Abstract
As an important step toward free access and, thus, impact of GoldenRice, a freedom-to-operate situation has been achieved for developing countries for the technology involved. Specifically, to carry the invention beyond its initial “proof-of-concept” status in a Japonica rice (Oryza sativa) cultivar, we report here on two transformed elite Indica varieties (IR64 and MTL250) plus one Japonica variety Taipei 309. Indica varieties are predominantly consumed in the areas with vitamin A deficiency. To conform with regulatory constraints, we changed the vector backbone, investigated the absence of beyond-border transfer, and relied on Agrobacterium tumefaciens-mediated transformation to obtain defined integration patterns. To avoid an antibiotic selection system, we now rely exclusively on phosphomannose isomerase as the selectable marker. Single integrations were given a preference to minimize potential epigenetic effects in subsequent generations. These novel lines, now in the T3 generation, are highly valuable because they are expected to more readily receive approval for follow-up studies such as nutritional and risk assessments and for breeding approaches leading to locally adapted variety development.
We reported previously on a genetically modified rice line, frequently termed GoldenRice, engineered to synthesize and accumulate pro-vitamin A (β-carotene) in the endosperm (Ye et al., 2000). Since then, the concept of genetic engineering-based nutritional enhancement of rice to contribute to a sustained reduction of vitamin A deficiency (VAD) has evoked strong expectations to develop in the public sector and to deliver a safe product in a short period of time. However, to carry the project from the scientific discovery to impact, i.e. to actually provide grain to subsistence farmers and urban poor in developing countries free of charge for the technology used, a large number of obstacles, not only scientific in nature, need to be dealt with. These are contractual and regulatory aspects.
Contractual issues have largely been solved to date, including a “freedom-to-operate” intellectual property situation for GoldenRice, the inventors, and their licensees. Free licenses have been granted by our license collaborators for patents used in this research designed to benefit resource-poor farmers in developing countries. The next step to be taken is to find regulatory acceptance in these countries, the prerequisite for most of the tasks ahead such as to allow grain production enabling feeding trials and to begin with diversified variety development.
GoldenRice as published (Ye et al., 2000) demonstrates the feasibility of the scientific approach but does not yet represent a product. Its development was based on the finding that the precursor geranylgeranyldiphosphate, which wild-type rice endosperm is capable of synthesizing, can be used by the subsequent enzyme phytoene synthase (PSY) when the latter is supplemented by transformation (Burkhardt et al., 1997). Providing the full supplement of all necessary biosynthetic genes resulted in yellow-colored grains, which contained carotenoids in significant quantities (up to 1.6 μg g-1) in the T1 generation. However, this most colored line we produced (zvit 11b) suffered from having undergone multiple integrations and recombination events (Ye et al., 2000), making it not ideal as the basis for obtaining deregulation and proceeding with development of stable and dependable rice food products.
To conform with regulatory requirements, the generation of defined DNA integrations along with the minimization of extraneous DNA from the vector backbone (Kononov et al., 1997) are necessary. Such integration events are likely to be straightforward to characterize and then to assess any risk associated with them. Therefore, the transfer of vector DNA into the genome of the transformants (beyond-border transfer) was monitored by PCR. In addition, to abandon the use of an antibiotic selectable marker is very desirable, especially from a regulatory perspective. Therefore, we instead established for rice (Lucca et al., 2001) the phospho-Man-isomerase (Bojsen et al., 1998; Bojsen et al., 1999) selection system and now rely exclusively on it. Agrobacterium tumefaciens-mediated transformation was used throughout because of the defined integration pattern this method produces, showing no secondary recombination events in most cases. Single integrations were to be favored to reduce the potential for deleterious epigenetic effects in subsequent generations. Moreover, we chose the modular pCambia vector series because of its worldwide use in laboratories in developing countries and by nonprofit organizations (Roberts et al., 2000).
To ultimately provide a useful product beyond the “proof-of-concept” stage, more issues needed to be addressed. The previously described lines were generated using the Japonica rice line Taipei 309 because it is relatively easy to transform. However, Indica cultivars, although much less amenable to transformation, are the most predominantly grown and consumed in countries where VAD prevails. Therefore, we set out to engineer the pathway into two elite Indica cultivars of Southeast Asia, namely, IR64 and MTL250. These are to be used in breeding approaches to transfer the trait into additional, locally important varieties. Alternatively, because breeding of the trait from Taipei 309 into Indica varieties is feasible, we decided to transform this cultivar also.
Here, we report on a Japonica variety line that conforms to these requirements. Further, as a next practical step, we also demonstrate the successful use of the GoldenRice technology in transformed Indica elite varieties. The present publication also represents our first progress report on the GoldenRice humanitarian project.
RESULTS
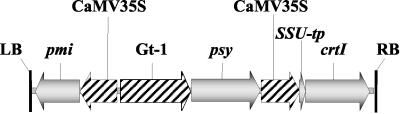
We transformed two Indica cultivars, namely IR64 and MTL250, and one Japonica cultivar (Taipei 309). IR64 is popular worldwide, whereas MTL250 is widely used in Vietnam. All these Indica cultivars possess a good agronomic background and good grain quality. The vector pCaCar (Fig. 1) was used throughout in the present study in the A. tumefaciens-mediated transformation. PMI was used as the selectable marker gene. The construct combines the bacterial phytoene desaturase gene crtI, fused with the pea (Pisum sativum) RuBisCo small subunit transit-encoding sequence (Misawa et al., 1993) and under the control of the constitutive 35S cauliflower mosaic virus (CaMV) promoter with the daffodil (Narcissus pseudonarcissus) psy, driven by the endosperm-specific Gt1 promoter. To use a combination of only these two structural genes coding for carotenoid bio-synthetic enzymes, omitting lcy, is the outcome of our previous findings showing that these were able to reconstitute the entire pathway, including the formation of α- and β-carotene and derived xanthophylls (Ye et al., 2000).
Figure 1.
Scheme of the vector insert of pCaCar. LB and RB, Left and right T-DNA borders; Gt1, glutelin promoter; pmi, phosphomannose isomerase; psy, phytoene synthase, crtI, bacterial phytoene desaturase; SSU-tp, transit peptide of the ribulose-bis-phosphate carboxylase small subunit.
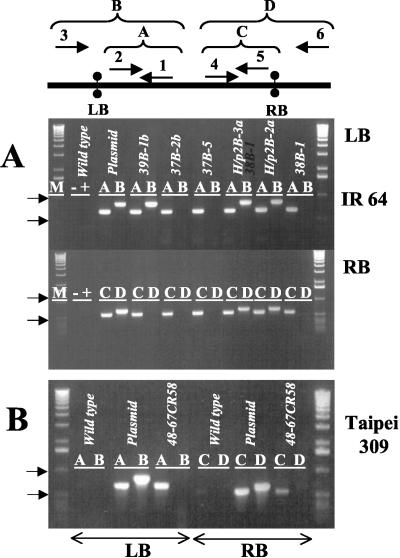
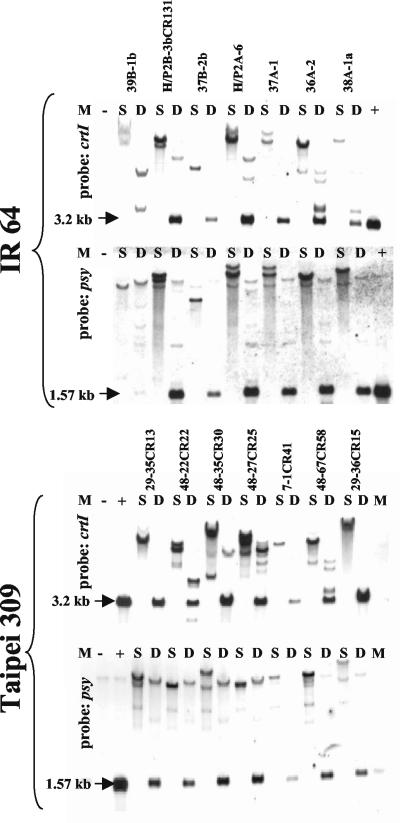
As given in Table I, the T1 seeds were first phenotypically selected by visible color after polishing. Segregating seed populations with notably yellow grains (about 20% of the transformants) were subjected to carotenoid analysis by HPLC and photometry (see below). The phenotypically best lines were then checked for the absence of beyond-border transfer by PCR (as shown in Fig. 2) for some representative examples; more than one-half of the lines examined were positive in this respect (Table I). The quality of integration and the integration number were judged by Southern hybridization with selected T0 transformants. The presence of the expected bands for the crtI cassette (3.2 kb) and psy cDNA (1.57 kb) is shown in Figure 3, taking some IR64 and Taipei 309 lines as an example; very similar patterns were observed for MTL250 (not shown, see Table I). The numbers of independent transgenic plants (T0) confirmed by Southern-blot analysis were 23 for IR64, seven for MTL250, and 27 for Taipei 309. A simple integration pattern of the transgenes (one-three copies) was predominant, and transgene recombination did not occur.
Table I.
Overview on generated transgenic To lines, with the resulting T1 and T2 seeds transformed with pCaCar
| Cultivar | No. of To Plants | T1 Selected by Visual Inspection and Carotenoid Analysis | Integration (n) beyond Border [l/r]a cat Presence (Yes/No/Not Tested [NT]) | T2 Seeds |
|---|---|---|---|---|
| Taipei 309 | 55 | 7-1 CR41 | (1) [−/−] | Yes |
| 25-11 CRNT | (2) [+/−] (NT) | |||
| 29-33 CRNT | (3) [−/−] (NT) | |||
| 29-35CR13 | (1) [−/+] (No) | Yes | ||
| 29-36CR15 | (1) [−/−] (NT) | |||
| 48-22CR22 | (2) [+/+] (Yes) | |||
| 48-23CR109 | (1) [+/+] (Yes) | |||
| 48-27CR25 | (3) [+/+] (No) | |||
| 48-33 CRNT | (2) [+/+] (Yes) | |||
| 48-35 CR30 | (3) [−/−] (NT) | |||
| 48-36CR33 | (3) [−/−] (NT) | |||
| 48-37CR53 | (1) [−/−] (NT) | |||
| 48-41 CR112 | (3) [+/−] (NT) | |||
| 48-42 CR36 | (3) [−/+] (NT) | |||
| 48-50CRNT | (2) [−/+] (No) | |||
| 48-67CR58 | (1) [−/−] (NT) | Yes | ||
| 52-34 CRNT | (1) [+/+] (Yes) | |||
| IR 64 | 35 | 36A2 | (2) [−/−] (NT) | Yes |
| 37A-1 | (3) [−/−] (NT) | Yes | ||
| 37B-2b | (1) [−/−] (NT) | Yes | ||
| 37B-5 | (5) [−/−] (NT) | Yes | ||
| 38A-1a | (1) [−/−] (NT) | Yes | ||
| 39B-1b | (2) [+/+] (Yes) | |||
| 39B-1a | (3) [−/−] (NT) | Yes | ||
| 61A-2 | (4) [+/+] Yes | |||
| H/P2B-2aCR128 | (3) [−/−] (NT) | Yes | ||
| H/P2B-3a | (2) [+/+] (Yes) | |||
| MTL 250 | 24 | 56A-1 | (1) [+/+] (Yes) | Yes |
| 56A-2 | (5) [+/+] (Yes) | Yes | ||
| 60B-12 | (3) [+/−] (Yes) | Yes |
>a1, Left border sequences detected ([+]) or not detected ([−]); r, right border sequences detected ([+]) or not detected ([−]).
Figure 2.
PCR analysis for beyond-border transfer. The scheme gives the primer combination used for the inside-border (reactions A and C) and outside-border amplicons (B and D). Primers were as given in “Materials and Methods.” A, Three examples for Indica IR 64 lines showing clean integration (37B-2b, 37B-5, and 38B-1) and some examples showing beyond-border transfer. B, No beyond-border transfer in the favorite Japonica Taipei 309 line 48-67CR58. For additional data, see Table I.
Figure 3.
Southern-blot analysis. Genomic DNA digested with KpnI as a single cutter (S) and with EcoRI as double cutters (D) was analyzed as given in “Materials and Methods.” The expected transgene band sizes are given with an arrow. The favorite lines IR 64 37B-2b and Taipei 309 48-67CR58 are given in the upper and lower panels, respectively.
Based on the sum of these data (see also Table I) and the carotenoid analysis done with the T1 seeds (see below), lines were selected to produce self-pollinated progeny (T2 seeds).
Analysis of T1 Seeds
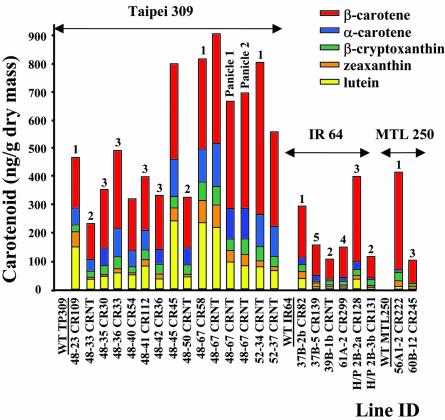
T1 segregating seeds (including white and colored) were subjected to carotenoid analysis. This was first done photometrically for quantification, followed by HPLC to determine the distribution of the individual carotenoids. However, because we soon noticed isoprenoid compounds other than carotenoids were changed relative to the controls (see below), internal standardization needed to be applied. Figure 4 shows an HPLC trace as an example allowing the simultaneous quantification of carotenoids, tocopherols/tocotrienols (vitamin E), and γ-oryzanol, the last representing a mixture of at least 10 steryl ferulate esters (Xu and Godber, 1999). The carotenoid amount in these T1 seeds was variable among the different lines (examples are given in Fig. 5), ranging from 0.1 to about 0.9 μg g-1 dry mass. Generally, higher values were obtained with the Japonica variety Taipei 309 than with the transformed Indica cultivars. Moreover, the amount seemed independent of the integration number of the transgenes, as evidenced when comparing line 48-42CR36 with 52-34-CRNT. This indicates that positional effects play a predominant role. The carotenoid pattern observed showed another difference between Japonica and Indica cultivars, with the β-carotene fraction comprising 39% to 67% and 61% to 82% of the total carotenoid complement, respectively. Considering all carotenoids exhibiting provitamin A activity (β-carotene, α-carotene, and β-cryptoxanthin), this translates into 56% to 87% and 81% to 93%, respectively. Different panicles from individual plants did not show much variation, neither in amount nor carotenoid pattern. At this point, it became evident already that—just as we saw before (Ye et al., 2000)—a combination of PSY and the bacterial desaturase alone was sufficient to install the entire biosynthetic pathway, including carotene cyclization, leading to α- and β-carotene and xanthophyll formation.
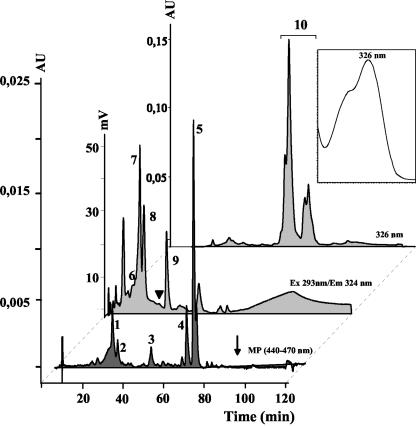
Figure 4.
HPLC analysis of embryo-free T2 rice seeds (Taipei 48-67-3). Lower trace, Carotenoids max-plot (MP; each compound displayed in its individual λmax). Arrow indicates the position of lycopene. Middle trace, Fluorescence detector response showing vitamin E compounds. Arrowhead indicates the position of α-tocopherol appearing in grains containing embryos. Upper trace, Detection of γ-oryzanol at 326 nm. Inset, γ-Oryzanol spectrum. Compounds detected: 1, lutein; 2, zeaxanthin; 3, β-cryptoxanthin; 4, α-carotene; 5, β-carotene; 6, γ-tocotrienol; 7, β/δ-tocotrienol; 8, α-tocotrienol; 9, α-tocopherol-acetate as internal standard; 10, γ-oryzanol.
Figure 5.
Carotenoid analysis of T1 seeds. Wild-type seeds were carotenoid free. The color code for individual carotenoids is given. Numbers above bars represent the integration number estimated by Southern-blot analysis. Dry mass refers to fully mature dry seeds.
Analysis of T2 Seeds
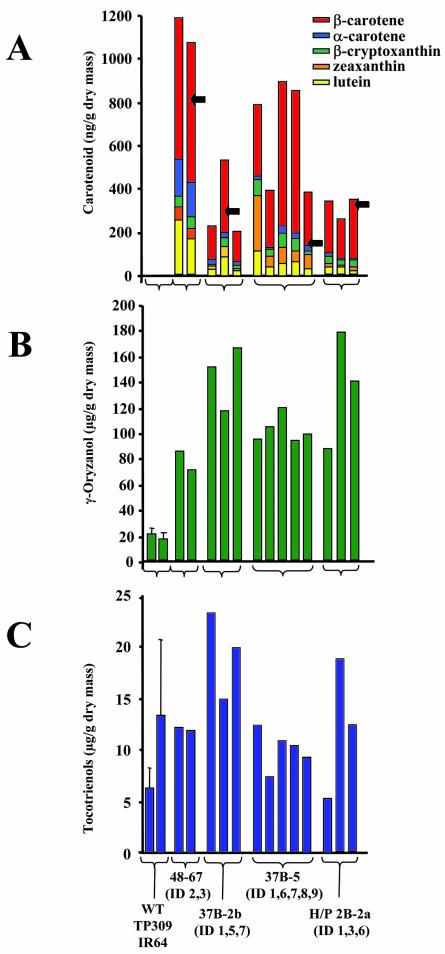
Promising lines (examples are shown in Fig. 6) were passed into T2 seed production (Table I) and re-analyzed in greater detail. Data on the best lines (underlined in Table I) are given in Figure 7. In these seeds, the carotenoid content was about the same or higher as compared with the parent T1 generation (Fig. 7A), as one would expect with a stable line. Some variation in both carotenoid content and pattern was observed, probably representing some degree of segregation and/or natural variability. As observed in T1, the carotenoid amount in the Japonica line (1.2 μg g-1 dry mass) exceeded that in the Indica lines (around 0.8 μg g-1 dry mass).
Figure 6.
Examples of some T2 seeds. A, IR64 wild type; B to F, transgenic IR64 lines; G, Taipei 309 wild type; H, transgenic Taipei 309 line.
Figure 7.
Analysis of T2 seeds. A, Carotenoid analysis. The color code for individual carotenoids is given. Arrows indicate the carotenoid amount in the parent T1 seeds; B, γ-oryzanol determination; and C, total tocotrienols.
A recurrent observation in our transformation experiments using the given transgene combination is a notable increase in most lines of a compound group that we identified as γ-oryzanol (see Fig. 7B). This valuable triterpenoid-ferulate antioxidant occurs in a very steep concentration gradient, with high concentrations in the outer layers and much lower concentrations in the endosperm, as seen in a time course of polishing (data not shown). A standardized polishing protocol (20-24 h, emery paper 200-220 rpm, for 40-60 seeds) was applied for the data set given in Figure 7B, showing very notable increases in most transformants. However, given the geometry of distribution in the grain, absolute values must be treated with caution.
Vitamin E compounds (tocopherols and tocotrienols) represent another class of isoprenoids present in rice grains. The presence of the embryo needs to be considered here because it contains large amounts of α-tocopherol. It is absent from the endosperm, where only tocotrienols are found. Analysis of embryo-deprived grains given in Figure 7C shows that this class of isoprenoid compounds, in contrast to γ-oryzanol, did not change significantly in the transgenic lines relative to the wild type.
DISCUSSION
Rice feeds nearly one-half of the world's population, but it is a poor source of many essential micro-nutrients. As a consequence, and due to poverty and limited access to more diversified foods, deficiencies of iron, zinc, iodine, and vitamin A prevail in rice-consuming developing countries. About 250 million people suffer from VAD (Underwood, 2000; UNICEF, 2000), causing increased mortality and growth retardation of children. VAD frequently impairs vision, leading to many thousands of blind children every year through xerophthalmia, and contributes to anemia by interfering with iron bioavailability (Gillespie and Haddad, 2001).
Naturally occurring vitamin A derives entirely from carotenoids with provitamin A activity. β-Carotene is the most important provitamin A for mammals. To be biologically active, β-carotene must be absorbed and then cleaved at the C15, C15′ double bond by an iron- and oxygen-dependent enzyme. Recently, the respective animal genes have been cloned from fruitfly (Drosophila melanogaster; von Lintig and Vogt, 2000) and mammals, including humans (Redmond et al., 2001; Yan et al., 2001). This food chain from plant to human for vitamin A is the basis of the “biofortified” GoldenRice, able to provide provitamin A by its own biosynthetic capacity, conferred to the rice grain by recombinant DNA technology. No breeding approach can substitute as an alternative to date, due to the present lack of parent cultivars exhibiting this valuable trait.
GoldenRice has, so far, only been an experimental Japonica line. Because the malnourished population lives mainly on Indica rice, it was essential to transfer the trait to Indica varieties. For this purpose, the locally important varieties in VAD-affected areas are currently being identified on the basis of needs assessments, as initiated in India by the Indo-Swiss Collaboration in Biotechnology.
In the current publication, we present three novel Indica and Japonica GoldenRice lines that have been specifically adapted to comply with regulatory requirements. To meet these standards by producing well-defined integration events will hopefully open the way to receiving approval for field testing and for bioavailability studies; such studies are currently in an advanced planning stage. We also expect that our new lines will facilitate the start of breeding approaches leading to variety development. Bioavailability studies especially are required to determine the uptake of the provitamin from the endosperm food matrix and, thus, evaluate the usefulness of the current lines to deal with the problem of VAD. Moreover, such data will provide guidance on the amount of the provitamin to be produced in future versions of GoldenRice. This effort is being accompanied by ex ante studies on projected effects (Zimmermann and Qaim, 2002).
Our three candidate rice lines now growing in the T3 generation in greenhouses are Taipei 309 (Japonica), 48-67CR58; and IR 64 (Indica), 37B-2b and 37B-5. These lines produced T2 grains with about 1.2, 0.4, and 0.8 μg carotenoid g-1 dry mass, respectively. Because in our chemical analyses we tested grains randomly and did not select yellow ones, these values will increase somewhat in later generations as these lines are bred to homozygosity. Anticipating the need to further increase the content to nutritionally required levels of the provitamin, we already have parallel investigations and rice transformations under way, aiming with several different approaches at increasing carotenoid production in the endosperm by a factor of 2 to 3.
The production of transgenic lines in different Indica varieties demonstrates the applicability of the technology to diverse rice genotypes; this has also been shown by biolistic approaches elsewhere (S. Datta, personal communication).
Consistent with the results on our published Japonica prototype lines (Ye et al., 2000), only two genes—coding for PSY and CrtI—are required to install the carotenoid biosynthetic pathway in rice. The additional enzymatic activities of lycopene cyclases yielding α- and β-carotene and hydroxylase activities yielding zeaxanthin and lutein observed in such transformants have not been genetically directly conferred by transformation. Preliminary evidence suggests rather a feedback signaling loop leading to the activation of the rice carotenoid biosynthesis genes. We assume that this phenomenon is initiated by the delivery of all-trans-configured lycopene by the CrtI gene product, in contrast to the two plant desaturases delivering poly-cis-lycopene (Bartley et al., 1999). This scenario is in line with transformation experience of others utilizing crtI, e.g. in tomato (Lycopersicon esculentum), where similar results have been obtained (Romer et al., 2000). Indicative for a potential crucial role of geometric carotene isomers is the recent discovery of a lycopene cis-transisomerase as an additional pathway enzyme in tomato (Isaacson et al., 2002) and Arabidopsis (Park et al., 2002).
Another novel observation we describe, namely the increased formation of γ-oryzanol (Xu and Godber, 1999), may be interpreted in these terms as well. Interestingly, an isoprenoid pathway (triterpenoid), although cytoplasmically localized, is involved together with the phenylpropanoid pathway in delivering the ferulate moiety. γ-Oryzanol is a healthy antioxidant compound, claimed to be more efficient than vitamin E (Xu et al., 2001; Huang et al., 2002). It remains to be investigated whether all this up-regulation takes place in the endosperm alone or in the outer layers of the fruit wall also, where γ-oryzanol occurs predominantly. In contrast, vitamin E compounds, which are plastid-localized isoprenoids just like carotenoids, remained largely unaffected. Taken together, these are unexpected but beneficial regulative side effects. To achieve a broader overview on the interactions involved here, expression profiling analyses are currently underway.
MATERIALS AND METHODS
Construction of the Binary Vector pCaCar
In this study, a single binary vector (pCaCar) was used for all transformation experiments. The vector pCaCar was constructed in two steps. First, the hygromycin phosphotransferase gene of pCAMBIA 1200 (Cambia, Canberra, Australia) was replaced by the PMI gene to yield pCamanose. For this purpose, the PMI gene was isolated from p-Man 19 (Lucca et al., 2001) by XhoI digestion and cloned into the corresponding sites under the control of the constitutive CaMV 35S promoter. Subsequently, the expression cassettes for psy under the control of the endosperm-specific glutelin (Gt1) and for crtI under the control of the constitutive CaMV 35S promoter were excised from pBaal2 (Ye et al., 2000) by using NotI, followed by a treatment with Klenow fragment, and finally by digesting with KpnI. The obtained fragment, containing both expression cassettes, was then ligated into HindIII-digested, Klenow fragment blunt-ended and KpnI-treated pCamanose to yield pCaCar. The binary vector was transformed into the competent cells of Agrobacterium tumefaciens strain LBA 4404 (Hoekema et al., 1984).
Plant Materials and Transformation
Immature embryos and embryogenic calli derived from immature embryos or mature seeds of the cultivars IR 64, MTL 250, and Taipei 309 were inoculated with A. tumefaciens strain LBA 4404 (Hoekema et al., 1984). The transformation procedures for immature embryos described by Aldemita and Hodges (1996) were followed. Callus transformations were performed as reported by Hoa et al. (2002). We followed the procedures for selection of transformed calli, regeneration, and rooting as described by Lucca et al. (2001) with the following modifications for Indica rice. We used low concentrations of Man: 2.5% (w/v) Man + 3% (w/v) Suc for the first selection, 2.5% (w/v) Man + 1.5% (w/v) Suc for the second selection, and 3.5% (w/v) Man + 0.5% (w/v) Suc for the third round. Man resistant rice plants were transferred to soil and grown in the greenhouse at 28°C (day) and 21°C (night) and 80% relative humidity.
DNA Isolation and Southern-Blot Analysis
Genomic DNA was isolated from frozen rice leaves following the method of McCouch et al. (1988). Ten micrograms of genomic DNA was digested with EcoRI to detect psy or crtI and with KpnI for copy number analysis. The digested DNA was electrophoretically fractionated through a 0.8% (w/v) agarose gel before capillary transfer and immobilization on nylon membranes (Hybond N+, Biosciences Europe, Freiburg, Germany). PCR-amplified, digoxygenin-labeled (Roche Applied Science, Mannheim, Germany) fragments of psy (primers: 5′ TACGTAGCAGGAACTG 3′ and 5′ CAAACAGGCCACCTGCTAGC 3′) and crtI (primers: 5′ GAGTGGGGCGTCTGGTT 3′ and 5′ TAACTGCCGCAACCTT 3′) were used as probes. Blotting, hybridization, washing, and detection were performed following the procedures as given in Wünn et al. (1996).
PCR to Examine beyond-Border Transfer
Genomic DNA was extracted from frozen rice leaves using the Nucleon extraction and purification kit (Amersham). PCR reactions were carried out using 100 to 200 ng of genomic DNA, 10 μμ each primer, 200 μm dNTPs, and 2.5 units of Taq-polymerase (Eurobiotaq, Laboratores EUROBIO, Les Ulis, France). After an initial denaturation step for 5 min, the PCR reactions were performed for 35 cycles including 1 min of denaturation at 95°C, 1 min of annealing at 58°C, and finally 1 min of polymerization at 72°C.
For monitoring beyond-border transfer, two PCR reactions were performed for each side (A and B, left border; and C and D, right border). PCR reactions A and C confirmed the transformation event and served as a positive control for checking the quality of the isolated genomic DNA and the PCR conditions. This PCR reaction should yield a product with all the transgenes, whereas PCR reactions B and D will lead to a product only in case of beyond-border transfer. Reactions A and B and C and D shared a primer that binds within the borders (see scheme in Fig. 2). For the left border, we used the following primers: reaction A, primer 1, 5′ CGCTATTGCTGAATGTGGTG 3′ (annealing site: 14,189-14,170 bp), primer 2, 5′ CGGGGGATCTGGATTTTAGT 3′ (annealing site: 13,522-13,541 bp); and reaction B, primer 1, 5′ CGCTATTGCTGAATGTGGTG 3′ (annealing site: 14,189-14,170 bp), primer 3, 5′ CTGCCTGTATCGAGTGGTGA 3′ (annealing site: 13,363-13,382 bp).
To examine the right border, we used the following primers: reaction C, primer 4, 5′ CAGCGTACTGATGCTCCAAG 3′ (annealing site: 6,432-6,451 bp), primer 5, 5′ TTTAAACTGAAGGCGGGAAA 3′ (annealing site: 6,983-6,964 bp); and reaction D, primer 4, 5′ CAGCGTACTGATGCTCCAAG 3′ (annealing site: 6,432-6,451 bp), primer 6: 5′ AAACCTTTTCACGCCCTTTT 3′ (annealing site: 7,077-7,058 bp).
In case of beyond-left-border transfer, a third PCR reaction was performed to confirm that the chloramphenicol resistance gene (CAT) had not also been transferred into the transgenic plants. For this purpose, the following primers were used: chloram, back, 5′ ATCACAAACGGCATGATGAA 3′ (annealing site: 12,307-12,326 bp); and chloram, forward, 5′ GCAGTCGCCCTAAAACAAAG 3′ (annealing site: 12,926-12907 bp). The reaction was performed as described above.
Segregation Analysis of the Transgenes
The self-pollinated seeds (T1 generation) of the transformants were grown in Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 3% (w/v) Suc and 3% (w/v) Man. The resistant plants scored 2 weeks after culture were transferred and grown in the greenhouse for further Southern-blot analysis, and T2 seeds were obtained.
Carotenoid Extraction and Analysis
Dehusked seeds were polished overnight on a shaker at 200 to 220 rpm in petri dishes equipped with emery paper. Alternatively, we used a commercial grain polisher (Kett, Tokyo) at a polishing time of 60 s. In contrast to the first method, this procedure removes the embryo from Taipei 309 seeds. Polished seeds (0.5 g) were ground for 60 s to a fine powder using a Micro-Dismembrator (Braun, Melsungan, Germany). The powder in the Teflon capsules was resuspended in 2 mL of acetone at room temperature and transferred to a glass test tube, and as an internal standard, 200 μg of α-tocopherol acetate was added from an appropriate acetone stock solution. This suspension was sonicated for 10 s, centrifuged, and the supernatant collected; the extraction procedure was repeated with another 2 mL to ensure complete carotenoid extraction. The combined extracts were dried in a Vortex-Evaporator (Haake-Buchler, Saddle Brook, NJ) and resuspended in 500 μL of acetone. A UV/VIS spectrum was read (700-350 nm), and the absorption was determined at 475 nm, corresponding to the side maximum of the carotenoid spectrum obtained. This was done to correct for a certain baseline drift occurring at shorter wavelengths. Given the relative uniformity of the carotenoid chromophore in transgenic rice (α- or β-carotene derivatives), the absorption at 450 nm could be calculated using a factor of 1.125. Total carotenoid amount was calculated using a molar extinction coefficient of ε = 134,500 L-1 mol-1 cm-1 averaging for both chromophores. The distribution of carotenoids was then determined by HPLC (see below). Alternatively, omitting photometry, the amount was determined by HPLC using the internal plus an external standard (echinenone, kindly provided by Hoffmann-La Roche, Basel, Switzerland). Samples were taken to dryness and dissolved in 70 μL of chloroform, of which 50 μL was injected into the HPLC system.
For HPLC, a Waters (Eschborn, Germany) system equipped with a photodiode array detector (model 996) and a fluorescence detector (model 474; excitation 293 nm, emission 324 nm for tocopherol detection) was used. The separation was performed with a C30-RP column (YMC Europe, Schermbeck, Germany) using the solvent systems B: MeOH/tert-butyl-methyl ether/water (60:12:12 [v/v]) and A: MeOH/tert-butylmethyl ether (1:1 [v/v]). The column was developed with a linear gradient from 100% B to 43% B within 45 min, then to 0% B within 35 min, then maintaining the final conditions for another 30 min. Using a maxplot (400-500 nm), carotenoid peaks were integrated at their individual λ max, whereas γ-oryzanol peaks were integrated at 326 nm. Tocopherols were integrated using the fluorescence detector signals. For normalization and quantification, all peaks were normalized relative to the internal α-tocopherol acetate standard to correct for extraction and injection variability. An external standard, echinenone, run separately, was then used to calculate carotenoid amounts. All carotenoid peaks underwent a second normalization to correct for their individual molar extinction coefficients relative to echinenone (=1), using: lutein (0.969), β-carotene (0.884), α-carotene (0.791), zeaxanthin (0.891), and β-cryptoxanthin (0.872).
Integrated tocopherol/tocotrienol fluorescence area units, with the aid of calibrated standard solutions (Sigma, Taufkirchen, Germany), were normalized with the following correction factors: α-tocopherol (474 ± 20), β-tocopherol (953 ± 13), γ-tocopherol (966 ± 5), δ-tocopherol (984 ± 12), α-tocotrienol (539 ± 2), β-tocotrienol (910 ± 4), γ-tocotrienol (1101 ± 11), and δ-tocotrienol (843 ± 3) ng-1, each. γ-Oryzanol was quantified using a standard solution extracted from a product available in U.S. vitamin shops (γ-oryzanol 200, GNC, Pittsburgh) applying a conversion factor of 1,831 area units ng-1.
Note Added in Proof
A related publication appeared recently (Dalta K, Baisakh N, Oliva N, Torrizo L, Abrigo E, Tan J, Rai M, Rehana S, Al-Babili S, Beyer P et al. Bioengineered golden indica rice cultivars with β-carotene metabolism in the endosperm with hygromycin and mannose selection systems. Plant Biotech J 1: 81-90).
Acknowledgments
We thank Prof. Wilhelm Gruissem (ETH Zurich) for his support. The skillful assistance of Ulricke Schneider-Ziebert (University of Freiburg), Katalin Konya (ETH Zurich), and Carmen Schubert (University of Freiburg) is gratefully acknowledged. We thank Dr. Katharina Jenny (Swiss Agency for Department and Cooperation, Berne) and Dr. Camilla Beech (Sygenta, Basel) for their very valuable discussions. Randall Cassada (University of Freiburg) corrected the English version of the manuscript.
This paper is dedicated to Dr. Adrian Dubock (Syngenta), who from early on was—and remains—a most selfless and effective supporter of the GoldenRice humanitarian project and an invaluable mediator between the public and private sectors.
This work was supported by the Rockefeller Foundation (New York).
References
- Aldemita RR, Hodges TK (1996) Agrobacterium tumefaciens-mediated transformation of Japonica and Indica rice varieties. Planta 199: 612-617 [Google Scholar]
- Bartley GE, Scolnik PA, Beyer P (1999) Two Arabidopsis thaliana carotene desaturases, phytoene desaturase and ζ-carotene desaturase, expressed in Escherichia coli, catalyze a poly-cis pathway to yield prolycopene. Eur J Biochem 259: 396-403 [DOI] [PubMed] [Google Scholar]
- Bosjen K, Donaldson I, Haldrup A, Joersboe M, Kreiberg JD, Nielsen J, Okkels FT, Peterson SG, Whenham RJ, inventors. November 30, 1999. Positive selection. United States Patent 5,994,629
- Bosjen K, Donaldson I, Haldrup A, Joersboe M, Kreiberg JD, Nielsen J, Okkels FT, Peterson SG, Whenham RJ, inventors. June 16, 1998. Mannose or xylose based positive selection. United States Patent 5,767,378
- Burkhardt PK, Beyer P, Wünn J, Kloti A, Armstrong GA, Schledz M, von Lintig V, Potrykus I (1997) Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J 11: 1071-1078 [DOI] [PubMed] [Google Scholar]
- Gillespie S, Haddad L (2001) Attacking the double burden of malnutrition in Asia and the Pacific. ADB Nutrition and Development Series No. 4. http://www.adb.org/Documents/Books/Nutrition/Malnutrition/default.asp
- Hoa TTC, Bong BB, Huq E, Hodges TK (2002) Cre/lox site-specific recombination controls the excision of the transgene from the rice genome. Theor Appl Genet 104: 518-525 [DOI] [PubMed] [Google Scholar]
- Hoekema A, Roelvink PW, Hooykaas PJJ, Schilperoort RA (1984) Delivery of T-DNA from the Agrobacterium tumefaciens chromosome into plant cells. EMBO J 3: 2485-2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Ou B, Hampsch-Woodill M, Flanagan JA, Deemer EK (2002) Development and validation of oxygen radical absorbance capacity assay for lipophilicantioxidants using randomply methylated β-cyclodextrin as the solubility enhancer. Agric Food Chem 50: 1815-1821 [DOI] [PubMed] [Google Scholar]
- Isaacson T, Ronen G, Zamir D, Hirschberg J (2002) Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of β-carotene and xanthophylls in plants. Plant Cell 14: 332-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononov ME, Bassuner B, Gelvin SB (1997) Integration of T-DNA binary vector “backbone” sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J 11: 945-957 [DOI] [PubMed] [Google Scholar]
- Lucca P, Ye X, Potrykus I (2001) Effective selection and regeneration of transgenic rice plants with mannose as selective agent. Mol Breed 7: 43-49 [Google Scholar]
- McCouch SR, Kochehert G, Yu ZH, Wanh ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 78: 815-829 [DOI] [PubMed] [Google Scholar]
- Misawa N, Yamano S, Linden H, de Felipe MR, Lucas M, Ikenaga H, Sandmann G (1993) Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J 4: 833-840 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog K (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473-497 [Google Scholar]
- Park H, Kreunen SS, Cuttriss AJ, DellaPenna D, Pogson BJ (2002) Identification of the carotenoid isomerase provides insight into carotenoid biosynthesis, prolamellar body formation, and photomorphogenesis. Plant Cell 14: 321-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond TM, Gentleman S, Duncan T, Yu S, Wiggert B, Gantt E, Cunningham FX Jr (2001) Identification, expression, and substrate specificity of a mammalian beta-carotene 15, 15′-dioxygenase. J Biol Chem 276: 6560-6565 [DOI] [PubMed] [Google Scholar]
- Roberts C, Rajagopal S, Smith LM, Nguyen TA, Yang W, Nugrohu S, Ravi KS, Vijayachandra K, Harcourt RL, Dransfield L et al. (2000) A comprehensive set of modular vectors for advanced manipulations and efficient transformation of plant GenBank accession numbers: pCAMBIA 1200 (AF234292), 1380 (AF234301); 1390 (AF234307). http://www.cambia.org/main/r_et_vman.htm
- Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM (2000) Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol 18: 666-669 [DOI] [PubMed] [Google Scholar]
- Underwood BA (2000) Overcoming micronutrient deficiencies in developing countries: Is there a role for agriculture? In N.S. Scimshaw, ed, Food and Nutrition Bulletin, Vol 21, No 4. United Nations University Press, Tokyo, Japan, pp 356-360. http://www.unu.edu/unupress/food/fnb21-4.pdf
- UNICEF (2000) The State of World's Children: Statistical Tables. UNICEF, New York
- von Lintig J, Vogt K (2000) Filling the gap in vitamin A research: molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem 275: 11915-11920 [DOI] [PubMed] [Google Scholar]
- Xu Z, Godber JS (1999) Purification and identification of components of γ-oryzanol in rice bran oil. J Agric Food Chem: 47: 2724-2728 [DOI] [PubMed] [Google Scholar]
- Wünn J, Klöti A, Burhardt PK, Ghosh Biswas GC, Launis K, Iglesias VA, Potrykus I (1996) Transgenic indica rice breeding line IR58 expressing a synthetic cryIA(b) gene from Bacillus thuringiensis provides effective insect pest control. Bio/Technology 14: 171-176 [DOI] [PubMed] [Google Scholar]
- Xu Z, Hua N, Godber JS (2001) antioxidant activity of tocopherols, tocotrienols, and γ-oryzanol components from rice bran against cholesterol oxidation allererated by 2,2′-azobis(2-methylpropronamidine)dihydrochloride. J Agric Food Chem: 49: 2077-2081 [DOI] [PubMed] [Google Scholar]
- Yan W, Jang GF, Haeseleer F, Esumi N, Chang J, Kerrigan M, Campochiaro M, Campochiaro P, Palczewski K, Zack DJ (2001) Cloning and characterization of a human beta, beta-carotene-15,15′-dioxygenase that is highly expressed in the retinal pigment epithelium. Genomics 72: 193-202 [DOI] [PubMed] [Google Scholar]
- Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid free) rice endosperm. Science 287: 303-305 [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Qaim M (2002) Projecting the benefits of Golden Rice in the Philippines. ZEF Discussion Papers on Development Policy No. 51, September 2002. ISSN: 1436-9931. Center for Development Research, Bonn. http://www.zef.de/download/zef_dp/zef_dp51.PDF