancy-breaking agents such as stratification or GAs (Koornneef and Karssen, 1994). To determine the degree of dormancy of 35S:FsPP2C1 seeds, we compared the germination percentage of the seeds harvested at the same time after different cold treatment periods (0, 24, and 120 h) with that of wild-type plants and ABA-related mutants that produce nondormant seeds (aba2-11, abi1-1, and abi2-1). As a result, all of the FsPP2C1 transgenic lines exhibited a reduced dormancy compared with the wild type, very similar to that of aba and abi mutants (Fig. 3C). In the absence of stratification at 4°C, FsPP2C1 transgenic seeds were able to germinate, reaching 100% germination after 1 d of treatment.
An additional seed germination assay was carried out in the presence of paclobutrazol (PCB), a well-known inhibitor of GA biosynthesis. GAs are antagonistic to ABA, and, therefore, seeds with reduced sensitivity to ABA (or diminished ABA levels), show PCB-resistant germination (Koorneef et al., 1998). In contrast to wild-type seeds, 35S:FsPP2C1 and aba2-11 and abi2-1 seeds were able to germinate and develop green cotyledons in medium supplemented with 10 μm PCB, indicating a reduced requirement for GAs at this developmental stage (Fig. 3D).
These results demonstrate that 35S:FsPP2C1 seeds exhibit a reduced degree of seed dormancy and insensitivity to inhibition of germination by exogenous ABA and PCB, suggesting that FsPP2C1 might be involved in ABA responsiveness in seeds.
35S:FsPP2C1 Seeds Exhibit Resistance to Salt and Osmotic Stresses
It has been previously suggested that ABA regulates many different stress responses. To test whether the ABA resistance induced by overexpression of FsPP2C1 is also effective against other ABA-mediated stresses that increases ABA levels, we analyzed the seed germination response of FsPP2C1 transgenic lines in the presence of high concentrations of NaCl and mannitol (Fig. 3D) and compared it with that of wild-type plants.
Seed germination under 400 mm mannitol and 200 mm NaCl leads to a severe delay in radicle emergence and further growth arrest in wild-type individuals; in contrast, 35S:FsPP2C1 seeds were able to germinate and develop green cotyledons under such conditions, even to a higher extent than abi2 and aba2 mutants do (Fig. 3D). These results indicate that FsPP2C1-expressing seeds are osmotolerant and resistant to inhibitory salt concentration in this germination assay.
Effect of FsPP2C1 Overexpression in Vegetative Tissues
To determine whether FsPP2C1 overexpression affected whole plant phenotypes, we analyzed ABA sensitivity in vegetative tissues of wild-type plants, ABA-related mutants (aba2-11, abi1-1, and abi2-1), and three 35S:FsPP2C1 transgenic lines.
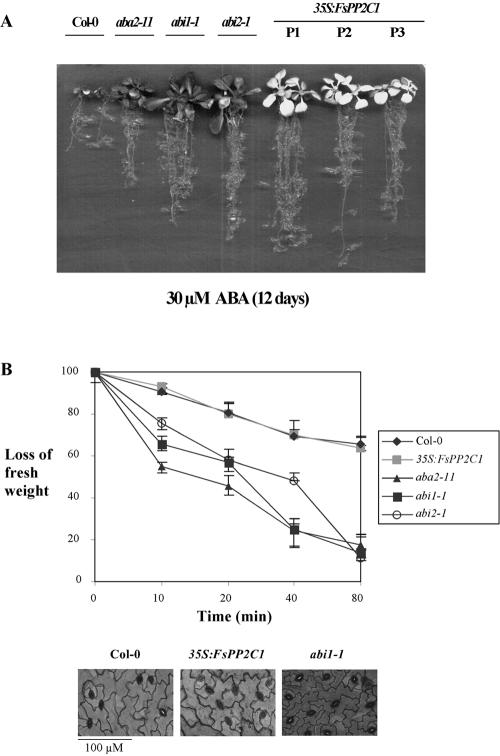
ABA has an inhibitory effect on root growth and consequently, ABA-insensitive mutants are resistant to this ABA-mediated process (Himmelbach et al., 1998). Twelve-day-old 35S:FsPP2C1 seedlings grown in the presence of 30 μm ABA showed a reduced inhibition of root growth compared with wild-type plants, and similar to that of abi mutants (Fig. 4A).
Figure 4.
A, Root growth assay for scoring ABA sensitivity. Growth of Col-0, aba2-11, abi1-1, abi2-1, and 35S:FsPP2C1 plants in medium supplemented with 30 μm ABA. The picture was taken after 12 d of the transfer of 5-d-old seedlings from Murashige and Skoog medium to plates containing 30 μm ABA. B, Micrographs showing the stomatal closure regulation of 35S:FsPP2C1 plants compared with wild-type plants and abi1-1. All pictures were taken at the same scale (Bar = 100 μm). Loss of fresh weight was measured in detached rosette leaves of Col-0, 35S:FsPP2C1, or ABA mutant (aba2-11, abi1-1, and abi2-1) plants. Data values represent one of three independent experiments with similar results.
However, prolonged culture of 35S:FsPP2C1 plants under 30 μm ABA led to growth arrest of the aerial part of the plant and yellowing of the leaves, whereas abi1-1 and abi2-1 mutants remained green and grew under these conditions (Fig. 4A). When grown in the absence of ABA, the FsPP2C1-overexpressing plants did not display any visible phenotypic alteration (data not shown).
ABA triggers stomatal closure and consequent reduction in water loss under drought conditions. The Arabidopsis ABA-insensitive mutants abi1-1 and abi2-1 are impaired in the ABA-induced stomatal closure and, therefore, in their ability to limit transpiration upon drought. On the contrary, FsPP2C1 overexpression did not affect stomatal regulation because detached leaves of 35S:FsPP2C1 plants showed similar rates of transpiration than wild type under ambient conditions (35% relative humidity) and clearly different from aba and abi mutants, which lost approximately 40% to 50% fresh weight after 40 min (Fig. 4B). Furthermore, preopened stomata of 35S:FsPP2C1 plants closed similarly to wild type (one representative line is shown in Fig. 4B, bottom) as compared with abi1 mutant where stomata failed to close, consistent with the inability of abi1 plants to reduce water loss upon drought.
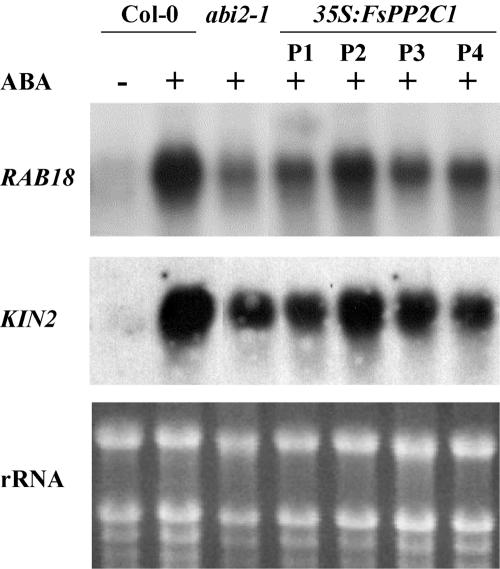
To examine whether the reduction in ABA sensitivity in transgenic plants was accompanied by altered expression of ABA-responsive genes, we compared the expression of RAB18 and KIN2 in 35S:FsPP2C1 with that of Columbia (Col-0) and abi2-1 (Fig. 5). RAB18 (Lang and Palva, 1992; Parcy et al., 1994) is an ABA-inducible gene whose expression is drastically inhibited in abi1-1 and abi2-1 mutants (Leung et al., 1997), and KIN2 is a cold-regulated gene previously described as ABA inducible (Wang et al., 1995) but down-regulated in the abh1 mutant (Hugouvieux et al., 2001). In that respect, 35S:FsPP2C1 plants behave as abi2-1 mutant as they showed a severe reduction (3- to 5-fold) in the expression of RAB18 and KIN2 upon ABA treatment.
Figure 5.
Expression of the ABA-regulated RAB18 and KIN2 genes in 35S:FsPP2C1 plants (P1 to P4) compared with wild-type (Col-0 transgenic plants with the kanamycin resistance gene) and abi2-1 controls. mRNA levels of the indicated genes were determined by northern-blot analysis using total RNAs (10 μg/line) isolated from mock-treated (-) or ABA-treated (+, 50 μm ABA for 3 h) plants. Bottom, ethidium bromide-stained gel showing rRNAs.
Taken together, these results indicate that expression of FsPP2C1 in vegetative tissues partially interferes with ABA signaling, affecting ABA-mediated inhibition of root growth and ABA-responsive gene expression but having no effect on stomatal regulation.
DISCUSSION
In seeds, ABA has been shown to play an important role in the formation, maintenance of dormancy, and inhibition of germination (Koornneef and Karssen, 1994; Bewley, 1997), and later on in the early seedling growth arrest under unfavorable environmental conditions (López-Molina et al., 2001; González-Guzmán et al., 2002).
Our work is focused on beech seed physiology (Nicolás et al., 1996) and on the mechanisms involved in ABA action during seed dormancy and germination (Nicolás et al., 1997, 1998; Lorenzo et al., 2001, 2002). In our previous reports (Lorenzo et al., 2001, 2002), we described two PP2Cs, FsPP2C1 and FsPP2C2, which represent the first PP2Cs described in woody plants (Kerk et al., 2002). FsPP2C1 catalytic domain showed high identity to the ABA-related PP2Cs ABI1, ABI2, and AtPP2CA, widely involved in ABA signaling in Arabidopsis, but divergences in the N-terminal extension were found. The amino acid sequence identity to AtPP2CA (Kuromori and Yamamoto, 1994) and NtPP2C1 (Vranova et al., 2000) in the noncatalytic N-terminal region is nearly 35%. The N-terminal extension has been suggested to facilitate the interaction with different substrates (Rodríguez, 1998), and N-terminal deletions of ABI1 and AtPP2CA led to enhanced PP2C activity, indicating a regulatory function for this domain (Sheen, 1998). Interestingly, evidence that AtPP2CA C-terminal (catalytic) domain is directly involved in the interaction with ATK2 (Chérel et al., 2002) and ATK3 (Vranova et al., 2001) has been provided.
In addition to their sequence similarity, FsPP2C1 is also similar to ABI1, ABI2, and AtPP2CA in their inducibility by ABA. The specificity of their function has been suggested to depend on their differential expression patterns. Thus, AtPP2CA is highly expressed in leaves (Tahtiharju and Palva, 2001), whereas ABI2 is mainly expressed in stems and roots, and ABI1 strongly expressed in leaves, roots, and stems (Leung et al., 1997). In the case of FsPP2C1, transcript expression is also tissue specific and was found to accumulate in ABA-treated seeds rather than in other vegetative tissues (Lorenzo et al., 2001), suggesting that their function might be restricted to this tissue. Beside FsPP2C1, no other PP2C with a seed-specific expression pattern has been reported. In fact, most of the phenotypes observed in the transgenic plants are related to the seed.
The features of FsPP2C1, that is, sequence similarity to ABA-related PP2Cs and ABA up-regulation in seeds, made it a logical candidate as a regulator of ABA signaling in seeds and, consequently, as a regulator of seed dormancy. Genetic evidence was necessary to assess whether FsPP2C1 functions as a positive or a negative regulator of seed dormancy and germination. In the present study, we used an overexpression approach in Arabidopsis to investigate FsPP2C1 function, due to the lack of genetic tools in beechnuts (Fig. 2). Studies of orthologous genes and functional tests in heterologous systems have shown that the ABA signal transduction pathway is mostly conserved among evolutionary distant plant species (for review, see Finkelstein et al., 2002). In case FsPP2C1 was a positive regulator of ABA signaling, constitutive expression in Arabidopsis could lead to enhanced seed responses to ABA and, consequently, to enhanced dormancy. Conversely, in case FsPP2C1 was a negative regulator of ABA signaling, constitutive expression would lead to diminished ABA-response in seeds and, consequently, to a reduced dormancy.
Seeds of 35S:FsPP2C1 transgenic plants displayed reduced dormancy, similar to the abi mutants (Fig. 3C), which is indicative of diminished responsiveness to endogenous ABA in seeds (Gosti et al., 1999). Whereas the germination of wild-type Arabidopsis seeds was suppressed by 3 μm ABA, FsPP2C1-overexpressing transgenic seeds emerged radicle and developed green and expanded cotyledons under these conditions (Fig. 3, A and B), as ABA-insensitive mutants do (Koornneef et al., 1984; Finkelstein, 1994). Taken together, these data are consistent with a role of FsPP2C1 as a negative regulator of ABA signaling in seeds. In addition to up-regulation by ABA, FsPP2C1 expression also increased after 2 weeks imbibition at 4°C, suggesting a role for this gene during the first weeks of stratification (Lorenzo et al., 2001). Taking into account that FsPP2C1 is a negative regulator of ABA signaling, blockade of the ABA response by FsPP2C1 might be a requisite to break dormancy during stratification. The reliability of the physiological role of FsPP2C1 is also reinforced by the germination assay in the presence of the GA biosynthesis inhibitor PCB (Fig. 3D). GAs and ABA play antagonistic roles in seed germination, and apparently, ABA-insensitive (or ABA-defective) individuals need less GAs during germination (Léon-Kloosterziel et al., 1996). Clearly, the GA requirement was reduced in 35S:FsPP2C1 seeds compared with wild-type seeds (Fig. 3D), suggesting that expression of FsPP2C1, through blockade of ABA signaling, positively regulates seed germination.
Another important role of ABA is the prevention of seed germination under unfavorable water conditions (González-Guzmán et al., 2002). Compelling evidence has shown that osmotic stress delays seed germination and arrests early seedling development. Several studies show that ABA plays an inhibitory role in these processes because ABA-insensitive and ABA-deficient mutants are able to bypass them (Werner and Finkelstein, 1995; Léon-Kloosterziel et al., 1996; López-Molina et al., 2001; González-Guzmán et al., 2002). As previously observed with aba2, the three FsPP2C1-expressing lines tested were able to germinate and carry out early growth in medium with the organic solute mannitol as well as inhibitory salt concentration (200 mm NaCl) compared with the incapability of wild-type plants (Fig. 3D). These results indicate that FsPP2C1-induced resistance is not restricted to a nonionic osmotic imposed stress, and further suggests a general insensitivity to osmotic stress. Although FsPP2C1 transcripts were not affected by water deficit in beechnuts (Lorenzo et al., 2001), FsPP2C1 overexpression in Arabidopsis overcome the inhibition of seed germination under low water potential conditions.
Interestingly, constitutive and ectopic expression of FsPP2C1 partially influences ABA responses in vegetative tissues of Arabidopsis. Thus, inhibition of early root growth by 30 μm ABA was notably reduced in 35S:FsPP2C1 seedlings compared with wild-type plants. However, FsPP2C1-overexpressing seedlings are more sensitive to ABA at further stages of development. After 12 d in 30 μm ABA, 35S:FsPP2C1 leaves showed wilting, whereas abi1-1 and abi2-1 mutant leaves remained green and turgid. (Fig. 4A). Another well-characterized ABA-mediated response is stomatal closure regulation. ABA promotes stomatal closure reducing water loss by transpiration during drought. Transpiration rate measured by the loss of fresh weight of detached rosette leaves in FsPP2C1-overexpressing plants, and the stomatal closure showed an ABA-response similar to wild-type plants, clearly different from abi mutants impaired in the ABA-induced stomatal closure in Arabidopsis and, therefore, in their ability to limit transpiration upon drought (Pei et al., 2000; Fig. 4B). In the same way, ABA-mediated drought responses were not affected by inhibition of AtPP2CA expression (Tahtiharju and Palva, 2001).
These results indicate that ABA responsiveness by FsPP2C1 overexpression is mainly restricted to seeds and they also suggest lack of the corresponding signaling component (substrate, activator, or partner) or instability of FsPP2C1 as a foreign protein in these tissues. In beech vegetative tissues, other PP2Cs may act in response to ABA, as the FsPP2C2 previously described by Lorenzo et al. (2002) with a broad range of expression.
The attenuation of the ABA signal in 35S:FsPP2C1 plants is further sustained by the diminished induction of the ABA-responsive genes RAB18 and KIN2. RAB18 is a dehydrin only found in ABA-treated plants and accumulates in Arabidopsis dry seeds (Nylander et al., 2001). abi1-1 and abi2-1 mutants show impaired induction of this gene (Leung et al., 1997) that is induced in antisense AtPP2CA plants (Tahtiharju and Palva, 2001). KIN2 is a cold-regulated protein whose transcriptional regulation and gene expression are induced by low temperature, ABA, osmoticum, and dehydration (Wang et al., 1995; Abe et al., 2003). The induction of KIN2 was severely reduced in abh1 mutant (Hugouvieux et al., 2001).
ABA responses depend on coordinated interactions between positive and negative regulators required for the proper control of this complex signaling pathway that operate in a cell. Some genes have been identified as negative regulators of the pathway, including the homeodomain protein, ATHB6 (Himmelbach et al., 2002), and the farnesyl transferase β-subunit, ERA1 (Cutler et al., 1996; Pei et al., 1998). Protein farnesylation of certain signaling proteins appears crucial for negative regulation of ABA signaling, as recently suggested in the case of ABI3 (Brady et al., 2003). In addition, genetics of abi1 and abi2 mutants, transient expression studies, as well as analysis of transgenic PP2CA antisense plants characterize PP2Cs as negative regulators of ABA signaling (Sheen, 1998; Gosti et al., 1999; Merlot et al., 2001; Tahtiharju and Palva, 2001). Therefore, a complex regulatory mechanism seems to have evolved to attenuate ABA signaling and avoid undesirable effects (i.e. inhibition of cell cycle) due to sustained activation of the ABA pathway.
Recent controversy on the role of ABI1 has arisen from the work of Wu et al. (2003). On the basis that ABI1 overexpression in Arabidopsis does not affect the ABA-signaling pathway and that microinjection of ABI1 protein had no apparent effect on ABA-induced RD29A-β-glucuronidase and KIN2-β-glucuronidase expression, the authors conclude that their results are not compatible with a negative regulatory role of ABI1 in ABA signaling. These results rise the question whether ABI1 catalyzes a rate-limiting step in ABA signaling more than questioning its role as a negative regulator of the pathway. However, the results of Wu et al. (2003) contrast with previous results reported by Sheen (1998), which showed that overexpression of ABI1 in maize (Zea mays) protoplasts led to a blockade of ABA-inducible gene expression. Nevertheless, additional overexpression experiments as well as loss-of-function studies with several PP2Cs are required to resolve the above controversy.
Results reported in this work with FsPP2C1 are in agreement with those reported by Sheen (1998) for ABI1 and PP2CA, and they show for the first time that overexpression of a PP2C in transgenic plants leads to a diminished response to ABA, therefore, we conclude that FsPP2C1 functions as a negative regulator of ABA signaling in seeds and early seedling growth. FsPP2C1 overexpression leads to a reduced seed dormancy, as compared with untransformed plants, and a promotion of germination under inhibitory concentrations of ABA or high-osmoticum media. This ABA-insensitive phenotype, together with the ABA-induced expression of FsPP2C1 transcripts specifically in seeds, suggests that this PP2C, through modulation of ABA signaling, is a factor contributing to promote the transition from seed dormancy to germination during early weeks of stratification.
MATERIALS AND METHODS
Plant Material
The Arabidopsis wild-type and transgenic plants used throughout this work were the Col-0 ecotype. They were routinely grown in a growth chamber under 40% humidity, a temperature of 22°C, and with a 16-h light/8-h dark photoperiod at 80 to 100 μE m-2 s-1 in pots containing a 1:3 vermiculite:soil mixture. For in vitro culture, seeds were surface sterilized in 70% (v/v) ethanol and 1% (v/v) Triton X-100 for 20 min, soaked for 10 min in 2.5% (v/v) bleach and 0.05% (v/v) Triton X-100, and finally, washed four times in sterile distilled water. Stratification of the seeds was conducted during 3 d at 4°C, otherwise as indicated. Afterward, seeds were sowed on Murashige and Skoog (1962) plates containing solid medium composed of Murashige and Skoog basal salts and 1% (w/v) Suc, solidified with 1% (w/v) agar and the pH was adjusted to 5.7 with KOH before autoclaving. Plates were sealed and incubated in a controlled environment growth chamber.
Vector Construction and Plant Transformation
To generate the construction, the coding region of the FsPP2C1 cDNA was excised from the pSKFsPP2C1 (Lorenzo et al., 2001) using an XhoI-XbaI double digestion and was subcloned into SmaI-Ecl136II doubly digested pBIN121 vector. The T-DNA region of the pBIN121-FsPP2C1 construct was transferred to Agrobacterium tumefaciens C58C1 (pGV2260; Deblaere et al., 1985) by electroporation. Arabidopsis plants (Col-0 ecotype) were transformed by the floral dip method (Clough and Bent, 1998). Seeds were harvested and plated on kanamycin selection medium (50 μg mL-1) to identify T1 transgenic plants. From 34 T1 kanamycin-resistant lines recovered, approximately 60% showed an ABA-insensitive phenotype in seed germination as compared with untransformed plant or plants transformed with the empty vector. Approximately 100 of the T2 seeds were plated on Murashige and Skoog kanamycin agar plates and transgenic lines with a 3:1 (resistant:sensitive) segregation ratio were selected. Southern-blot analysis was performed as described by Lorenzo et al. (2002) to select lines carrying a single T-DNA copy. T3 progenies homozygous for kanamycin resistance were used for further studies.
Germination Assays
To measure ABA sensitivity, seeds were plated on solid medium composed of Murashige and Skoog basal salts, 1% (w/v) Suc, and different concentrations of ABA (0.5, 1, or 3 μm). For the dormancy assay, seed lots to be compared were harvested on the same day from individual plants grown in identical environmental conditions and were stratified during 0, 1, and 5 d at 4°C. Each value represents the average germination percentage of 80 to 100 seeds with the ses of at least three replicates.
To determine sensitivity to inhibition of germination by high osmoticum, the medium was supplemented with 400 mm mannitol and by salt with 200 mm NaCl. To measure PCB sensitivity, seeds were plated on medium containing 10 μm PCB. The percentage of seeds that had germinated and developed fully green expanded cotyledons was determined in all the assays after 7 d of sowing.
Root Growth and Transpiration Assays
The root growth assay for scoring ABA sensitivity was done by measuring root growth after 5 d of the transfer of 7-d-old seedlings onto vertical Murashige and Skoog plates containing 30 μm ABA. For the transpiration assays, the loss of fresh weight of excised leaves was measured at room temperature. To this end, four leaves at the same developmental stage and size from single 3-week-old plants were excised and fresh weight was determined at ambient conditions after the indicated periods of time.
In the stomatal study, leaves from 4-week-old plants with preopened stomata were detached, and paradermal sections of abaxial epidermis were fixed during 6 min in Romeis solution (44.8% alcohol, 10% formol, and 2% acetic acid, all v/v), washed with water during 15 min, stained for 5 min in Giemsa (Giemsa-Lösung; Merck, West Point, PA), and washed in running water during 15 min. Sections of the epidermis were dehydrated at 37°C until a color change was observed, and were then dehydrated with xilol for 3 min twice and Entellan (Merck). Observations and photographs were done on a light microscope (Ax70; Olympus, Melville, NY, with photographic digital system Apogee).
Northern-Blot Analysis
Approximately 10 to 12 7-d-old seedlings were transferred from Murashige and Skoog plates to 125-mL flasks containing 25 mL of Murashige and Skoog solution and 1% (w/v) Suc. The flasks were shaken at 130 rpm under cool fluorescent light. After 10 d, seedlings were mock-treated or treated with 50 μm ABA. Plant material was collected and frozen in liquid nitrogen. Total RNA was extracted as described (González-Guzmán et al., 2002), separated on formaldehyde-agarose gels, and blotted onto a nylon membrane. Blots were hybridized with random-priming 32P-labeled probes (30 μCi). Full-length cDNAs probe of FsPP2C1 was prepared as described previously (Lorenzo et al., 2001). The RAB18 probe was prepared by PCR amplification from genomic DNA of wild-type Col-0 plants with the primers described previously (González-Guzmán et al., 2002) and KIN2 probe was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus; expressed sequence tag 224O21). Blots were exposed for 24 h in a Phosphorimager screen (Molecular Dynamics, Sunnyvale, CA).
Acknowledgments
We thank Dr. Roberto Solano for critical reading of the manuscript and stimulating discussions. We also thank Elena Cid and Dr. Jose Aijón for technical assistance in the stomatal micrographs.
This work was supported by the Ministerio de Ciencia y Tecnología (grant no. BFI2000-1361) and by Junta de Castilla y León (grant no. SA010/02). P.L.R. was supported by a Ramón y Cajal research contract.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley JD (1997) Seed germination and dormancy. Plant Cell 9: 1055-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P (2003) The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J 34: 67-75 [DOI] [PubMed] [Google Scholar]
- Chérel I, Michard E, Platet N, Mouline K, Alcon C, Sentenac H, Thibaud JB (2002) Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell 14: 1133-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt (1996) A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273: 1239-1241 [DOI] [PubMed] [Google Scholar]
- Deblaere R, Bytebier B, De Greve H, Deboeck F, Schell J, Van Montagu M, Leemans J (1985) Efficient octopine Ti plasmid-derived vectors for Agrobacterium-mediated gene transfer to plants. Nucleic Acids Res 13: 4777-4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765-771 [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell Suppl 14: S15-S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Verhey SD, Holappa LD, Shen Q, Ho THD, Walker-Simmons MK (1999) An abscisic acid induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone. Proc Natl Acad Sci USA 96: 1767-1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Zentella R, Walker-Simmons MK, Ho THD (2001) Gibberellin/abscisic acid antagonism in barley aleurone cells: site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13: 667-769 [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Belles JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833-1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11: 1897-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey SJ, Bacon A, Burnett E, Neill SJ (1997) Abscisic acid signal transduction in epidermal cells of Pisum sativum L. Argenteum: both dehydrin mRNA accumulation and stomatal responses require protein phosphorylation and dephosphorylation. Planta 202: 85-92 [Google Scholar]
- Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E (2002) Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J 21: 3029-3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach A, Iten M, Grill E (1998) Signaling of abscisic acid to regulate plant growth. Philo Trans R Soc Lond B Biol Sci 353: 1439-1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Kwak JM, Schroeder JI (2001) An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106: 477-487 [DOI] [PubMed] [Google Scholar]
- Kerk D, Bulgrien J, Smith DW, Barsam B, Veretnik S, Gribskov M (2002) The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol 129: 908-925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ (1998) Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148: 885-992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Bentsink L, Hilhorst H (2002) Seed dormancy and germination. Curr Opin Plant Biol 5: 33-36 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM (1994) Seed dormancy and germination. In EM Meyerowitz, CR Somerville, eds, Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 313-334
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61: 377-383 [Google Scholar]
- Kuromori T, Yamamoto M (1994) Cloning of cDNAs from Arabidopsis thaliana that encode putative protein phosphatase 2C and a human Dr1-like protein by transformation of a fission yeast mutant. Nucleic Acids Res 22: 5296-5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, DeLong A, Schroeder JI (2002) Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic acid insensitivity in Arabidopsis. Plant Cell 14: 2849-2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang V, Palva ET (1992) The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol Biol 20: 951-962 [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10: 655-661 [DOI] [PubMed] [Google Scholar]
- Leung J, Bouvier-Durand M, Morris PC, Guerrier D, Chefdor F, Giraudat J (1994) Arabidopsis ABA response gene ABI1: features of a calcium-modulated protein phosphatase. Science 264: 1448-1452 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 19: 199-222 [DOI] [PubMed] [Google Scholar]
- Leung J, Merlot S, Giraudat J (1997) The Arabidopsis ABSCISIC ACID- INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatase 2C involved in abscisic acid signal transduction. Plant Cell 9: 759-771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang XQ, Watson MB, Assman SM (2000) Regulation of abscisic acid-induced stomatal closure and anion channels by guard cells AAPK kinase. Science 287: 300-303 [DOI] [PubMed] [Google Scholar]
- López-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782-4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Nicolás C, Nicolás G, Rodríguez D (2002) Molecular cloning of a functional protein phosphatase 2C (FsPP2C2) with unusual features and synergistically up-regulated by ABA and calcium in dormant seeds of Fagus sylvatica. Physiol Plant 114: 482-490 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Rodríguez D, Nicolás G, Rodríguez PL, Nicolás C (2001) A new protein phosphatase 2C (FsPP2C1) induced by abscisic acid is specifically expressed in dormant beechnut seeds. Plant Physiol 125: 1949-1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fedoroff N (2000) A mutation in the Arabidopsis HYL gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin and cytokinin. Plant Cell 12: 2351-2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie EA (2002) Evidence for a role for protein tyrosine phosphatase in the control of ion release from the guard cell vacuole in stomatal closure. Proc Natl Acad Sci USA 99: 11963-11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte WR Jr, Guiltinan MJ, Quatrano RS (1992) ABA-regulated gene expression: cis-acting sequences and trans-acting factors. Biochem Soc Trans 20: 93-97 [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signaling pathway. Plant J 25: 295-303 [DOI] [PubMed] [Google Scholar]
- Meyer K, Leube MP, Grill E (1994) A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264: 1452-1455 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco. Physiol Plant 15: 473-497 [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089-3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolás C, Nicolás G, Rodríguez D (1996) Antagonistic effects of abscisic acid and gibberellic acid on the breaking of dormancy of Fagus sylvatica seeds. Physiol Plant 96: 244-250 [Google Scholar]
- Nicolás C, Nicolás G, Rodríguez D (1998) Transcripts of a gene, encoding a small GTP-binding protein from Fagus sylvatica, are induced by ABA and accumulated in the embryonic axis of dormant seeds. Plant Mol Biol 36: 487-491 [DOI] [PubMed] [Google Scholar]
- Nicolás C, Rodríguez D, Poulsen F, Eriksen EN, Nicolás G (1997) The expression of an abscisic acid-responsive glycine-rich protein coincides with the level of seed dormancy in Fagus sylvatica. Plant Cell Physiol 38: 1303-1310 [DOI] [PubMed] [Google Scholar]
- Nylander M, Svensson J, Palva ET, Welin BV (2001) Stress-induced accumulation and tissue-specific localization of dehydrins in Arabidopsis thaliana. Plant Mol Biol 45: 263-279 [DOI] [PubMed] [Google Scholar]
- Pandey S, Tiwary SB, Upadhyaya KC, Sopory SK (2000) Calcium signaling: linking environmental signals to cellular functions. Crit Rev Plant Sci 19: 291-318 [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567-1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI (1998) Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282: 287-290 [DOI] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI (1997) Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 9: 409-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI (2000) Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406: 731-734 [DOI] [PubMed] [Google Scholar]
- Rock CD, Quatrano RS (1995) The role of hormones during seed development. In PJ Davies, ed, Plant Hormones. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 671-697
- Rodríguez PL (1998) Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol Biol 38: 919-927 [DOI] [PubMed] [Google Scholar]
- Rodríguez PL, Benning G, Grill E (1998) ABI2, a second protein phosphatase involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett 421: 185-190 [DOI] [PubMed] [Google Scholar]
- Schmidt C, Schelle I, Liao YJ, Schroeder JI (1995) Strong regulation of slow anion channels and abscisic acid signaling in guard cells by phosphorylation and dephosphorylation events. Proc Natl Acad Sci USA 92: 9535-9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J (1998) Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proc Natl Acad Sci USA 95: 975-981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju S, Palva T (2001) Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J 26: 461-470 [DOI] [PubMed] [Google Scholar]
- Vranova E, Langebartels C, Van Montagu M, Inze D, Van Camp W (2000) Oxidative stress, heat shock and drought differentially affect expression of a tobacco protein phosphatase 2C. J Exp Bot 51: 1763-1764 [DOI] [PubMed] [Google Scholar]
- Vranova E, Tahtiharju S, Sriprang R, Willekens H, Heino P, Palva ET, Inze D, Van Camp W (2001) The AKT3 potassium channel protein interacts with the AtPP2CA protein phosphatase 2C. J Exp Bot 52: 181-182 [PubMed] [Google Scholar]
- Wang H, Datla R, Georges F, Loewen M, Cutler AJ (1995) Promoters from kin1 and cor6.6, two homologous Arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature ABA, osmoticum and dehydration. Plant Mol Biol 28: 605-617 [DOI] [PubMed] [Google Scholar]
- Werner WE, Finkelstein R (1995) Arabidopsis mutants with reduced response to NaCl and osmotic stress. Physiol Plant 93: 659-666 [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ (1997) A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol 15: 444-447 [DOI] [PubMed] [Google Scholar]
- Wu Y, Kuzma J, Marechal E, Graeff R, Lee HC, Foster R, Chua NH (1997) Abscisic acid signaling through cyclic ADP-ribose in plants. Science 278: 2126-2130 [DOI] [PubMed] [Google Scholar]
- Wu Y, Sánchez JP, López-Molina L, Himmelbach A, Grill E, Chua NH (2003) The abi1-1 mutation blocks ABA signaling downstream of cADPR action. Plant J 34: 307-315 [DOI] [PubMed] [Google Scholar]
- Xiong L, Gong Z, Rock CD, Subramanian S, Guo Y, Xu W, Galbraith D, Zhu JK (2001) Modulation of abscisic acid signal transduction and biosynthesis by an Sm-like protein in Arabidopsis. Dev Cell 1: 771-781 [DOI] [PubMed] [Google Scholar]