Abstract
We have cloned a SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) gene from Medicago truncatula (MtSERK1) and examined its expression in culture using real time PCR. In the presence of the auxin 1-naphthaleneacetic acid (NAA) alone, root differentiation occurs from the proliferating calli in both the cultured highly embryogenic seed line (2HA) and a low to nonembryogenic seed line (M. truncatula cv Jemalong). Auxin stimulated MtSERK1 expression in both 2HA and M. truncatula cv Jemalong. Embryo induction in proliferating calli requires a cytokinin in M. truncatula and unlike root formation is substantively induced in 2HA, not M. truncatula cv Jemalong. On embryo induction medium containing NAA and the cytokinin 6-benzylaminopurine (BAP), expression of MtSERK1 is elevated within 2 d of initiation of culture in both M. truncatula cv Jemalong and 2HA. However, MtSERK1 expression is much higher when both NAA and BAP are in the medium. BAP potentiates the NAA induction because MtSERK1 expression is not up-regulated by BAP alone. The 2HA genotype is able to increase its embryo formation because of the way it responds to cytokinin, but not because of the cytokinin effect on MtSERK1. Although the studies with M. truncatula indicate that somatic embryogenesis is associated with high SERK expression, auxin alone does not induce somatic embryogenesis as in carrot (Daucus carota) and Arabidopsis. Auxin in M. truncatula induces roots, and there is a clear up-regulation of MtSERK1. Although our analyses suggest that MtSERK1 is orthologous to AtSERK1, which in Arabidopsis is involved in somatic embryogenesis, in legumes, MtSERK1 may have a broader role in morphogenesis in cultured tissue rather than being specific to somatic embryogenesis.
The development of somatic embryos in vitro was first demonstrated by both Reinert (1958) and Steward et al. (1958). Research into somatic embryogenesis intensified as plant regeneration in vitro came to be widely used in transformation. Some general principles for the induction of somatic embryogenesis have emerged. Usually, an auxin is required to induce somatic embryogenesis, and subsequent auxin withdrawal or lowering of the auxin concentration is required for embryo maturation (Dudits et al., 1991). In addition to the hormone regime, key variables are explant type and developmental stage, nutrition regime, and genotype (Ammirato, 1983; Rose et al., 1999). However, the molecular mechanisms involved in the induction of somatic embryogenesis are not well understood.
There are many reports of genes involved in the regulation of the later stages of somatic embryogenesis (Zimmerman, 1993; Schmidt et al., 1997), but much less is known of the induction process. Ectopic expression of a number of different genes in Arabidopsis has been shown to induce vegetative cells to become embryogenic. Ectopic expression of the genes LEAFY COTYLEDON 1 (LEC1; Lotan et al., 1998) and LEAFY COTYLEDON 2 (LEC2; Stone et al., 2001), BABY BOOM (BBM; Boutilier et al., 2002), and WUSCHEL (WUS; Zuo et al., 2002) in Arabidopsis cause spontaneous formation of somatic embryos on intact plants or explants. The LEC genes (Lotan et al., 1998; Stone et al., 2001), the BBM gene (Boutilier et al., 2002), and the WUS gene (Zuo et al., 2002) encode transcription factors. To date, the most substantive work has been carried out on the SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) gene, a Leu-rich repeat, receptor-like kinase (LRR-RLK), which has been shown to be a marker of embryogenic cells in carrot (Daucus carota), Dactylis glomerata, and Arabidopsis (Schmidt et al., 1997; Somleva et al., 2000; Hecht et al., 2001). The first SERK gene (DcSERK) was identified in carrot (Schmidt et al., 1997). DcSERK expression is first detectable in embryogenic cultures after 7 d of growth in the presence of 2,4-D and is expressed in cells that develop into somatic embryos. Expression continues until the 100-cell stage of the globular somatic embryo. No expression of DcSERK is detected in nonembryogenic cultures. DcSERK is also expressed in zygotic embryos, again only up to the early globular stage (Schmidt et al., 1997). A correlation between SERK expression and somatic embryogenesis was also demonstrated in cultured tissue of D. glomerata (Somleva et al., 2000).
The most extensively characterized SERK gene is AtSERK1 from Arabidopsis. Four other SERK genes have also been identified in Arabidopsis (AtSERK 2-5) based on their sequence similarity to AtSERK1 (Baudino et al., 2001), indicating that in Arabidopsis, SERK genes exist as a gene family. Families of SERK genes exist in other plants with three SERK genes identified in maize (Zea mays; Baudino et al., 2001) and four partial sequences for Helianthus sp. SERK genes on the GenBank database (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/). The identification of SERK genes in maize and also in D. glomerata (Somleva et al., 2000) indicates that SERK functions in monocotyledonous plants as well as dicotyledonous plants. AtSERK1 is expressed in ovules before fertilization and during early embryo development until the heart stage. A low level of AtSERK1 expression was also observed in vascular tissue of seedlings and mature plants (Hecht et al., 2001). In culture, AtSERK1 is expressed in embryogenic structures that form on callus tissue. Nonembryogenic cultures obtained from the embryogenic cultures through selective subculture gradually lost AtSERK1 expression (Hecht et al., 2001).
A SERK-induced effect on somatic embryogenesis has been demonstrated (Hecht et al., 2001). Plants transformed with the AtSERK1 gene, under the control of the cauliflower mosaic virus 35S promoter showed a marked increase in embryogenic capacity over wild-type cultures, but a lower embryogenic capacity than cultures from the altered meristem program (amp1) mutant. The amp1 (or primordia timing [pt]) mutant, shows an enlargement of the shoot apical meristem and readily forms embryogenic cultures when seedlings are germinated in the presence of 2,4-D (Mordhorst et al., 1998). In these cultures, the AtSERK1 transcript is expressed at a very low level in cultures from wild-type plants but is expressed at high levels in cultures from AtSERK1-transformed plants and amp1 mutants, with expression higher in embryogenic cultures than in nonembryogenic cultures.
The general structure of SERK proteins places them in the LRR-RLK family of plant protein kinases (Walker, 1994). A putative signal peptide is present, then the extracellular domain, consisting of a putative Leu zipper (ZIP), five LRRs, and then a Pro-rich domain (SPP; Hecht et al., 2001). Adjacent to the SPP domain is a single transmembrane domain, then the intracellular kinase domain, and a C-terminal Leu-rich domain that may be involved in protein-protein interactions (Schmidt et al., 1997).
Medicago truncatula Gaertn. (barrel medic) has become a model legume for the study of plant-microbe interactions (Cook, 1999; Frugoli and Harris, 2001), because it has the advantages of being a diploid, autogamous annual species with a small genome (Blondon et al., 1994). Regeneration protocols established usually supply both auxin and cytokinin in the embryo induction medium (Nolan et al., 1989; Chabaud et al., 1996; Nolan and Rose, 1998; das Neves et al., 1999; Iantcheva et al., 2001). The highly embryogenic seed line of M. truncatula, Jemalong 2HA (2HA) was developed from M. truncatula cv Jemalong (Nolan et al., 1989; Rose et al., 1999) and so 2HA can be considered to be an isogenic, highly embryogenic mutant of M. truncatula cv Jemalong.
In this study, we have used 2HA and M. truncatula cv Jemalong to study expression of a SERK gene (MtSERK1) in response to auxin and cytokinin in cultured tissue, with the view that an understanding of this system may provide some insight into the regulation of somatic embryogenesis and the recalcitrant nature of legume regeneration. Both M. truncatula cv Jemalong and 2HA cultures incubated with auxin alone produce numerous roots but not embryos. Auxin alone, but not cytokinin alone, stimulates MtSERK1 expression in both M. truncatula cv Jemalong and 2HA. When cytokinin is supplied in conjunction with auxin, the level of expression of MtSERK1 is synergistically increased in comparison with the up-regulation on auxin alone. Under these conditions, 2HA explants form highly embryogenic callus cultures. The presence of cytokinin is essential for embryo formation. Data obtained in M. truncatula suggest MtSERK1 expression is not specifically associated with somatic embryogenesis and may play a broader role in morphogenesis in cultured tissue of legumes.
RESULTS
The MtSERK1 Sequence
The M. truncatula SERK gene (MtSERK1) was cloned from 2-week-cultured tissue from the highly embryogenic seed line, 2HA. The cloning strategy used degenerate primers from the DcSERK cDNA sequence followed by 3′-RACE and a modified 5′-RACE procedure as described. The sequence was then verified by obtaining a full-length cDNA in a single PCR reaction and resequencing. The MtSERK1 cDNA was then isolated from callus cultures of the M. truncatula cv Jemalong seed line, amplified in a single PCR reaction, and sequenced. The M. truncatula cv Jemalong sequence was identical to the 2HA sequence.
The MtSERK1 mRNA sequence obtained (GenBank accession no. AY162176) is 2,392 bp in length, with a predicted amino acid sequence of 627 amino acids, a calculated molecular mass of 69.125 kD, and a predicted pI of 5.56. On a nucleotide level, the MtSERK1 mRNA sequence showed the highest identity with AtSERK1 (59%), ZmSERK2 (58%), and AtSERK2 (57%; GenBank accession nos. A67827, AJ277703, and AF384969, respectively). Amino acid similarity is 92% with AtSERK1, 93% with AtSERK2, and 88% with both ZmSERK1 and ZmSERK2 (GenBank protein identification nos. CAB42254, AAK68073, CAC37638, and CAC37639).
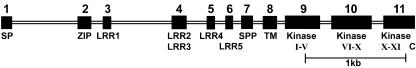
The amino acid alignment is shown in Figure 1, and it is clear that MtSERK1 aligns closely with SERK genes from other species. Furthermore, genomic sequencing of MtSERK1 (GenBank accession no. AY162177) shows that it contains 11 exons (Fig. 2), with the conserved exon/intron structure identified in other SERK genes, as well as the tendency for different domains of the protein to be encoded by separate exons (Fig. 2; Baudino et al., 2001). More detailed analysis using PSORT (Nakai and Kanehisa, 1992) for definition of a signal peptide and Scan Prosite (Hofmann et al., 1999) for identity of other specific regions identified regions characteristic of SERK protein structure.
Figure 1.
Alignment of the deduced amino acid sequence of MtSERK1 with AtSERK1 from Arabidopsis, ZmSERK1 from maize, and DcSERK from carrot. GenBank protein identification numbers: MtSERK1, AAN64293; AtSERK1, CAB42254; ZmSERK1, CAC37638; and DcSERK, AAB61708.
Figure 2.
Intron/exon structure of MtSERK1 from ATG start codon to stop codon. Large rectangles are exons (1-11). Small rectangles are introns. Specific areas of the protein coded by each exon are shown beneath rectangles. SP, Signal peptide; ZIP, Leu-Zipper; LRR, Leu-Rich Repeat; SPP, Pro-Rich Region; TM, transmembrane region; I-XI, kinase domains; C, C-terminal region.
A hydrophobic amino acid signal peptide sequence is present, with a possible cleavage site between positions 28 and 29. This is followed by a ZIP sequence. There are five LRRs, as defined by Hecht et al. (2001). Between the LRRs and the single transmembrane region is an SPP containing a repeated SPP motif, characteristic of SERK proteins (Hecht et al., 2001). The intracellular kinase domain contains the 11 subdomains of conserved amino acid sequences as described by Hanks et al. (1988). A putative protein kinase ATP-binding region signature is present at position 310 to 332 in the kinase domain. The 29-amino acid residue activation loop (A-loop) in subdomains VII and VIII, which was shown to be the active site of AtSERK1 (Shah et al., 2001c), is present, with 100% identity in MtSERK1 at position 448 to 577. MtSERK1 contains an active-site signature of Ser/Thr protein kinases between residues 427 to 439 in subdomain VI. Following the kinase region is a Leu-rich C-terminal domain as described by Schmidt et al. (1997). There are six putative N-glycosylation sites, two in LRR2, two in LRR4, one in LRR5, and one in domain V of the kinase region.
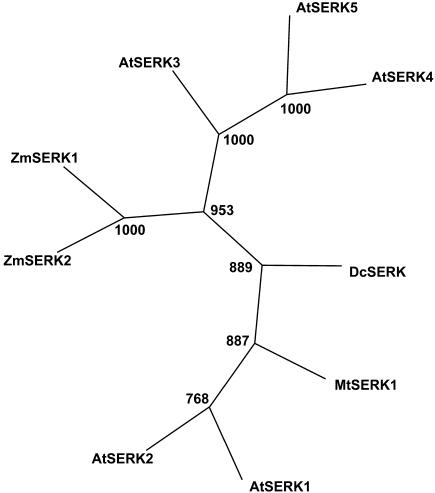
The amino acid sequence alignment and the protein structure of MtSERK1 are consistent with MtSERK1 being an ortholog of AtSERK1, which is involved in somatic embryogenesis in Arabidopsis. This is further supported by the phylogenetic analysis of amino acid sequences using an unrooted tree (Fig. 3). MtSERK1 is clustered together with At-SERK1 and AtSERK2. The only Arabidopsis SERK protein that has to date been directly implicated in somatic embryogenesis is AtSERK1, which has a high homology with AtSERK2.
Figure 3.
A phylogenetic tree of the known SERK full-length amino acid sequences. MtSERK1 is clustered together with AtSERK1 and AtSERK2. The numbers at the forks indicate the number of times the group consisting of the species that are distal to that fork occurred among the trees, out of 1,000 trees. SERK sequences used and GenBank protein identification numbers are: MtSERK1, AAN64293; AtSERK1, CAB42254; AtSERK2, AAK68073; AtSERK3, AAK68074; AtSERK4, AAD28318; AtSERK5, AAD28319; ZmSERK1, CAC37638; ZmSERK2, CAC37639; and DcSERK, AAB61708.
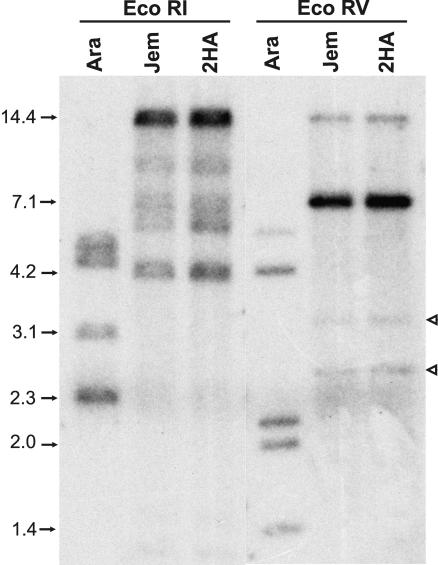
Given that AtSERK1 is part of a small gene family, the question arises as to whether there are other M. truncatula genes that are more closely related to At-SERK1. Southern hybridization using a sequence coding for part of the kinase domain and the C-terminal domain of MtSERK1 as a probe was carried out on 2HA and M. truncatula cv Jemalong DNA digested with EcoRI or EcoRV, neither of which has a restriction site within the MtSERK1 sequence. The results are consistent with five copies of the SERK gene, with five bands apparent in the EcoRI digest and four bands in the EcoRV digest (Fig. 4). The MtSERK1 probe also hybridized to Arabidopsis DNA giving four bands with the EcoRI digest and five bands with the EcoRV digest. There are five SERK genes in Arabidopsis, and these results indicate that this probe can detect all of these genes. Hybridization with a probe from the 3′-untranslated region of MtSERK1 indicated that the strongly hybridizing bands in Figure 4 (14.4 kb in the EcoRI digest and 7.1 kb in the EcoRV digest) are the bands corresponding to the MtSERK1 sequence.
Figure 4.
Southern analysis of SERK in M. truncatula cv Jemalong (Jem) and 2HA, and Arabidopsis (Ara). Genomic DNA from leaves was digested with EcoRI or EcoRV and probed with an 865-bp sequence containing the coding region for part of the kinase domain and the C-terminal domain from MtSERK1. Band patterns are identical in 2HA and M. truncatula cv Jemalong. The EcoRI digest shows five M. truncatula SERK hybridizing bands between 4.2 and 14.4 kb in size. The EcoRV digest shows four bands, two of which are faint (white arrowheads). The EcoRV digest indicates the five SERK genes in Arabidopsis. In the M. truncatula cv Jemalong and 2HA lanes, the 14.4-kb band in the EcoRI digest and the 7.1-kb band in the EcoRV digest correspond to the MtSERK1 genes.
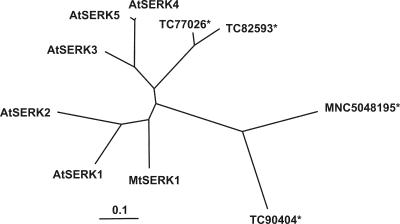
To obtain further information on this point, phylogenetic analysis was conducted using four partial mRNA sequences, which were selected by sequence similarity to AtSERK1 as other putative SERK genes in M. truncatula. Each of these sequences contained sequence for part of the kinase domain and the remainder of the coding region, which contains the C-terminal domain in SERK sequences. The C-terminal domain is considered to be a SERK-specific region (Baudino et al., 2001). Phylogenetic analyses of these sequences and the corresponding regions of MtSERK1 and AtSERK1-5 are shown in Figure 5. MtSERK1 is the M. truncatula sequence closest to AtSERK1. This again suggests that MtSERK1 and AtSERK1 are orthologs.
Figure 5.
A phylogenetic tree of four candidate M. truncatula SERK EST sequences (*) from The Institute for Genomic Research (TIGR) Medicago truncatula Gene Index (MtGI v7.0) and the CCGBMtDB2.0 databases and the aligning regions from MtSERK1 and AtSERK1-5. TC90404, TC77026, and TC82593 are sequences from TIGR MtGI. MNC5048195 is a sequence from CCGB-MtDB2.0, which matches the shorter sequence, TC89589, from TIGR MtGI. GenBank accession numbers of the other sequences are AY162176 (MtSERK1), A67827 (AtSERK1), AF384969 (AtSERK2), AF384970 (AtSERK3), locus F13J11.14 on AC006436 (AtSERK4), and AY094412 (AtSERK5). MtSERK1 is closer to AtSERK1 than the other M. truncatula sequences.
Expression Studies
High (2HA) and Low (M. truncatula cv Jemalong) Regenerators Incubated with Auxin and Cytokinin (1-Naphthaleneacetic Acid [NAA] + 6-benzylaminopurine [BAP])
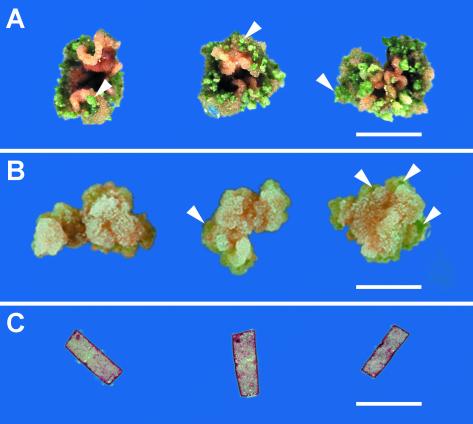
In the 2HA/M. truncatula cv Jemalong culture system used in this study, leaf explants rapidly form callus tissue in the presence of the hormones NAA and BAP. Initially, there are no visible differences between the responses of the 2HA and M. truncatula cv Jemalong seed lines. At 3 weeks, abscisic acid is routinely added to the culture medium in addition to NAA and BAP because this has been shown to increase the number of embryos that form (Nolan and Rose, 1998). After 3.5 to 5 weeks in culture, somatic embryos begin to emerge from the 2HA tissue, and this development continues over subsequent weeks, with the continual formation of new embryos (Fig. 6A). In contrast the M. truncatula cv Jemalong explants rarely form somatic embryos (Fig. 6B).
Figure 6.
A, 2HA calli at 8 weeks after culture in light. Numerous green somatic embryos (such as the three arrowed) have formed on the calli. B, M. truncatula cv Jemalong calli at 8 weeks after culture in light. Some green areas (arrows) of embryogenic-like tissue are visible on calli. C, 2HA leaf explants after 2 weeks of culture on medium containing no hormones. Bar = 1 cm.
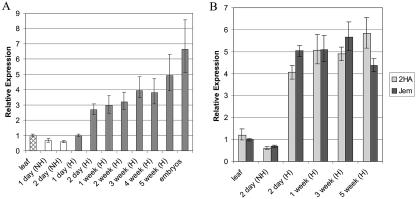
A time course of MtSERK1 expression in leaf cultures from the highly embryogenic 2HA seed line is shown in Figure 7A. In the presence of the hormones supplied in the embryo induction medium, there was a clear increase in MtSERK1 expression 2 d after culture initiation. The expression continued to slowly increase and remained elevated over the 5-week time period examined. The expression of MtSERK1 was also elevated in embryos of various stages, which were collected at 8 weeks. Tissue grown on medium without hormones showed a decrease in MtSERK1 expression within 2 d, indicating that the presence of hormones in the culture medium was inducing MtSERK1 expression (Fig. 7A). The presence of hormones was also essential for explant survival, because tissue grown without hormones became necrotic in appearance within 1 to 2 weeks of culture (Fig. 6C). The level of expression of MtSERK1 was measured in cultures from the low embryogenic seed line M. truncatula cv Jemalong and was compared with expression in the highly embryogenic 2HA (Fig. 7B). M. truncatula cv Jemalong cultures displayed the same level of MtSERK1 expression as 2HA cultures in both the presence and absence of hormones. Again, expression was stimulated at 2 d from the initiation of culture and remained high over the 5-week period examined. The reduced embryogenic capacity of M. truncatula cv Jemalong was not characterized by lower expression of MtSERK1. These trends were verified by using RNA extracted from two sets of culture experiments and by northern hybridization (data not shown). Although M. truncatula cv Jemalong calli infrequently form somatic embryos, when calli are cultured in the light, they will form small green areas, which appear embryogenic, but usually no further development occurs from these areas (our observations; Fig. 6B).
Figure 7.
Expression of MtSERK1, measured using real time PCR in cultured leaf explants from 2HA and M. truncatula cv Jemalong. Tissue grown on hormones (H) was cultured for 3 weeks on P4 10:4 and then transferred to P4 10:4:1. All expression is normalized to the expression of MtGAPDH. Each point represents the mean of three or four replicates with error bars representing the sd. A, Relative expression of MtSERK1 over time in cultured 2HA leaf explants. Gray bars represent tissue cultured in the presence of hormones (H); white bars, tissue cultured in the absence of hormones (NH). The starting explant tissue (leaf) is hatched. Relative expression is calibrated to MtSERK1 expression in 2HA leaf. B, Comparison of MtSERK1 expression in cultured leaf explants from 2HA and M. truncatula cv Jemalong plants over time under normal culture conditions (H) and in the absence of hormones (NH). Relative expression is calibrated to MtSERK1 expression in M. truncatula cv Jemalong leaf tissue.
High (2HA) and Low (M. truncatula cv Jemalong) Regenerators Incubated with Auxin (NAA) and Cytokinin (BAP) Alone
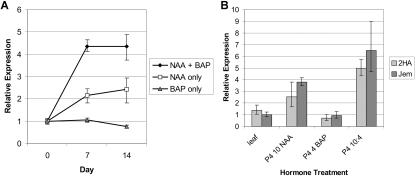
In the 2HA and M. truncatula cv Jemalong study, the increased MtSERK1 expression observed by 48 h was the result of the addition of auxin plus cytokinin. To further dissect the hormone response shown for NAA + BAP, the effects of NAA alone and BAP alone were investigated in 2HA. MtSERK1 expression was induced by NAA alone, but not by BAP alone (Fig. 8A). The level of MtSERK1 expression in the presence of BAP alone showed a slight decrease in expression over a 2-week culture period. However, NAA alone is not responsible for the total increase in MtSERK1 expression under normal culture conditions, because the response of tissue cultured with NAA alone was only about one-half of the response seen with tissue cultured in medium containing both NAA and BAP (Fig. 8A). This indicates that BAP acts synergistically with NAA to further increase MtSERK1 expression in culture.
Figure 8.
Expression of MtSERK1, measured using real time PCR in cultured leaf explants. All expression is normalized to the expression of MtGAPDH. Each point represents the mean of three or four replicates with error bars representing the sd. A, Relative expression of MtSERK1 in 2HA leaf explants cultured under varying hormone treatments for 2 weeks. Day 0, MtSERK1 expression in starting leaf explant tissue before culture. Relative expression is calibrated to MtSERK1 expression in 2HA leaf tissue. B, Comparison of MtSERK1 expression in 2HA and M. truncatula cv Jemalong leaf cultures after 2 weeks of culture under varying hormone treatments. Relative expression is calibrated to MtSERK1 expression in M. truncatula cv Jemalong leaf tissue.
The hormone-induced MtSERK1 expression shown for 2HA was also examined in M. truncatula cv Jemalong. As shown in Figure 8B, there was no difference between M. truncatula cv Jemalong and 2HA, with the MtSERK1 expression being auxin dependent.
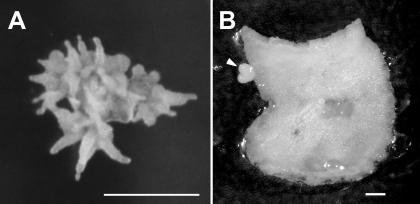
When cultured on NAA + BAP, before the appearance of embryos, explants from both 2HA and M. truncatula cv Jemalong proliferated to form callus tissue. When cultured on NAA alone, both 2HA and M. truncatula cv Jemalong leaf explants initially responded by forming callus tissue at a slower rate than observed in the presence of NAA + BAP. Root primordia begin to emerge from the callusing tissue of both seed lines after 2 weeks of culture. After 4 weeks, numerous roots are present (Fig. 9A). Explants from both seed lines appeared identical in their development. Tissue cultured on BAP alone did not show any notable callus formation. The explants enlarge, some curl, and some gradually thicken. After a period of 3.5 to 5 weeks, some of the 2HA explants developed embryo-like structures on the edge of explants (Fig. 9B). M. truncatula cv Jemalong tissue did not show any similar development, again emphasizing the increased propensity for totipotency in 2HA somatic cells.
Figure 9.
A, 2HA tissue showing extensive root formation after culture for 4 weeks on medium with 10 μm NAA only. Bar = 1 cm. B, A heart-shaped embryo (arrow) growing from the edge of 2HA tissue after 26 d of culture on medium containing 4 μm BAP. Bar = 1 mm.
The results here show that MtSERK1 expression is increased under conditions that produce a morphogenetic proliferating callus. Interestingly, unlike carrot and Arabidopsis, in both 2HA and M. truncatula cv Jemalong in the presence of NAA alone, root differentiation occurs in the proliferating calli. When the cytokinin BAP is combined with the auxin, in the case of 2HA but not M. truncatula cv Jemalong, high rates of somatic embryo differentiation occur in the proliferating calli. The expression of MtSERK1 in conditions favoring callus and somatic embryo formation (NAA + BAP) is double the expression in conditions favoring callus and root formation (NAA only). Treatment with BAP alone does not induce any substantial callus formation or MtSERK1 expression, although interestingly structures, which appear to be somatic embryos, develop on the edge of the 2HA tissue.
Expression Information Based on the M. truncatula Expressed Sequence Tag (EST) Database
Using the relation between cDNA libraries and their EST composition, it is possible to obtain additional information on the expression of MtSERK1. The MtSERK1 mRNA sequence corresponds to TC78763 on the TIGR MtGI database. TC78763 is partly made up of ESTs from a variety of cDNA libraries, with the rest of the sequence coming from MtSERK1 sequence on the National Center for Biotechnology Information database (GenBank Accession nos. AY162176 and AY162177). Table I lists the cDNA libraries that were the sources of these ESTs. From Table I, it can be seen that MtSERK1 is expressed throughout the plant with ESTs originating from libraries obtained from stem, leaf, root, and flower tissue.
Table I.
A list of the sources of ESTs belonging to MtSERK1 on the TIGR MtGI
The MtSERK1 mRNA sequence corresponds to the contig TC78763 on the TIGR MtGI database. Part of this contig was assembled from ESTs, which came from the seven cDNA libraries listed. These show that MtSERK1 is expressed in a broad range of tissues within the plant. Further information on the cDNA libraries is available from TIGR MtGI (http://www.tigr.org/tdb/tgi/mtgi/).
| cDNA Library | Description |
|---|---|
| DSIR | ESTs from roots after inoculation with Phytophthora medicaginis |
| DSIL | ESTs from leaves after inoculation with Colletotrichum trifolii |
| MHAM | ESTs from roots after colonization with Glomus versiforme |
| NF-FL Developing flower | ESTs from pooled flowers |
| NF-ST Developing stem | ESTs from a mixture of internodal stem segments |
| KV3 | ESTs from roots 72 h after inoculation with Rhizobium sp. |
| MtBA | ESTs from nitrogen-starved M. truncatula root tips |
DISCUSSION
In this study, a SERK gene (MtSERK1) has been cloned from cultured tissue of M. truncatula, and its expression has been investigated in root-forming, embryogenic, and nonembryogenic cultures under varying hormone regimes. This is the first report, to our knowledge, in legumes of cloning of a SERK gene and the study of its expression. Legumes are known for their recalcitrant nature in respect to plant regeneration and transformation. An understanding of the regulation of somatic embryogenesis in culture will assist in the development of more effective regeneration protocols, but the data from the M. truncatula system studied here also show that in vitro studies in legumes are valuable for studying differentiation.
The MtSERK1 Gene
The predicted amino acid sequence of MtSERK1, like other SERKs, indicates that it functions as an LRR-RLK (Walker, 1994). The plant RLKs resemble the receptor protein kinases (RPKs), which play an essential role in animal development. The model for RPK function is that the RPKs exist as monomers until binding of an extracellular signal molecule induces receptor dimerization. This brings the intracellular kinase domains of the individual monomers into close proximity, allowing transphosphorylation, which activates the kinase domains and causes the regulation of a cellular response (Becraft, 1998). The RLKs encoded by the CLAVATA (CLV) genes appear to act similarly in plant meristems (Clark et al., 1997; Rojo et al., 2002).
Of the SERK proteins identified to date, the Arabidopsis AtSERK1 has been most extensively studied, with elucidation of some of the roles played by the different regions of the protein (Hecht et al., 2001; Shah et al., 2001a, 2001c, 2002). The high sequence similarity of MtSERK1 with AtSERK1 (92%) suggests that the corresponding regions in MtSERK1 would play a similar role. The ZIP region has been shown to be involved in oligomerization of AtSERK1 proteins (Shah et al., 2001a), indicating that this region may play a role in dimerization. LRRs in general are involved in protein-protein interactions (Kobe and Deisenhofer, 1995) and are a candidate site for ligand binding. The LRRs of AtSERK1 are required for the correct targeting of the protein to the plasma membrane, with glycosylation of sites within this region probably targeting the protein to the plasma membrane (Shah et al., 2001a). Within the LRR region of both AtSERK1 and MtSERK1 are five putative N-glycosylation sites, two in LRR2, two in LRR4, and one in LRR5. The SPP domain between the LRRs and transmembrane region contains the SPP motif, which has been suggested to act like a hinge providing flexibility to the extracellular part of the receptor or as a region for interaction with the cell wall (Hecht et al., 2001). The single transmembrane region divides the extracellular domains described above, with the intracellular region containing the kinase and C-terminal domains. DcSERK and AtSERK1 have both been shown to be capable of functioning as kinases (Shah et al., 2001b, 2001c). In AtSERK1, two Thr residues lying within a putative A-loop were identified as important for both autophosphorylation and phosphorylation of artificial substrates (Shah et al., 2001c). The 100% identity of the corresponding region in MtSERK1 with the AtSERK1 A-loop indicates that this region in both proteins would serve the same function.
The similarity between MtSERK1 and AtSERK1 of the amino acid sequence and protein structure suggests that the MtSERK1 and AtSERK1 genes are orthologs. The phylogenies based on amino acid sequences (Fig. 3) and ESTs (Fig. 5) support the ortholog interpretation. Furthermore, MtSERK1 was isolated from 2HA cultures during the embryo initiation phase. We note that AtSERK1 and AtSERK2 are closely related and suggest that they are recent duplications, and it would be of interest to know whether they have similar or different functions.
Expression of MtSERK1 in High and Low Regenerators Incubated with Auxin (NAA) and Cytokinin (BAP)
There was no significant difference in MtSERK1 expression between the highly embryogenic 2HA and M. truncatula cv Jemalong, which shows little or no embryo formation. The tissue was cultured in our standard regeneration medium with NAA + BAP (Nolan and Rose, 1998). MtSERK1 expression was induced rapidly, within 2 d of culture, which was long before the first appearance of somatic embryos, which are first visible to the eye after 3.5 to 5 weeks. In closer correlation with the first increase in MtSERK1 expression was cell proliferation resulting in callus formation, which was evident after 1 week in both 2HA and M. truncatula cv Jemalong. There was no significant difference in the expression of MtSERK1 between 2HA and M. truncatula cv Jemalong cultures. These results differ from those obtained from carrot, D. glomerata, and Arabidopsis, where SERK expression was detected in embryogenic calli but not in nonembryogenic calli (Hecht et al., 2001; Schmidt et al., 1997; Somleva et al., 2000). However, in maize cultures, neither ZmSERK1 nor ZmSERK2 displayed an embryo-specific expression pattern with both genes expressed in both embryogenic and nonembryogenic cultures (Baudino et al., 2001). However, MtSERK1 could still be involved in the initiation of somatic embryogenesis in M. truncatula. It is possible that the embryogenic pathway is initiated in M. truncatula cv Jemalong cultures, but this pathway is blocked somewhere post-MtSERK1 expression. The formation of small green areas of embryogenic appearance when M. truncatula cv Jemalong calli are cultured in the light may be an indication of failed embryo development. Also, no histological differences can be detected between 2-week-old M. truncatula cv Jemalong and 2HA cultures (K.E. Nolan, R.R. Irwanto, and R.J. Rose, unpublished data). In D. glomerata cultures, there are many more SERK-expressing cells than the number of somatic embryos that form, suggesting that embryo development is arrested after SERK expression in some of these cells (Somleva et al., 2000).
Expression of MtSERK1 in High and Low Regenerators in the Presence of Auxin (NAA) and Cytokinin (BAP) Alone
Culture of tissue on either NAA or BAP alone showed that MtSERK1 expression is clearly induced by the auxin NAA and not induced by the cytokinin BAP. However, when both hormones are present, as in the usual regeneration procedure, the expression levels are synergistically increased to almost double that on NAA alone. In the treatments showing increased MtSERK1 expression, there are both cell proliferation and differentiation, i.e. morphogenetic callus. The highest rate of cell proliferation occurs on medium containing both hormones, which also has the highest expression of MtSERK1. Tissue on BAP alone, which exhibits little or no callus formation, has the lowest expression of MtSERK1. NAA alone, which has callus production in between the other two treatments, shows a corresponding rate of MtSERK1 expression between the other two.
Differentiation with NAA is in the form of roots with both 2HA and M. truncatula cv Jemalong. This is quite different from carrot and Arabidopsis (Schmidt et al., 1997; Hecht et al., 2001). Therefore both somatic embryos and roots develop in conditions of increased MtSERK1 expression in M. truncatula. It is worthy of note that 2HA calli producing roots with a unipolar meristem have one-half the MtSERK1 expression of calli producing the bipolar embryo. This is suggestive of a broader role of SERK in organ formation, rather than embryogenesis alone in M. truncatula.
A correlation between SERK and embryogenesis has been well established (Hecht et al., 2001; Schmidt et al., 1997; Somleva et al., 2000), but a possible role of SERK during other forms of differentiation, to our knowledge, has not been studied. All of the reports on SERK expression during culture to date have been in the context of auxin-induced somatic embryogenesis. It was noted in D. glomerata cultures that induction of root formation occurred within the first 4 d of culture, but the first SERK-expressing cells were not visible, using in situ hybridization, for 5 d (Somleva et al., 2000). These results appear to indicate that SERK is not expressed during rhizogenesis in these cultures. However, it is possible that SERK is expressed and that the method of detection used was not sensitive enough to identify a lower level of expression, such as the lower level of MtSERK1 expression observed during rhizogenesis in this study. Alternatively, these results may indicate a different role played by SERK in legumes.
Differentiation, in the form of direct somatic embryogenesis, also occurred in 2HA but not M. truncatula cv Jemalong when cultured on BAP alone. In this case, somatic embryos are forming in the treatment that does not show increased MtSERK1 expression. The induction of somatic embryogenesis by cytokinin alone is very unusual, but it has been previously reported in M. truncatula (Iantcheva et al., 1999). There appears to be a varied response in the cytokinin induction of somatic embryogenesis, because we previously cultured 2HA tissue in the presence of BAP (as in this study), and there was no development of somatic embryos (Nolan and Rose, 1998). It is worth noting here that the 2HA tissue used in the earlier study was at a time when the 2HA seed line was in the process of being developed, and so it was not as highly selected as the tissue used in this study. If MtSERK1 is playing a role in somatic embryo induction, it is either doing so only in the context of somatic embryogenesis induced in an environment of callus formation, or the cells that express MtSERK1 during direct somatic embryogenesis are so few in number that the level of MtSERK1 mRNA in the total amount of tissue is relatively low. Conversely, the structures that form on 2HA tissue in the presence of cytokinin alone may be shoots, not embryos. However, this is not likely because there is histological evidence (Iantcheva et al., 1999) and morphological evidence (Fig. 9B) that the structures that form on cytokinin alone are somatic embryos. SERK expression during direct somatic embryogenesis has been shown to occur in D. glomerata cultures. During the early stages of auxin-induced somatic embryogenesis in these cultures, a single SERK-expressing cell arises from a dividing mesophyll cell (Somleva et al., 2000).
The Mechanism of High Regenerability in 2HA
The data obtained from the current study have shown that both M. truncatula cv Jemalong and 2HA cultures respond in the same way to auxin by forming roots and by having the same pattern of up-regulation of MtSERK1 expression (Fig. 8B). Both 2HA and M. truncatula cv Jemalong have the same MtSERK1 sequence. The dramatic difference in 2HA regenerability appears in the presence of cytokinin when it is added to the auxin-containing medium (Figs. 6, A and B). While cytokinin potentiates the MtSERK1 expression (Fig. 8), it does this in both M. truncatula cv Jemalong and 2HA. The 2HA genotype is able to respond to cytokinin in a way that M. truncatula cv Jemalong cannot, but it is most likely downstream of MtSERK1 expression. We have also shown that cytokinin alone will stimulate direct somatic embryogenesis in 2HA but not M. truncatula cv Jemalong (Fig. 9B). Cytokinin is essential for somatic embryogenesis in M. truncatula (Nolan and Rose, 1998).
The Role of SERK in Development
SERK genes have been shown to be specifically expressed during embryo induction and early somatic embryogenesis in carrot, D. glomerata, and Arabidopsis (Schmidt et al., 1997; Somleva et al., 2000; Hecht et al., 2001) and are considered to be a marker for cells competent to form somatic embryos (Schmidt et al., 1997). Overexpression of AtSERK1 has been shown to induce somatic embryo formation in Arabidopsis cultures (Hecht et al., 2001), supporting the view that SERK plays a role in conferring embryogenic competence in culture. Recently, it was also demonstrated that ectopic expression of the BBM and WUS genes could initiate somatic embryogenesis in Arabidopsis tissues without any external plant hormones (Boutilier et al., 2002; Zuo et al., 2002). BBM is a transcription factor, but little is known about the role it plays (Boutilier et al., 2002). WUS is a central regulator of stem cell fate in Arabidopsis shoot and floral meristems (Mayer et al., 1998). WUS expression is negatively regulated by the CLV genes, and together, the WUS/CLV system interacts to maintain meristem homeostasis (Schoof et al., 2000). It is unclear whether WUS and AtSERK1 act in the same pathway or in different pathways. Under the embryogenic conditions of ectopic WUS expression in Arabidopsis, there were no detectable changes in AtSERK1 expression. However, this may be because AtSERK1 is acting upstream of WUS (Zuo et al., 2002). It appears that another negative feedback loop may operate between AtSERK1 and AMP1, with AMP1 acting to suppress the expression of AtSERK1 after germination (Hecht et al., 2001). AMP1 encodes a putative Glu carboxypeptidase, which is thought to play a role in cleaving small signaling peptides and other molecules (Helliwell et al., 2001), suggesting that AMP1 may play a role in regulating the ligand of AtSERK1. The theory that CLV and AMP1 are involved in suppressing embryogenic pathways is supported by the fact that mutations in these genes enhance the ability of germinating Arabidopsis seedlings to form embryogenic cultures (Mordhorst et al., 1998).
The data that we have obtained from M. truncatula show that somatic embryogenesis can be associated with high rates of MtSERK1 expression. However, the results obtained here also raise the possibility that the connection between SERK and somatic embryogenesis in legumes differs from other species. This putative difference in the role of SERK in M. truncatula may provide some insight into the recalcitrance of this species, which requires cytokinin and a specific genotype for somatic embryogenesis. The auxin data clearly suggest that in M. truncatula up-regulation of MtSERK1 can also be associated with root growth. This is of particular interest given the regulation by auxin of lateral root formation (Larkin et al., 1996; Casimiro et al., 2001). The data overall suggest that MtSERK1 may play a broader role in morphogenesis in cultured tissue rather than being specific to somatic embryogenesis. M. truncatula cv Jemalong, with its ability to produce roots in culture but rarely somatic embryos, and 2HA, with its ability to produce roots or somatic embryos in culture, provide a useful system to further investigate signaling in hormone-induced differentiation. Studies with maize and the MtSERK1 EST expression data (Table I) also support the idea that SERK genes closely related to AtSERK1 and AtSERK2 have a broader developmental role than embryogenesis alone (Baudino et al., 2001). In the case of legumes, the discovery of the involvement of a CLV1-like gene in the regulation of nodule number (Krusell et al., 2002; Nishimura et al., 2002; Searle et al., 2003) points to the subtle way related receptor kinases can influence development. The full understanding of the function of the member genes of the SERK gene family in legumes should provide novel insights into legume and plant development and their precise role in somatic embryogenesis.
MATERIALS AND METHODS
Tissue Culture and RNA Extraction
Tissue cultures originated from Medicago truncatula plants from either the highly regenerable seed line 2HA (Rose et al., 1999) or M. truncatula cv Jemalong. Seeds of M. truncatula cv Jemalong were obtained from the National Medicago Collection (Northfield Research Laboratories, Adelaide, South Australia). The youngest expanded leaves on individual shoots were selected for culture. Leaves were sterilized for 30 s in 70% (v/v) ethanol and then for 10 min in White King bleach diluted 1 in 8 in distilled water. This was followed by two rinses in sterile distilled water. Petioles were removed to separate each foliole, and the edges of the foliole were trimmed off to form a rectangular piece of tissue with the midvein running down the center. Cuts were made perpendicular to the midvein to obtain between two and four pieces of tissue measuring approximately 2 × 4 mm. Leaf pieces were plated with abaxial side down in 9-cm plastic petri dishes containing approximately 20 mL of agar medium. The basal medium used was P4, which is based on Gamborg's B5 medium (Gamborg et al., 1968) as described by Thomas et al. (1990). In the usual culture procedure, leaf explants were plated onto P4 medium containing 10 μm NAA and 4 μm BAP (P4 10:4) for 3 weeks and then transferred to P4 medium with 10 μm NAA, 4 μm BAP, and 1 μm abscisic acid (P4 10:4:1) with subculture to fresh P4 10:4:1 medium every 4 weeks. Cultures were incubated in the dark. This culture procedure has been optimized to maximize embryo number and quality in 2HA cultures (Nolan and Rose, 1998). For some experiments, tissue was cultured on P4 medium with various hormone treatments as described in the text. RNA extractions were carried out using the RNeasy Plant Mini Kit (Qiagen, Clifton Hill, Victoria, Australia) according to the manufacturer's instructions.
Obtaining the MtSERK1 Transcript
An initial 680-bp fragment of the MtSERK1 transcript was obtained by degenerate PCR of cDNA from 2-week-old 2HA callus tissue. Degenerate primers were derived from the amino acid sequences at the sites of the carrot (Daucus carota) SERK-specific primers of Schmidt et al. (1997). The degenerate primer sequences were 5′-ATGGCNTTYGCNTGGTGG-3′ and 5′-YTCNGANGAYTTNCCNGT-3′, where N = A, T, G, or C, and Y = T or C. The PCR products were cloned into the PCR Script Amp SK(+) cloning vector (Stratagene, La Jolla, CA) and electroporated into Epicurian Coli XL1-Blue MRF′ competent cells (Stratagene). Colonies were lysed and bound to positively charged nylon filters (Sambrook and Russell, 2001) and screened by three rounds of colony hybridization using a carrot SERK probe made from cDNA from carrot callus cultures with the primers described by Schmidt et al. (1997). The 3′ end of MtSERK1 was obtained using 3′-RACE of cDNA from 2-week-old 2HA callus cultures. cDNA was synthesized using the 3′-RACE Adaptor primer (Invitrogen, Mount Waverley, Victoria, Australia) and the Superscript Preamplification System for First Strand cDNA synthesis kit (Invitrogen). Primers used for the first RACE PCR reaction were the gene-specific primer SF1, 5′-TCACTGGTTGCTGTCAAAAGA-3′, and Abridged Universal Amplification Primer (Invitrogen). The gene-specific primer for the second nested PCR reaction was SF4, 5′-GGTGGGGAGCTTCAGTTTCAG-3′. 5′-RACE of MtSERK1 was conducted using the 5′-RACE System for Rapid Amplification of cDNA ends, v2 (Invitrogen). Reverse transcription was carried out on RNA from 2-week-old 2HA callus cultures, using the gene-specific primer SR1, 5′-AATCCAAAATCCCCCACAA-3′. The nested gene-specific primers used for subsequent PCR reactions were SR2, 5′-TGGCCAATCTAGTGGTTCTTGAT-3′, and SR8, 5′-CAGTGAACCGTCAGCCAAAC-3′. 5′-RACE gave 361 bp more of the 5′ sequence of the gene but not the entire 5′ end. More of the 5′ end was obtained by degenerate PCR using a primer designed by Baudino et al. (2001), which is targeted to the ZIP region. The remainder of the sequence was obtained from an EST (TC48098), which was on an earlier version of the TIGR MtGI (http://www.tigr.org/tdb/tgi/mtgi/). The current version of the TIGR MtGI (v7.0) contains the full-length MtSERK1 mRNA sequence. The full-length cDNA from 2HA callus cultures and M. truncatula cv Jemalong callus cultures was amplified in one PCR reaction using the primers 5′-TTGGGGGTTAGGGTTTATG-3′ and 5′-CAAGGAACCAAAAGAAGAAGA-3′ and was sequenced. The corresponding genomic DNA was amplified from 2HA leaf tissue and was sequenced.
Sequence Comparison
Database searches were performed at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) using the BLAST network service (Altschul et al., 1997) or at the TIGR MtGI (http://www.tigr.org/tdb/tgi/mtgi/). Sequence alignments and comparisons were performed using ClustalW (accurate) on BioManager by ANGIS (http://www.angis.org.au). Protein predictions were performed using PSORT (http://psort.nibb.ac.jp/; Nakai and Kanehisa, 1992) and Scan Prosite (http://www.expasy.org/prosite/; Hofmann et al., 1999).
Phylogenetic Analysis
Phylogenetic analyses of the full-length amino acid sequences of MtSERK1, AtSERK1-5, ZmSERK1, ZmSERK2, and DcSERK were performed on BioManager by ANGIS using the programs ClustalW (fast) (Thompson et al., 1994), Seqboot, Prodist, Fitch, and Consense (Felsenstein, 1989). Trees were drawn using Tree View (v1.6.6; Page, 1996).
To find other putative SERK mRNA sequences in M. truncatula, a BLASTN search of the TIGR MtGI database was conducted with the AtSERK1 mRNA sequence (GenBank accession no. AF384969). The top hit was the MtSERK1 sequence (TC78763). The next four highest matches (TC77026, TC90404, TC82593, and TC89589) were each aligned near the 3′ end of the gene. These four (non-MtSERK1) sequences were chosen as the other most likely candidates on the TIGR MtGI database to be the other SERK genes in M. truncatula, with each having an E value of ≤3e-52. More sequence information on one of these sequences was found by checking the M. truncatula Centre for Computational Genomics and Bioinformatics (CCGBMtDB2.0) database (http://www.medicago.org/MtDB/; Lambin et al., 2003). TC89589 was part of a larger sequence (MNC5048195) from CCBGMtDB2.0. This new sequence information was added to the sequences obtained from TIGR MtGI. These sequences and the mRNA sequences from MtSERK1 and AtSERK1-5 were aligned using ClustalW (accurate). A region of between 814 and 832 bp in length where all of these sequences aligned was used for phylogenetic analysis. Phylogenetic analysis was performed on BioManager by ANGIS using the maximum likelihood method, DNAML (Felsenstein, 1989). Trees were drawn using Tree View (v1.6.6; Page, 1996).
Southern-Blot Analysis
To investigate whether SERK genes in addition to MtSERK1 were present in M. truncatula, total DNA was probed with an 865-bp MtSERK1 fragment using Southern blotting and hybridization. DNA was extracted from fresh tissue by the cetyl-trimethyl-ammonium bromide method of Murray and Thompson (1980). The Southern blotting and hybridization were essentially according to Sambrook and Russell (2001). DNA fragments from EcoR1 and EcoRV restrictions were transferred to positively charged nylon membranes (Amersham Biosciences, Sydney) for hybridization to the probe labeled with [α-32P]dCTP using a Multiprime DNA labeling system (Amersham Biosciences). The probe was made by PCR using the primers 5′-TCACTGGTTGCTGTCAAAAGA-3′ and 5′-AATCCAAAATCCCCCACAA-3′. Hybridization was carried out overnight in a mini hybridization oven (Hybaid, Ashford, UK) at 65°C. The membrane was washed with gentle shaking twice for 30 min in buffer 1 (1 mm EDTA, 40 mm NaHPO4, and 5% [w/v] SDS, pH 7.2) at 65°C. The membrane was then washed twice in buffer 2 (1 mm EDTA, 40 mm NaHPO4, and 5% [w/v] SDS, pH 7.2) for 30 min at 65°C. After washing, the membrane was blotted between Whatman 3 MM paper (Whatman International, Maidstone, UK) and autoradiographed overnight at -80°C. The MtSERK1 probe was from the 3′ end of the coding region containing part of the kinase domain and the C-terminal domain. Another hybridization was conducted as above using a 165-bp probe made from the 3′-untranslated region of the MtSERK1 gene. The probe was made by PCR using the primers 5′-TGATGAAAAGAAAAATGAATGGAA-3′ and 5′-CGACCCCTATGCACAAATGTA-3′. Neither probe contained an EcoRI or EcoRV restriction site.
Expression Studies
The relative expression of MtSERK1 was measured using real time PCR on the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Scoresby, Victoria, Australia), using a Taq-Man probe and primers (Applied Biosystems). A gene coding for a M. truncatula glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as the endogenous control. To prevent amplification of any contaminating genomic DNA, the Taq-Man probes for each of MtSERK1 and MtGAPDH were designed over an exon/intron boundary in the cDNA sequence. A suitable GAPDH mRNA sequence (TC85222) was obtained from the TIGR MtGI database, and genomic DNA from this region was amplified by PCR and sequenced to identify exon/intron boundaries. Taq-Man probe and primers were designed using Primer Express software (Applied Biosystems). The primer and probe sequences used were: SERK probe, 5′-FAM-TGCCTCCTGTTTAAGAGAGCGTCCTCC-TAMRA-3′; SERK forward primer, 5′-TCCCTACATGGCTAATGGAAGTG-3′; SERK reverse primer, 5′-TGGCCAATCTAGTGGTTCTTGAT-3′; GAPDH MGB probe, 5′-VIC-CCACCCTTCAAATGA-3′; GAPDH forward primer, 5′-ACAAACATGGGAGCATCCTTACTAG-3′; and GAPDH reverse primer, 5′-GTTTTTACCGACAAGGACAAAGCT-3′. Real time PCR results were analyzed using the Comparative CT method with appropriate validation experiments performed beforehand (Applied Biosystems, User Bulletin #2, http://home.appliedbiosystems.com/).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammirato PV (1983) Embryogenesis. In DA Evans, WR Sharp, PV Ammirato, Y Yamada, eds, Handbook of Plant Cell Culture, Vol 1. Techniques in Propagation and Breeding. MacMillan, New York, pp 82-123
- Baudino S, Hansen S, Brettschneider R, Hecht VRG, Dresselhaus T, Lorz H, Dumas C, Rogowsky PM (2001) Molecular characterisation of two novel maize LRR receptor-like kinases, which belong to the SERK gene family. Planta 213: 1-10 [DOI] [PubMed] [Google Scholar]
- Becraft PW (1998) Receptor kinases in plant development. Trends Plant Sci 3: 384-388 [Google Scholar]
- Blondon F, Marie D, Brown S, Kondorosi A (1994) Genome size and base composition in Medicago sativa and M. truncatula species. Genome 37: 264-270 [DOI] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang LM, Hattori J, Liu CM, van Lammeren AAM, Miki BLA et al. (2002) Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14: 1737-1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalero RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inzé D, Sandberg G, Casero PJ et al. (2001) Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13: 843-852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, Larsonneau C, Marmouget C, Huguet T (1996) Transformation of barrel medic (Medicago truncatula Gaertn) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the Mtenod12 nodulin promoter fused to the Gus reporter gene. Plant Cell Rep 15: 305-310 [DOI] [PubMed] [Google Scholar]
- Clark SE, Williams RW, Meyerowitz EM (1997) The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89: 575-585 [DOI] [PubMed] [Google Scholar]
- Cook DR (1999) Medicago truncatula: a model in the making! Commentary. Curr Opin Plant Biol 2: 301-304 [DOI] [PubMed] [Google Scholar]
- das Neves LO, Duque SRL, de Almeida JS, Fevereiro PS (1999) Repetitive somatic embryogenesis in Medicago truncatula ssp. Narbonensis and M. truncatula Gaertn cv. Jemalong. Plant Cell Rep 18: 398-405 [Google Scholar]
- Dudits D, Bogre L, Gyorgyey J (1991) Molecular and cellular approaches to the analysis of plant embryo development from somatic cells in vitro. J Cell Sci 99: 475-484 [Google Scholar]
- Felsenstein J (1989) PHYLIP: Phylogeny Inference Package (Version 3.2). Cladistics 5: 164-166 [Google Scholar]
- Frugoli J, Harris J (2001) Medicago truncatula on the move! Plant Cell 13: 458-463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151-158 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241: 42-52 [DOI] [PubMed] [Google Scholar]
- Hecht V, Vielle-Calzada JP, Hartog MV, Schmidt EDL, Boutilier K, Grossniklaus U, de Vries SC (2001) The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol 127: 803-816 [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Chin-Atkins AN, Wilson IW, Chapple R, Dennis ES, Chaudhury A (2001) The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13: 2115-2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P, Falquet L, Bairoch A (1999) The PROSITE database: its status in 1999. Nucleic Acids Res 27: 215-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iantcheva A, Vlahova M, Bakalova E, Kondorosi E, Elliott MC, Atanassov A (1999) Regeneration of diploid annual medics via direct somatic embryogenesis promoted by thidiazuron and benzylaminopurine. Plant Cell Rep 18: 904-910 [Google Scholar]
- Iantcheva A, Vlahova M, Trinh TH, Brown SC, Slater A, Elliot MC, Atanassov A (2001) Assessment of polysomaty, embryo formation and regeneration in liquid media for various species of diploid annual Medicago. Plant Sci 160: 621-627 [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J (1995) Proteins with leucine-rich repeats. Curr Opin Struct Biol 5: 409-416 [DOI] [PubMed] [Google Scholar]
- Krusell L, Madsen LH, Sato S, Aubert G, Genua A, Szczyglowski K, Duc G, Kaneko T, Tabata S, de Bruijn F et al. (2002) Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature 420: 422-426 [DOI] [PubMed] [Google Scholar]
- Lamblin AFJ, Crow JA, Johnson JE, Silverstein KAT, Kunau TM, Kilian A, Benz D, Stromvik M, Endre G, VandenBosch KA et al. (2003) MtDB: a database for personalized data mining of the model legume Medicago truncatula transcriptome. Nucleic Acids Res 31: 196-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin PJ, Gibson JM, Mathesius U, Weinman JJ, Gartner E, Hall E, Tanner GJ, Rolfe BG, Djordjevic MA (1996) Transgenic white clover: studies with the auxin-responsive promoter, GH3, in root gravitropism and lateral root development. Transgenic Res 5: 325-335 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195-1205 [DOI] [PubMed] [Google Scholar]
- Mayer KFX, Schoof H, Haecker A, Lenhard M, Jurgens G, Laux T (1998) Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805-815 [DOI] [PubMed] [Google Scholar]
- Mordhorst AP, Voerman KJ, Hartog MV, Meijer EA, van Went J, Koornneef M, de Vries SC (1998) Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149: 549-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Thompson WF (1980) Rapid isolation of high-molecular-weight plant DNA. Nucleic Acids Res 8: 4321-4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14: 897-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R, Hayashi M, Wu G-J, Kouchi H, Imaizumi-Anraku H, Murakami Y, Kawasaki S, Akao S, Ohmori M, Nagasawa M et al. (2002) HAR1 mediates systemic regulation of symbiotic organ development. Nature 420: 426-429 [DOI] [PubMed] [Google Scholar]
- Nolan KE, Rose RJ (1998) Plant regeneration from cultured Medicago truncatula with particular reference to abscisic acid and light treatments. Aust J Bot 46: 151-160 [Google Scholar]
- Nolan KE, Rose RJ, Gorst JE (1989) Regeneration of Medicago truncatula from tissue culture: increased somatic embryogenesis from regenerated plants. Plant Cell Rep 8: 278-281 [DOI] [PubMed] [Google Scholar]
- Page RDM (1996) TREEVIEW: an application to display phytogenetic trees on personal computers. Comput Appl Biosci 12: 357-358 ' [DOI] [PubMed] [Google Scholar]
- Reinert J (1958) Morphogenese und ihre kontrolle an gewebekulturen aus carotten. Naturwissenchaften 45: 344-345 [Google Scholar]
- Rojo E, Sharma VK, Kovaleva V, Raikhel NV, Fletcher JC (2002) CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14: 969-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose RJ, Nolan KE, Bicego L (1999) The development of the highly regenerable seed line Jemalong 2HA for transformation of Medicago truncatula: implications for regenerability via somatic embryogenesis. J Plant Physiol 155: 788-791 [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, 3rd ed, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schmidt EDL, Guzzo F, Toonen MAJ, de Vries SC (1997) A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124: 2049-2062 [DOI] [PubMed] [Google Scholar]
- Schoof H, Lenhard M, Haecker A, Mayer KFX, Jurgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635-644 [DOI] [PubMed] [Google Scholar]
- Searle IR, Men AE, Laniya TS, Buzas DM, Iturbe-Ormaetxe I, Carroll BJ, Gresshoff PM (2003) Long-distance signalling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299: 109-112 [DOI] [PubMed] [Google Scholar]
- Shah H, Gadella TWJ, van Erp H, Hecht V, de Vries SC (2001a) Subcellular localization and oligomerization of the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 protein. J Mol Biol 309: 641-655 [DOI] [PubMed] [Google Scholar]
- Shah K, Russinova E, Gadella TWJ, Willemse J, de Vries SC (2002) The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev 16: 1707-1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah K, Schmidt EDL, Vlak JM, de Vries SC (2001b) Expression of the Daucus carota somatic embryogenesis receptor kinase (DcSERK) protein in insect cells. Biochimie 83: 415-421 [DOI] [PubMed] [Google Scholar]
- Shah K, Vervoort J, de Vries SC (2001c) Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J Biol Chem 276: 41263-41269 [DOI] [PubMed] [Google Scholar]
- Somleva MN, Schmidt EDL, de Vries SC (2000) Embryogenic cells in Dactylis glomerata L. (Poaceae) explants identified by cell tracking and by SERK expression. Plant Cell Rep 19: 718-726 [DOI] [PubMed] [Google Scholar]
- Steward FC, Mapes MO, Mears K (1958) Growth and organized development of cultured cells: II. Organization in cultures grown from freely suspended cells. Am J Bot 45: 705-708 [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON 2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806-11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MR, Johnson LB, White FF (1990) Selection of interspecific somatic hybrids of Medicago by using Agrobacterium-transformed tissues. Plant Sci 69: 189-198 [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC (1994) Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol 26: 1599-1609 [DOI] [PubMed] [Google Scholar]
- Zimmerman JL (1993) Somatic embryogenesis: a model for early development in higher plants. Plant Cell 5: 1411-1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo JR, Niu QW, Frugis G, Chua NH (2002) The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J 30: 349-359 [DOI] [PubMed] [Google Scholar]