Abstract
Mutant characterization has demonstrated that ABI4 (Abscisic Acid [ABA] Insensitive 4), ABI5 (ABA Insensitive 5), and CTR1 (Constitutive Triple Response 1) genes play an important role in the sugar signaling response in plants. The present study shows that the transcripts of these three genes are modulated by glucose (Glc) independently of the developmental arrest caused by high Glc concentrations. ABI4 and ABI5 transcripts accumulate in response to sugars, whereas the CTR1 transcript is transiently reduced followed by a rapid recovery. The results of our kinetic studies on gene expression indicate that ABI4, ABI5, and CTR1 are regulated by multiple signals including Glc, osmotic stress, and ABA. However, the differential expression profiles caused by these treatments suggest that distinct signaling pathways are used for each signal. ABI4 and ABI5 response to the Glc analog 2-deoxy-Glc supports this conclusion. Glc regulation of ABI4 and CTR1 transcripts is dependent on the developmental stage. Finally, the Glc-mediated regulation of ABI4 and ABI5 is affected in mutants displaying Glc-insensitive phenotypes such as gins, abas, abi4, abi5, and ctr1 but not in abi1-1, abi2-1, and abi3-1, which do not show a Glc-insensitive phenotype. The capacity of transcription factors, like the ones analyzed in this work, to be regulated by a variety of signals might contribute to the ability of plants to respond in a flexible and integral way to continuous changes in the internal and external environment.
Sugars play a central role as signaling molecules that modulate the metabolism, development, and physiology of plants (Koch, 1996; Sheen et al., 1999; Smeekens, 2000; Coruzzi and Zhou, 2001; Rolland et al., 2002). Despite their central function, the molecular mechanisms underlying sugar signaling are still poorly understood. Evidence based on the use of sugar analogs such as 6-deoxy-Glc, 2-deoxy-Glc (2DG), and 3-O-methyl-Glc, shows that sugar-mediated regulation occurs through distinct signaling pathways (Sheen et al., 1999; Smeekens, 2000). One of these pathways responds exclusively to Suc (Chiou and Bush, 1998; Loreti et al., 2000); others respond to hexoses, such as Glc and Fru. Some signaling systems rely only on hexose sensing without any catabolism (hexokinase [HXK] independent), whereas others require hexose phosphorylation or even further hexose metabolism (Sheen et al., 1999; Fujiki et al., 2000; Smeekens, 2000; Xiao et al., 2000; Rolland et al., 2002).
At present, few components of the sugar signaling network are known. Evidence exists that, similar to yeast (Saccharomyces cerevisiae), the plant enzyme involved in hexose phosphorylation, HXK, functions as a primary sugar sensor (Jang and Sheen, 1997; Rolland et al., 2002; Moore et al., 2003). Protein phosphorylation and dephosphorylation also have been implicated in sugar regulation (Takeda et al., 1994; Ehness et al., 1997; Halford and Hardie, 1998; Fujiki et al., 2000; Smeekens, 2000; Halford et al., 2003).
Genetic approaches also have been used to identify components of the plant sugar signaling cascade (Smeekens, 2000; Rolland et al., 2002). Sugar response mutants have been isolated based on the effects caused by high or low sugar levels during germination or early seedling development. Others have been selected by screening transgenic plants with altered expression of sugar-regulated promoters. Interestingly, several sugar-insensitive mutants affect known genes involved in hormone biosynthesis or signaling (Coruzzi and Zhou, 2001; Gazzarrini and McCourt, 2001; Finkelstein and Gibson, 2002; Leon and Sheen, 2003). Mutants such as gin1 and gin5 (Glc-insensitive), isi4 (impaired Suc induction), and sis4 (Suc insensitive) affect genes involved in the biosynthesis of abscisic acid (ABA; Zhou et al., 1998; Arenas-Huertero et al., 2000; Laby et al., 2000; Rook et al., 2001; Cheng et al., 2002). GIN1/ISI4/SIS4 correspond to the ABA2 (ABA-deficient 2) gene (Laby et al., 2000; Rook et al., 2001; Cheng et al., 2002; Seo and Koshiba, 2002) and the GIN5 to the ABA3 gene (Arroyo et al., 2001).
An additional set of sugar-insensitive mutants revealed that components of ABA signaling also participate in sugar responses during early seedling development. Four independent sugar-insensitive mutants (gin6, sun6, sis5, and isi3) are allelic to the ABA-insensitive mutant abi4 (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001), which exhibits altered ABA sensitivity in seeds and seedlings (Finkelstein, 1994; Söderman et al., 2000). ABI4 belongs to the family of AP2 (APETALA 2) transcription factors (Finkelstein et al., 1998; Riechmann and Meyerowitz, 1998). Like abi4, the mutant abi5 displays a sugar-insensitive phenotype (Arenas-Huertero et al., 2000; Laby et al., 2000; Brocard et al., 2002), which shows that this factor also plays a role during sugar signaling. The product of the ABI5 gene corresponds to the basic Leu zipper class of transcriptional regulators (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Jakoby et al., 2002). ABI5 expression is subject to complex regulation by different factors including ABA, drought, salt, and Glc, which are positive regulators (Lopez-Molina et al., 2001; Brocard et al., 2002). However, the ABI5 protein in vegetative tissues appears to be present only during a narrow developmental window, shortly after seed germination, and is proposed to be a key player in monitoring environmental conditions during early seedling growth (Lopez-Molina et al., 2001).
In addition to strengthening the connection between ABA and sugar signaling, the analysis of the gin1 mutant revealed an antagonistic relation between sugar and ethylene signaling pathways (Zhou et al., 1998). This is supported by the finding that two sugar-insensitive mutants, gin4 and sis1, are alleles of the constitutive ethylene response mutant, ctr1 (Gibson et al., 2001; Cheng et al., 2002). Also, it has been demonstrated that the etr1 (ethylene resistant 1) and ethylene-insensitive mutants (ein2, ein3, and ein6) display sugar oversensitive phenotypes (Zhou et al., 1998; Cheng et al., 2002).
The emerging picture supports the notion that sugar signaling during early seedling development involves extensive interactions with the hormones ABA and ethylene (Leon and Sheen, 2003). Nevertheless, to understand in detail the molecular roles that ABA and ethylene play in sugar signaling, the function and regulation that known factors play in these signaling cascades must be addressed. In the present work, we performed a detailed analysis of the regulation of the ABI4 gene by Glc. We found that the ABI4 transcript accumulates shortly after Glc exposure and that this response depends on sequences located in the 5′-upstream region. We also have extended our analyses to two other genes, ABI5 and CTR1, mutations in these loci(s) result in Glc-insensitive phenotypes. We found that the ABI5 and CTR1 transcript levels are also affected by Glc. Interestingly, the ABI4 transcript response to Glc is only observed in a short developmental window that correlates with the susceptibility of seedlings to arrest development by sugars. Although transcript levels of these genes are also affected by osmotic stress and ABA, specific regulation by Glc is supported by the transcript accumulation pattern profile and by the use of the Glc analog 2DG. We also show that ABI4 and ABI5 Glc regulation is affected in gin, aba, abi, and ctr1 mutants, supporting the participation of ABA and ethylene in sugar responses.
RESULTS
Regulation of ABI4 by Glc
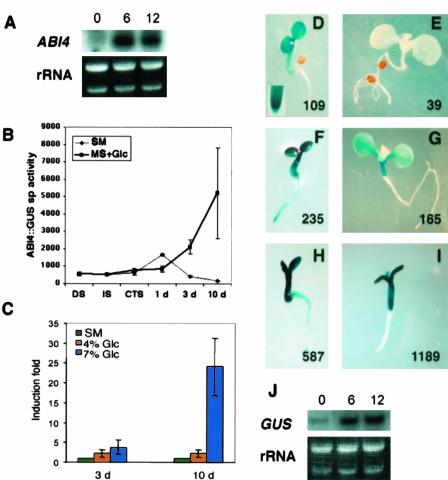
It has been shown previously that the ABI4 transcript accumulates in wild-type seedlings grown in the presence of Murashige and Skoog 7% (w/v) Glc but not in those grown on Murashige and Skoog 2% (w/v) Glc (Arenas-Huertero et al., 2000; Cheng et al., 2002). Because the expression of ABI4 gene is high in dry seeds (Söderman et al., 2000), and seedling growth is arrested by high Glc (Arenas-Huertero et al., 2000), it was important to determine if ABI4 transcript accumulation in Glc-treated seedlings was residual from the seed. The accumulation of the ABI4 transcript was followed in seedlings germinated on standard medium and then exposed to Murashige and Skoog 7% (w/v) Glc media for different incubation periods. We used Murashige and Skoog 1% (w/v) Suc as standard media because it has identical effects on ABI4 expression as 2% (w/v) Glc; also, Suc is the preferred carbon source used by plants. As shown in Figure 1A, in 2-d-old seedlings, the ABI4 transcript accumulates at higher levels shortly after Glc treatment (6 h), and this accumulation is maintained for at least 12 h. These results support the idea that the ABI4 transcript accumulates de novo in response to Glc.
Figure 1.
Glc induction of the ABI4 transcript and spatial expression pattern. A, Glc regulation of the ABI4 gene. Northern blot from Columbia-0 (Col-0) plants grown for 2 d on standard liquid media and exposed to Murashige and Skoog (MS) 7% (w/v) Glc for 0, 6, and 12 h. Each lane contains 8 μg of total RNA. The complete ABI4 cDNA was used as a probe. B, β-Glucuronidase (GUS)-specific activity was measured in dry seeds (DS), 1-h-imbibed seeds (IS), 3-d cold-treated seeds (CTS), or germinating seedlings in standard media (SM) or Murashige and Skoog 7% (w/v) Glc for 1, 3, and 10 d. Each point represents the mean of four independent lines and four biologically independent experiments, expressed as nanomoles of methylumbelliferone per microgram of total protein per minute. C, GUS activity of Glc-treated seedlings relative to the GUS level of their corresponding standard media grown plants, normalized to 1 in each case. Bars = sd (sometimes smaller than the scale). GUS staining of ABI4::GUS seedlings grown on standard media for 3 (D) or 10 (E) d. GUS staining of ABI4::GUS seedlings grown on MS 4% (w/v) Glc for 3 (F) or 10 (G) d. GUS staining of ABI4::GUS seedlings grown on MS 7% (w/v) Glc for 3 (H) or 10 (I) d. The value of a representative experiment of the GUS-specific activity of transgenic lines grown in each condition is shown. J, Glc regulation of the GUS transcript level. Transgenic plants carrying the ABI4::GUS expression cassette were grown in standard liquid media for 2 d followed by the exposure to Murashige and Skoog 7% (w/v) Glc. Samples were taken at 0, 6, and 12 h. Five micrograms of total RNA from each sample were loaded in each lane and hybridized with a 600-bp fragment of the GUS gene.
To determine the spatial, temporal, and developmental expression pattern of ABI4 Glc-mediated accumulation, we used transgenic plants expressing the GUS reporter under the control of the 3-kb region upstream of the start codon of the ABI4 gene. This construct was described previously and was shown to confer high GUS expression during seed development, as expected for the ABI4 gene (Söderman et al., 2000). Because the GUS protein is very stable (Jefferson et al., 1987), the experiments performed with these transgenic lines were done under continuous growth on either standard or Glc media. Using fluorometric analysis performed in the linear range of product accumulation, the specific activity of GUS was measured in standard media grown seeds and seedlings and compared with those grown in the presence of Glc. A considerable level of GUS expression is detected in dry, 1-h-imbibed, and 3-d cold-treated seeds; however, no difference was observed between standard media and Glc media (Fig. 1B). Although GUS-specific activity increased transiently in seedlings grown on standard media, 3-d-old seedlings grown in either 4% or 7% (w/v) Glc had 2- to 4-fold higher GUS activity than those grown on standard media (Fig. 1, B and C). This difference is even higher in the 10-d-old seedlings grown in 4% or 7% (w/v) Glc, as the result of the lower levels found in standard media, and clearly reflects an induction above the levels found in dry seeds (Fig. 1, B and C). Similar results were obtained with four independent transgenic lines, which demonstrate that the ABI4 5′ regulatory region used in these constructs includes at least part of the cis-acting elements involved in the response to Glc.
The expression pattern of the ABI4 transcript upon Glc treatment was determined by GUS histochemical analysis. As shown in Figure 1D in 3-d-old seedlings grown on standard media, GUS staining is mostly confined to the cotyledons and hypocotyl region and is not observed in roots, with the exception of the root cap and the quiescent center. Interestingly, detectable GUS staining in older seedlings is lost (Fig. 1E), which is consistent with the ABI4 mRNA analysis (see below) and with previously reported data (Söderman et al., 2000). This expression pattern was reproducible in independent lines showing that ABI4 is clearly regulated by a developmental program (compare Fig. 1, D with E). When plants were germinated in the presence of 4% or 7% (w/v) Glc, the levels of GUS staining increased with Glc concentration. In the presence of Glc, GUS activity was detected not only in 3-d-old seedlings (Fig. 1, D versus F and H) but also in 10-d-old plants (Fig. 1, E versus G and I). The GUS expression patterns in Glc and standard media were qualitatively similar, except in the root of 3-d-old seedlings, where the presence to either 4% or 7% (w/v) Glc expanded the domain of GUS expression (Fig. 1, D versus F and H). These results suggest that the presence of Glc might not only increase the levels of the ABI4 transcript but also could modify its expression pattern.
Because most of our analysis has been carried out in arrested seedlings, we explored whether Glc also induced the ABI4 transcript in a nonarrested developmental condition. Although 10-d-old seedlings grown on standard media had no detectable GUS activity (Fig. 1E), those grown in the presence of 4% (w/v) Glc exhibited GUS activity even though these seedlings were not arrested (Fig. 1G). The GUS staining pattern of these plants, which is restricted to the hypocotyl and cotyledons, resembles that of the 3-d-old seedlings grown on standard media (Fig. 1D). Interestingly, no staining was ever detected in the first pair of true leaves. We corroborated by northern-blot analysis that the increase in GUS activity represents an induction of GUS mRNA accumulation in response to Glc in 2-d-old seedlings (Fig. 1J), which correlates with the endogenous ABI4 mRNA accumulation shown in Figure 1A.
Glc Affects the ABI5 and CTR1 Transcript Levels
The results described above indicate that Glc induces the expression of the ABI4 transcript, a factor required for proper sugar response during postgermination development. Similar results were recently found for genes involved in sugar signaling, such as ABA2 and other ABA biosynthetic genes, whose transcript levels are regulated by Glc (Cheng et al., 2002). Thus, Glc appears to modulate the transcript level of genes involved in its own signaling pathway. In addition to ABI4 and the ABA genes, the molecular characterization of other sugar-insensitive mutants has permitted the identification of additional factors required for a normal sugar response. The abi5 mutant also displays resistance to high sugar concentrations (Arenas-Huertero et al., 2000; Laby et al., 2000), whereas overexpression of ABI5 results in hypersensitivity to sugar (Brocard et al., 2002; Finkelstein et al., 2002). However, the participation of ABI5 during sugar seedling response is still under debate because the resistance of the abi5 mutant seems not to be as strong as the abi4 mutant (Gazzarrini and Mc-Court, 2001). In addition, two other independent sugar mutants (sis1 and gin4) have been found to affect the ethylene signaling CTR1 gene (Gibson et al., 2001; Cheng et al., 2002).
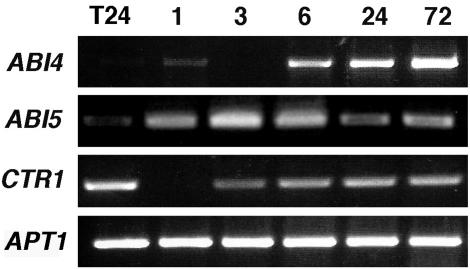
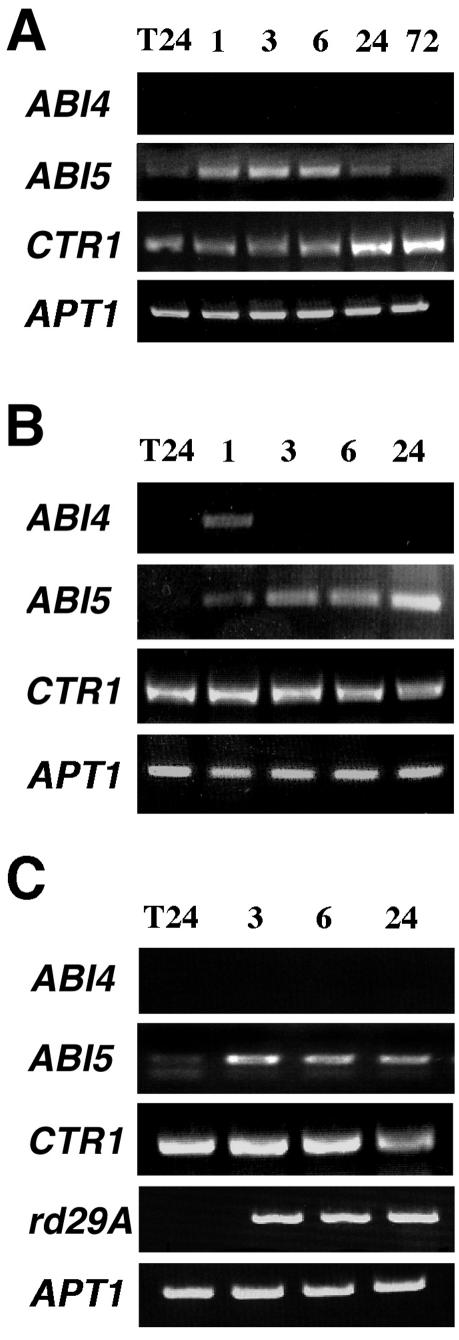
To determine whether or not, or under what conditions, the ABI5 and CTR1 genes are regulated by Glc, we followed the transcript accumulation pattern of ABI4, ABI5, and CTR1 genes in 3-d-old wild-type seedlings after transfer to 7% (w/v) Glc media. In this case, semiquantitative reverse transcriptase (RT)PCR analysis was used at different times after transfer to Glc treatment. As shown in Figure 2, a strong accumulation of ABI4 transcript is detected 6 h after transfer, in comparison with the undetectable levels found at the beginning of the treatment (Fig. 2). This result is in accordance with our northern data (Fig. 1A) that corroborate that this is a reliable method to analyze the other genes. Interestingly, we found that the levels of the ABI5 and CTR1 transcripts are also affected by Glc. ABI5 accumulates in response to Glc even faster than ABI4 because it is detected shortly (1 h) after transfer to Glc. The CTR1 expression pattern is more complex because a decrease of the transcript level is observed reproducibly 1 h after transfer to Glc, but its level subsequently recovers. The Glc response of these genes is not due to a stress caused by transferring the plant as it was corroborated by biologically independent transfer experiments to standard media (see following sections). We observed that the expression of the ABI5 gene in response to Glc is not maintained at the same levels between 24 and 72 h of the Glc treatment. As important developmental changes occur in this period, it is possible that this fluctuation maybe related to the intrinsic plant development.
Figure 2.
ABI4, ABI5, and CTR1 transcript accumulation is regulated by Glc. RT-PCR analysis of total RNA from 3-d-old wild-type (Wassilewskija [Ws]) seedlings grown in standard media and transferred to Murashige and Skoog 7% (w/v) Glc for 1, 3, 6, 24, and 72 h. The PCR product of APT1 was used as a cDNA loading control. Specific primers were used to amplify ABI4, ABI5, CTR1, and APT1 gene transcripts. The lengths of the PCR products are 974, 183, 353, and 478 bp for ABI4, ABI5, CTR1, and APT1, respectively. The linear phase of the exponential PCR reaction was corroborated for each gene (data not shown). A representative experiment from three biologically independent experiments is shown, including only the 24-h transference (T24) control for simplification. The means ± se of all three experiments and their corresponding transfer controls are included in Figure 4, D to F.
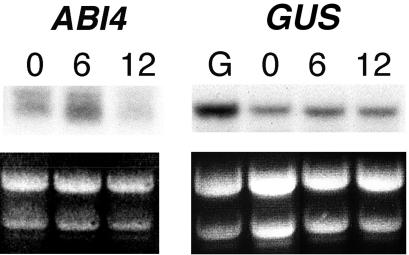
To discriminate whether the response of ABI4, ABI5, and CTR1 gene transcripts corresponds to a Glc signal or to an osmotic effect caused by the Glc concentration used in the experiments, we analyzed gene expression in response to isosmotic concentrations of mannitol. Initially, we followed the ABI4 response to 7% (w/v) mannitol by northern-blot analysis in wild-type and ABI4::GUS transgenic seedlings, using the same conditions described in Figure 1, A and J. As shown in Figure 3, the levels of ABI4 and ABI4::GUS gene transcripts slightly increase in the presence of mannitol but do not show the substantial sustained increases observed in the presence of Glc (Figs. 1A versus 3). Meanwhile, the ABI4 transcript level is maintained high in Glc over the period analyzed (12 h), it is reduced in the mannitol treatment (Fig. 3). In the case of the ABI4::GUS gene transcript, we did not observe a decrease in the transcript level at 12 h, and this may be due to the intrinsic stability of the GUS mRNA compared with the ABI4 transcript. Thus, these results led us to conclude that the ABI4 transcript accumulation observed in the Glc treatment is not entirely due to an osmotic response.
Figure 3.
ABI4 response to osmotic stress. Northern blots from wild-type (Col-0) and ABI4::GUS transgenic 2-d-old seedlings grown on standard liquid media and exposed to Murashige and Skoog 7% (w/v) mannitol for 0, 6, and 12 h or Murashige and Skoog 7% (w/v) Glc for 12 h (G). Each lane contains 8 μg of total RNA. The complete ABI4 cDNA and a 600-bp GUS fragment were used as probes. rRNAs were used as loading control.
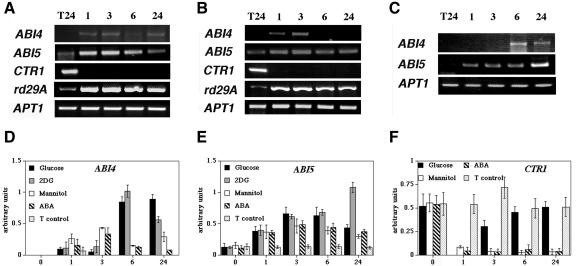
To fully analyze the osmotic response during shorter and longer treatment periods for ABI4, ABI5, and CTR1 genes, RT-PCR experiments were performed. Interestingly, the ABI5 and CTR1 genes are also responsive to osmoticum. The ABI4 and ABI5 transcripts are positively regulated by mannitol, whereas CTR1 transcript level is reduced (Fig. 4A). A specific accumulation of ABI4 transcript in response to Glc begins at 6 h (Figs. 2 and 4D). In response to osmotic stress, ABI4 transcript accumulation is observed earlier, but the levels are notably lower than in response to Glc (Fig. 4D), in agreement with the data from the northern blot (Figs. 1A and 3). Once again, no detectable levels of ABI4 transcript were found at 0 h of treatment (Fig. 4D). Under our conditions, the ABI5 expression pattern is similar in both Glc and mannitol treatments (Figs. 2 and 4, A and E). Thus, with these data, it is still not possible to discriminate whether Glc directly regulates ABI5 gene expression. In the case of the CTR1 transcript, mannitol treatment results in a permanent repression in contrast to Glc, where a transient repression is followed by a recovery (Fig. 4F). Thus, we suggested that Glc is capable of overriding the osmotic signal generated in these conditions, which led us to conclude that Glc regulates CTR1 transcript levels.
Figure 4.
Glc regulation of ABI4, ABI5, and CTR1 genes is not due to the osmotic effect of Glc. Three-day-old WT (Ws) seedlings grown in standard media were transferred to standard media plus either 7% (w/v) mannitol (A), 100 μm ABA (B), or 0.5 mm 2DG (C) for 1, 3, 6, and 24 h. For simplification, only the 24-h transfer (T24) control was included. RT-PCR was performed as described in Figure 2. The rd29A gene was used as control of ABA and osmotic treatments. Comparative histograms of the densitometric quantification of the PCR products from biologically independent experiments are shown for ABI4 (D), ABI5 (E), and CTR1 (F) in response to 7% (w/v) Glc, 0.5 mm 2DG, 7% (w/v) mannitol, and 100 μm ABA and including the transfer controls (T control) for each time point. In the case of ABI4, where no detectable transcript levels were found, such as 0 h and most of the T control times, no bar is observed (D). The APT1 RT-PCR product was used as internal control in each reaction, and the value of each condition and the relative units of the amplification signals obtained by the densitometric analysis of each gene are normalized to the amplification signal of the control APT1 gene (arbitrary units). The data on the graphic correspond to the mean ± se of three separate experiments.
We have demonstrated previously that seedlings grown on Glc contain higher levels of ABA, required for normal Glc signaling, and the participation of ABA in osmotic responses is also very well established (Zeevaart and Creelman, 1988; Arenas-Huertero et al., 2000). In addition, it has been reported that the ABI5 gene is induced in young seedlings by ABA, salt, Glc, and drought treatments (Lopez-Molina et al., 2001; Brocard et al., 2002). Therefore, it was important to determine if ABA also regulates the ABI4 and CTR1 transcripts. We tested the accumulation of ABI4, ABI5, and CTR1 genes in response to the addition of 100 μm ABA (Fig. 4B). The rd29A gene was used as control because it is known to be ABA and osmotic responsive (Yamaguchi-Shinozaki and Shinozaki, 1994). The levels of the three transcripts were modulated in response to ABA (Fig. 4B). In general terms, the transcript accumulation profile for all three regulatory genes after ABA treatment correlates with their responses to mannitol (Fig. 4, D-F), suggesting that at least part of the observed osmotic response might be ABA dependent, similar to the rd29A control gene (Fig. 4, A and B). As in the case of the mannitol treatment, ABI4 accumulation in response to ABA is notably lower than that observed in response to Glc, when compared with the APT1 internal control gene (Fig. 4D).
To further investigate the specificity of Glc regulation over the ABI5 gene, inductions were performed using the phosphorylable Glc analog 2DG. 2DG has been shown to act as a potent sugar signal for the regulation of a variety of Glc-regulated genes in yeast and plants. Glc-mediated gene regulation by 2DG has been detected when used at much lower concentrations (0.5-2 mm) than Glc (Graham et al., 1994; Jang and Sheen, 1994), which avoid an osmotic effect. 2DG is known to be very toxic to plant cells; thus, the initial experiments were performed with increasing concentrations of this analog to establish the lowest effective concentration (data not shown). It was observed that a concentration of 0.5 mm 2DG gave detectable responses of the Glc-regulated ABI4 gene without affecting the levels of the APT1 gene used as internal control (Fig. 4C). Similar to ABI4, the ABI5 gene is responsive to 2DG (Fig. 4C), supporting a direct regulation by Glc independent of the osmotic responses. Based on the present analysis, we conclude that Glc has a direct and specific regulatory role over the transcript accumulation levels of ABI4, ABI5, and CTR1, which can be differentiated from their osmotic regulation. In summary, our data point to a complex regulation of the transcript accumulation of ABI4, ABI5, and CTR1 genes by multiple factors.
Glc Regulation of ABI4 and CTR1 Transcript Accumulation Is Affected by Developmental Signals
It has been shown that the susceptibility of young seedlings to developmental arrest in response to sugars is restricted to a short period of time after germination (Gibson et al., 2001). We examined whether the regulation of the ABI4, ABI5, and CTR1 genes by sugar observed in cotyledon stage seedlings was also limited to a developmental period. The accumulation of ABI4, ABI5, and CTR1 transcripts in response to Glc (Fig. 5A) was examined in 6-d-old seedlings containing the first pair of true leaves. Interestingly, the response to Glc of ABI4 and CTR1 is different from that observed in younger plants. Unlike in 3-d-old seedlings, ABI4 induction by Glc was undetectable in 6-d-old plants (Fig. 5A); similar results were observed in ABI4::GUS transgenic lines by GUS protein activity (data not shown). The capacity of these plants to respond is lost not only for the Glc signal but also for ABA and even for the potent sugar analog 2DG (Fig. 5). We occasionally observed a very slight band after a 1-h transfer to 2DG treatment; however, this fluctuation was not reproducible in all experiments. Hence, this variation was considered to be within the experimental fluctuation and not a 2DG-specific effect. In the case of CTR1 gene, the transient repression observed in the first hour of Glc treatment in 3-d-old seedlings (presumably due to osmotic stress) is lost at this older stage. However, the level of this gene is induced after 24 to 72 h of Glc treatment. Thus, it appears that CTR1 is still responsive to Glc but with a different expression pattern than the one observed early in development (Fig. 4A). It is interesting that 2DG did not reproduce the Glc induction (Fig. 4B), suggesting that either a signal derived from the further metabolism of Glc (Xiao et al., 2000) or from additional developmental signals might be responsible for this response. Finally, a different CTR1 response is also observed with a direct ABA treatment (Fig. 5C).
Figure 5.
Glc regulation of ABI4 and CTR1 transcript accumulation is affected by developmental signals. Six-day-old WT (Ws) seedlings grown in standard media and transferred to Murashige and Skoog 7% (w/v) Glc (A), standard media plus 0.5 mm 2DG (B), or 100 μm ABA (C) for the indicated times. For simplification, the 24-h transfer (T24) control is included, but similar levels to this control were observed in the rest of the control times (data not shown). The PCR product of APT1 was used as a cDNA loading control. Samples were collected and used for RT-PCR analysis as described. This picture is representative of three biologically independent experiments.
Surprisingly, in 6-d-old seedlings, the ABI5 transcript is regulated by Glc (Fig. 5A), 2DG (Fig. 5B), and ABA (Fig. 5C), with a similar pattern to the one found at 3 d. These results are consistent to those recently reported by Brocard et al. (2002) but contrast with the report that the ABI5 protein induction in response to ABA is restricted to a narrow developmental window (Lopez-Molina et al., 2001). We also confirmed that none of the responses was a result of the physical transfer of the plants (data not shown).
ABI4 and ABI5 Glc Regulation Is Affected in Mutants Displaying Altered Glc Responses
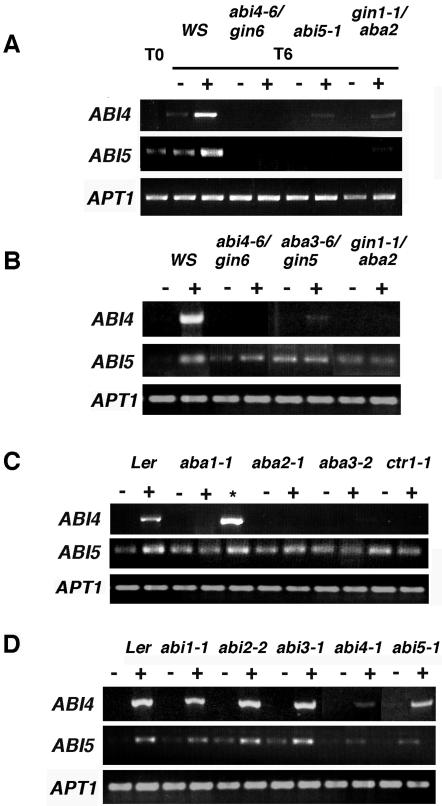
With the aim to further analyze the Glc-mediated regulation of ABI4 and ABI5 genes, we explored the levels of their transcripts in different mutant backgrounds that display a Glc-insensitive phenotype. Because the CTR1 Glc response is complex, this analysis was carried out only with ABI4 and ABI5 genes. Thus, the ABA-deficient (aba3-6/gin5, aba1-1, gin1-1/aba2, and aba3-2), the ABA-insensitive (abi4-6/gin6 and abi5-1), and ethylene signaling ctr1-1 mutants were chosen for this study. Initially, the abundance of ABI4 and ABI5 transcripts was determined in the abi4-6/gin6, abi5-1 and gin1-1/aba2 mutants grown for 3 d in standard media and transferred to 7% (w/v) Glc media for 6 h. Figure 6A shows that ABI4 and ABI5 transcript accumulation in response to Glc is dramatically reduced in these three mutants. A very similar response was observed when these mutants were grown continuously in the presence of Glc (Fig. 6, B and D). Given that these three mutants showed no difference when grown continuously in Glc, the rest of the mutants were also analyzed under these conditions. Figure 6B shows that the induction of ABI4 transcript in response to Glc is dramatically reduced in the gin mutants analyzed. Also, an important reduction of the Glc-mediated transcript accumulation for the ABI5 gene is observed for this set of mutants with the exception of abi4-6/gin6, in which a slight induction is still observed. It is possible that the level of the ABI4 factor in this mutant, which affects the promoter region of the ABI4 gene (Arenas-Huertero et al., 2000), is sufficient to preserve some regulation that it is detectable under the continuous Glc growth condition. ABI4 and ABI5 transcript accumulation in response to Glc in aba mutants was also compared with their corresponding wild-type ecotype (Fig. 6C). Similar to the gin mutants, ABI4 transcript induction by Glc is basically eliminated in the aba mutants. The ABI5 expression pattern is also affected in this set of mutants, although to a lesser extent than in aba2-1 (Fig. 5C). Our previous report demonstrated that the addition of nanomolar concentrations of ABA restores Glc sensitivity in aba mutants (Arenas-Huertero et al., 2000). In accordance with this finding, the addition of 100 nm ABA to the Glc media restores transcript accumulation in the aba1-1 plant of ABI4 and also in ABI5, where the observed induction is not as high as ABI4, probably due to the existing basal levels of this gene (Fig. 6C).
Figure 6.
ABI4 and ABI5 Glc regulation is affected in ABA-deficient, -insensitive, and ethylene signaling mutants. A, Indicated genotypes were grown in standard media for 3 d (T0) and transferred to the same media (-) or Murashige and Skoog 7% (w/v) Glc (+) for 6 h (T6). gin (B), aba and ctr1 (C), and abi (D) plants were grown on standard media (-), Murashige and Skoog 7% (w/v) Glc (+), or Murashige and Skoog 7% (w/v) Glc plus 100 nm ABA (*) during 18 d. Col-0 ecotype (aba2-1 and ctr1-1 genetic background) has exactly the same response as the Landsberg erecta (Ler) ecotype under these conditions. For simplification, the Col-0 ecotype PCR is omitted in this figure. Samples were harvested and used for RT-PCR analysis as described. The PCR product of APT1 was used as a cDNA loading control. A representative experiment from three biologically independent experiments is shown.
Finally, we followed ABI4 and ABI5 transcript response to Glc in ethylene and ABA signaling mutants. The response of these two transcripts was affected in the ctr1-1 mutant (Fig. 6C). We have shown previously that only two of the reported ABA-insensitive mutants (abi4-1 and abi5-1) display altered Glc sensitivity, whereas the remainder (abi1-1, abi2-1, and abi3-1) show a wild-type response (Arenas-Huertero et al., 2000). As shown in Figure 6D, transcript accumulation of ABI4 and ABI5 in response to Glc is altered only in abi4-1 and abi5-1 plants but not in abi mutants with normal Glc sensitivity (abi1, abi2-1, and abi3-1).
DISCUSSION
The increasing number and diversity of genes regulated by sugars reflects the importance of sugar signaling (Koch, 1996; Rolland et al., 2002). Such regulation has profound effects on plant growth, development, and stress response (Koch, 1996; Smeekens, 1998). The mechanisms by which sugars modulate gene expression are variable. It has been shown that the transcription of several genes is induced or repressed by sugars, but other levels of regulation have also been reported (Koch, 1996; Chan and Yu, 1998; Rook et al., 1998; Cheng et al., 1999; Cotelle et al., 2000; Toyofuku et al., 2000). Very little is known about the effects that sugars have on the expression of genes that participate in the sugar signaling cascade. It has been reported that a putative sugar signaling component (AtSR2) that belongs to the SNF1-related protein family is induced by Suc, Glc, and Fru (Chikano et al., 2001). Recently, it has been found that the expression level of several genes involved in ABA biosynthesis and in the Glc post-germination response is also modulated by Glc (Cheng et al., 2002). During the characterization of the gin6 mutant, we observed that the ABI4 transcript accumulated when plants were grown in the presence of Glc, suggesting a possible regulation of this gene by sugars (Arenas-Huertero et al., 2000). In this work, it is shown that the transcript levels of ABI4 and two additional factors that affect sugar signaling, ABI5 and CTR1, are modulated upon Glc addition. The induction strategy used in this work permitted to demonstrate that the modulation of transcript levels by Glc is independent of the developmental arrest produced by the continuous presence of sugars.
Loci Implicated in Sugar Response Are Regulated by Glc, Osmotic Stress, and ABA
ABI4 mutant alleles have been isolated in sugar mutant screens on multiple occasions, using different approaches and different mutant collections (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001). This finding may be indicative of the central role that ABI4 plays in Glc responses during early seedling development and prompted us to analyze several aspects of its regulation in further detail. The ABI4 transcription factor, in addition to its role in sugar signaling, is well known to be required in seed development and salt responses (Finkelstein et al., 1998, 2002; Quesada et al., 2000). ABI4 is expressed in seeds, and its highest levels are found at seed maturity, but very low levels of this transcript (that disappear shortly after germination) have been reported in vegetative tissues (Söderman et al., 2000). In agreement with this report, under our standard growth conditions, we found that after germination (3 d), the ABI4 transcript declines to levels undetectable by RT-PCR. The very low levels of GUS activity detected in 10-d-old ABI4::GUS plants probably reflect the intrinsic stability of the GUS protein. However, Glc has the capacity to increase ABI4 transcript levels. This is true not only in the developmentally arrested seedlings but also under conditions that do not result in developmental arrest. Although transcriptional regulation by sugars is the most studied process (Smeekens, 2000), posttranscriptional sugar regulatory events might also be responsible for such transcript accumulation. In either case, because the accumulation is also observed in ABI4::GUS transgenic plants, at least part of the region involved in this sugar-mediated regulation seems to be located within the 3-kb upstream of the initiation codon of the ABI4 gene.
Based on the GUS expression pattern of different independent transgenic lines grown in 4% and 7% (w/v) Glc, the tissues where the ABI4 transcript is detected are similar to those of younger seedlings when grown in low sugar (standard media). A notable exception is the primary root, where the detectable expression in 3-d-old standard media-grown seedlings was confined to the root tip, whereas in those grown in Glc, the GUS reporter is detected in most of the root tissues. Thus, it is possible that Glc not only increases the transcript levels of the ABI4 gene but also changes its expression pattern. It is interesting that in abi4, abi5, and three ABA biosynthetic mutants, an altered lateral root formation in response to nitrate is observed (Signora et al., 2001). These findings suggest that ABA, ABI4, and ABI5 might have a role in the lateral root development, regulated by the antagonism between N and C (Zhang et al., 2000). Thus, the expression of the ABI4 gene along the root under high Glc concentrations might be related to such interaction. Similarly, ABI5::GUS expression was reported recently to be localized to both primary and lateral root tips under unstressed conditions, with expression increased and expanded throughout 3-d-old seedling roots in response to ABA, Glc, and other stresses (Brocard et al., 2002). The mechanism of ABI4 and ABI5 action in this interaction remains to be explored in the future.
In addition to ABI4, two additional sugar signaling factors, ABI5 and CTR1, have been shown to affect Glc responses in plants. Mutations of these two genes cause Glc-insensitive phenotypes (Arenas-Huertero et al., 2000; Laby et al., 2000; Gibson et al., 2001; Cheng et al., 2002) in addition to their respective reported roles in ABA and ethylene signaling (Finkelstein, 1994; Stepanova and Ecker, 2000). Previous results had already shown that ABA, salt, and drought triggers the accumulation of the ABI5 protein (Lopez-Molina et al., 2001). We observed that the ABI5 and CTR1 transcript levels are modulated by Glc. Similar to ABI4, the ABI5 transcript accumulates in wild-type seedlings transferred to Glc. During the time that this analysis was performed, it was reported that GUS-specific activity in ABI5::GUS transgenic plants increases in response to several stresses, including a Glc treatment for 2 d (Brocard et al., 2002). In this work, we further demonstrate that ABI5 transcript accumulation is detectable 1 h after Glc addition. This response is even faster than the one observed for the ABI4 transcript (6 h), which might suggest a different time frame requirement for these two factors during Glc response and/or a different mechanism of induction.
Distinct Glc and Osmotic Regulation of Sugar Response Loci
Several lines of evidence allow us to suggest that the observed Glc regulation of the ABI4, ABI5, and CTR1 genes is specific and not a pure osmotic response. First, even though ABI4 responds to osmoticum and ABA, the transcript accumulation kinetics for both signals are different from Glc induction, and the levels are considerably lower than in Glc-treated seedlings by northern and RT-PCR detection methods. Also, the CTR1 osmotic and ABA responses are completely opposite to the one observed in Glc. Second, ABI4 and ABI5 genes are responsive to 0.5 mm 2DG and, when compared with the 388 mm Glc corresponding to 7% (w/v), exert a negligible osmotic stress. Third, Glc-regulation of ABI4 and ABI5 is altered in mutants that affect Glc signaling but not in ABA response mutants that have a normal sugar response (abi1, abi2, and abi3). Thus, our results strongly support the idea that even though these three genes respond to multiple factors including osmoticum and ABA, they do have a specific Glc response. The regulation of the transcript levels by Glc might represent a positive feedback loop, important to ensure a more sustain Glc response. Also, the induction of these factors by Glc could result in the modulation of other signaling pathways in which these factors play a role.
Based on the Glc-resistant phenotype of abi4-6/gin6 and abi5 mutants, it is clear that both transcription factors are required for Glc responses in seedlings. This requirement could be accomplished by a direct induction of the transcript level by Glc. In fact, the present analysis demonstrates that the levels of both transcripts do increase upon Glc treatment. The fact that the ctr1 mutation cause Glc insensitivity indicates that the presence of CTR1 is also necessary for Glc signaling (Zhou et al., 1998; Gibson et al., 2001; Cheng et al., 2002). Surprisingly, the CTR1 transcript in the presence of 7% (w/v) Glc is rapidly and transiently reduced probably as an osmotic response, but it subsequently recovers. This recovery is specific for Glc and overrides the permanent repression caused by osmoticum or ABA treatments observed at early seedling development. Because CTR1 acts as a negative regulator responsible for blocking the ethylene cascade, its presence should allow the increase in ABA level required during Glc response (Arenas-Huertero et al., 2000; Cheng et al., 2002) that again could result in a positive feedback loop of the Glc signaling. Such observations are in agreement with the proposed opposite roles of ABA and ethylene in regulating Arabidopsis growth (Beaudoin et al., 2000; Ghassemian et al., 2000; Gazzarrini and McCourt, 2001; Finkelstein and Gibson, 2002).
It is noteworthy that the osmotic response for all three genes is similar to the pattern in ABA, suggesting that this response might be at least in part ABA dependent (Leung and Giraudat, 1998). The regulation of these genes by mannitol is not surprising, considering that the ABI5 protein is induced by drought and that alleles of the abi4 and abi5 mutants (sañ5 and nem1, respectively) result in resistance to salt and osmotic stress inhibition of germination (Quesada et al., 2000; Lopez-Molina et al., 2001; Carles et al., 2002). Moreover, all mutants with a Glc-insensitive phenotype, including abi, aba, and ctr1, are also more resistant to the inhibition of growth observed in wild type by mannitol (400 mm), sorbitol (400 mm), and NaCl (150 mm; Laby et al., 2000; A. Arroyo and P. León, unpublished data). Thus, these data further reinforce the participation of ABI4 and ABI5 during various aspects of the vegetative growth such as stress and developmental responses. However, Glc-sensitive abi mutants (abi1, abi2, and abi3) also show some resistance to salt and osmotic stress at germination (Werner and Finkelstein, 1995; R.R. Finkelstein, unpublished data), supporting the notion that responses to Glc and osmotic stress involve overlapping but different mechanisms.
The essential role of ABA during sugar regulation is further reinforced because all mutants with low ABA levels are affected in the transcript regulation of the ABI4 and ABI5 transcription factors, but the exogenous addition of low concentrations of ABA restores Glc regulation. It is clear that the Glc effect does not depend on ABA alone because the ABA treatment does not reproduce the response on the transcript levels observed by the sugar signal. This correlates with our previous results in which mutants such as gin6/abi4 displayed a Glc-insensitive phenotype despite that ABA levels are induced by the presence of Glc (Arenas-Huertero et al., 2000).
It has been shown that gin1-1/aba2 and gin5/aba3-6 mutants genetically interact with HXK, leading to the proposal that these genes participate in the HXK-mediated pathway (Zhou et al., 1998; Arenas-Huertero et al., 2000). It was observed that Glc-mediated regulation of ABI4 and ABI5 genes is altered in these mutants, suggesting that such regulation might be, at least in part, dependent on hexose phosphorylation. This possibility is further supported by the finding that ABI4 and ABI5 are positively regulated by 2DG, which has been used widely to dissect the HXK dependence in the Glc signal transduction mechanism of a variety of genes (Rolland et al., 2002). Thus, these results together suggest that HXK might play a direct role on at least part of the regulation of ABI4 and ABI5 transcript levels reported in this work, although direct proof is still required.
Developmental Controls of Glc Responsiveness
We observed that the transcript levels of these three genes in response to Glc, 2DG, and ABA are affected by developmental signals. For the conditions analyzed, ABI4 is responsive during a very restricted developmental window of early seedling development (before 6 d), whereas different CTR1 responses occur at distinct developmental stages. A different story was found for the ABI5 transcript, which was still responsive in the 6-d-old seedlings. This finding agrees with that recently obtained by Brocard et al. (2002), but contrasts with another report where it was found that the ABA-dependent accumulation of the ABI5 protein is restricted to the first 2 d of seedling development (Lopez-Molina et al., 2001). Thus, despite the accumulation of the transcript in response to ABA in these developmental stages, a posttranscriptional regulation is critical for the accumulation of ABI5 factor (Lopez-Molina et al., 2003). It would be very interesting to see whether a similar mechanism operates in response to Glc. Lopez-Molina et al. (2001) proposed that ABI5 might be a key player for a checkpoint period when the environment is monitored. This developmental period might be the same as the sugar-mediated arrest, where ABI4 and ABI5 seem to play key functions. So far, no physical interactions between these two factors have been detected (Nakamura et al., 2001), which opens the possibility that these two transcriptional factors modulate the expression of a different set of genes. In response to Glc, this might also be true because a different time frame of expression patterns is observed between the two factors as has been mentioned already. It has been suggested that ABI5 and some of its homologous basic Leu zippers might function with some redundancy in ABA, sugar, and other stress responses (Kang et al., 2002; Leon and Sheen, 2003). However, ABI4 could play a more critical role in monitoring sugar status, at least during early developmental stages, because no apparent homologs are found, at least in the Arabidopsis genome. In conclusion, these results indicate that these genes are finely regulated during plant development by a variety of signals, and different sugar signaling molecules possibly are acting simultaneously at different tissues and stages of plant development. These results also raise questions about the molecules involved in Glc responses in later developmental stages. For example, undetectable levels of ABI4 transcript were found both by northern and RT-PCR analysis in 6-d-old seedlings, and similar results have been reported (Söderman et al., 2000). Thus, the requirement of this factor in later developmental stages is a subject that is presently analyzed by our group.
In summary, this work provides direct evidence that the transcript abundance of three factors, ABI4, ABI5, and CTR1, is specifically regulated by Glc. These factors participate in a variety of plant responses to environmental conditions, hormonal signaling, and sugar regulation during early seedling development. Thus, these molecules should be integrating a set of external signals and metabolic signals through the interaction with specific components. The regulation of these three factors by Glc might provide an important regulatory loop that allows a sustained Glc response. In addition, this regulation might integrate the fluctuations of the endogenous sugar concentrations with other signaling pathways in which these factors have a role, such as ABA and ethylene signaling. Glc regulation over these genes is accomplished at least in part on their transcript levels that either modulate transcription or affect its mRNA stability. However, further analysis is needed to determine whether the changes observed in this work are also reflected at the protein level.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis plants were routinely grown under sterile conditions in controlled growth chambers (24°C, 16-h-light/8-h-dark photoperiod). Seeds were surface sterilized and plated on media containing 1× Murashige and Skoog basal salt mixture (GIBCO-BRL, Grand Island, NY) supplemented with B5 vitamins, 0.05% (w/v) MES, and 0.7% (w/v) phytoagar. All media contained either 1% (w/v) Suc (standard media; Murashige and Skoog 1% [w/v] Suc) or Glc at the indicated concentrations as carbon sources. For induction experiments in liquid, plants were grown on Murashige and Skoog 1% (w/v) Suc media, then replaced with Murashige and Skoog 7% (w/v) Glc or 7% (w/v) mannitol. For inductions in solid media, plants were grown over nylon filters on standard media, and the filter was then transferred to Murashige and Skoog 7% (w/v) Glc or Murashige and Skoog 1% (w/v) Suc media plus 2DG (0.5 mm) or 7% (w/v) mannitol or 100 μm ABA of (±) cis-trans-isomer (Sigma, St. Louis). With this procedure, we avoided touching the seedling during transfer. In all experiments, seeds were incubated at 4°C for 4 d to break dormancy.
The aba, abi, and ctr1-1 mutants and wild-type plants Ws, Ler, and Col-0 were obtained from the Arabidopsis Biological Resource Center. The gin1-1 mutant was kindly provided by Dr. Jen Sheen (Department of Genetics, Harvard Medical School, and Department of Molecular Biology, Massachusetts General Hospital, Boston, MA). The aba1-1, aba3-2, abi1-1, abi2-1, and abi3-1 mutants are in the Ler ecotype, whereas aba2-1, abi4-1, and ctr1-1 are in Col-0, and abi5-1 and gin1-1 are in Ws. The ABI4::GUS transgenic plants (Col-0 background) were reported previously (Söderman et al., 2000). Homozygous plants from four independent ABI4::GUS transgenic lines were used to perform the GUS analysis in this work.
Expression Analysis
Total RNA was isolated using standard protocols (Ausubel et al., 1989) from seedlings grown as indicated. For northern blots, total RNA was fractionated by electrophoresis in 1.2% (w/v) agarose gels and transferred onto Hybond N+ nylon membrane (Amersham Corporation, Arlington Heights, IL). Hybridizations and washes were performed at high-stringency conditions according to standard procedures, using 32P-radiolabeled probes (Church and Gilbert, 1984).
For RT-PCR analysis, cDNA was synthesized with Super Script II RT (GIBCO-BRL) using 5 μg of total RNA by oligo(dT)-primed reverse transcription. A fraction (1/80) of the first strand cDNA was used as a template for the PCR. Linearity phase of the exponential PCR reaction was corroborated for each gene by comparisons of the PCR product at different cycles (18, 25, 28, 30, 33, 35, and 40). The selected cycles for each gene used in the experiments were as follows: 40 cycles for ABI4, 33 for ABI5, 30 for CTR1, and 30 for APT1. Each set of RT-PCR data reported in this work were repeated in at least three biologically independent experiments. The graphic representation of densitometric quantification of the RT-PCR experiments was done using the National Institutes of Health Image 1.61 program (National Institutes of Health, Bethesda, MD) and normalized in each case by the APT1 control gene. The sequence of the primers used, the corresponding size products, and Gene Bank accession numbers are as follows: ABI4, 5′-ATGGACCCTTTAGCTTCCCA-3′ and 5′-AAGATGGGATCAATAAAATC-3′, 974 bp, AF040959; ABI5, 5′-GCA-TAT-ACA-GTG-GAA-TTGGA-3′ and 5′-CGG-GTT-CCT-CAT-CAA-TGT-CC-3′, 183 bp, AC006921.5; CTR1, 5′-GGT-CTC-TCG-CGA-TTG-AAG-GC-3′ and 5′-GAG-CGG-TTGGGC-GGA-GGA-AC-3′, 353 bp, L08790; rd29A, 5′-CGG-CGA-GAA-GTGATG-ATG-TG-3′ and 5′-CTT-CTC-CAC-TTC-CTT-TGT-CG-3′, 518 bp, D13044; and APT1, 5′-TCCCAGAATCGCTAAGATTGCC-3′ and 5′-CCTTTCCCTTAAGCTCTG-3′, 478 bp, BT000370.
ABI4 Histochemical and Fluorometric Analyses
GUS histochemical analysis was performed in homozygous plants as described previously (Jefferson, 1987). The plant tissue was incubated with the GUS substrate at 37°C overnight (12 h). Clearing was accomplished by 30-min incubations with an acetone:methanol (3:1 [v/v]) solution. Plants were observed under bright-field microscopy (Nikon Type 104, Nikon, Tokyo). Fluorometric assays of GUS activity were performed according to Jefferson (1987). Seeds or seedlings were collected and ground in 2 volumes of GUS extraction buffer. Total protein extracts were centrifuged at 14,000 rpm twice for 20 min at 4°C, and protein content was quantified using Bradford reagent (Bio-Rad Laboratories, Hercules, CA). For each assay, 4.5 μg of total protein was incubated with 4-methylumbelliferyl-d-Glucuronide (Sigma) at 37°C, and samples were taken at different time points to corroborate linearity. GUS activity was determined using a DyNAQuant 200 fluorometer (Hoefer, San Francisco, CA) and reported as GUS-specific activity expressed as nanomoles of methyl-umbelliferone per microgram of total protein per minute. The fold induction is calculated as the ratio of the GUS-specific activity from seedlings grown in Glc media divided by the GUS-specific activity from plants grown in standard media.
Acknowledgments
We would like to thank Araceli Cantero, Martha Trujillo, and Carolina San Roman for technical support; Alma Lidia Martínez for computer support; and Paul Gaytan and Eugenio López for oligo production. We thank Drs. Jen Sheen, Crisanto Gutierrez, Elena Ramirez Parra, Arturo Guevara, and Miguel A. Villalobos for discussion and helpful comments. Furthermore, we thank Drs. Kenneth Luehrsen, Rosalia De Necochea, Stewart Gillmor, and Stuart Reichler for critical reading of the manuscript. We also thank the Arabidopsis Biological Resource Center for providing different seed stocks.
This work was supported by Consejo Nacional de Ciencia y Tecnológia (grant no. 31791-N and fellowship to A.A.), by the Dirección General de Asuntos para el Personal Académico (grant no. IN210200), by Howard Hughes (grants), and by DGEP (fellow-ship to A.A.).
References
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14: 2085-2096 [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Arenas F, Jiménez A, Cantero A, León P (2001) The participation of ABA in the glucose-mediated regulation in Arabidopsis. Presented at 11th International Conference on Arabidopsis Research, June 23-27, 2001, Madison, WI
- Ausubel FM, Brent R, Kingston RE, Moore DD, Siedman JG, Smith JA, Struhl K (1989) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Beaudoin N, Serizet C, Gosti F, Giraudat J (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard IM, Lynch TJ, Finkelstein RR (2002) Regulation and role of the Arabidopsis ABA-insensitive 5 gene in ABA, sugar and stress response. Plant Physiol 129: 1533-1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C, Bies-Etheve N, Aspart L, Léon-Kloosterziel KM, Koornneef M, Echeverria M, Delseny D (2002) Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J 30: 373-383 [DOI] [PubMed] [Google Scholar]
- Chan MT, Yu SM (1998) The 3′ untranslated region of a rice alpha-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc Natl Acad Sci USA 95: 6543-6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, Penney J, Chen H-C, Arroyo A, Leon P, Nambara E, Asami T, Seo S et al. (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis abscisic acid biosynthesis and glucose signaling. Plant Cell 14: 2723-2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WH, Taliercio EW, Chourey PS (1999) Sugars modulate an unusual mode of control of the cell-wall invertase gene (Incw1) through its 3′ untranslated region in a cell suspension culture of maize. Proc Natl Acad Sci USA 96: 10512-10517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikano H, Ogawa M, Ikeda Y, Koizumi N, Kusano T, Sano H (2001) Two novel genes encoding SNF-1 related protein kinases from Arabidopsis thaliana: differential accumulation of AtSR1 and AtSR2 transcripts in response to cytokinins and sugars, and phosphorylation of sucrose synthase by AtSR2. Mol Gen Genet 264: 674-681 [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Bush DR (1998) Sucrose is a signal molecule in assimilate partitioning. Proc Natl Acad Sci USA 95: 4784-4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991-1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi GM, Zhou L (2001) Carbon and nitrogen sensing and signaling in plants: emerging “matrix effects.” Curr Opin Plant Biol 4: 247-253 [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SE, Provan F, Milne FC, Morrice N, MacKintosh C (2000) 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. EMBO J 19: 2869-2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehness R, Ecker M, Godt DE, Roitsch T (1997) Glucose and stress independently regulate source and sink metabolism and defense mechanisms via signal transduction pathways involving protein phosphorylation. Plant Cell 9: 1825-1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR (1994) Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J 5: 765-771 [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15-S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI (2002) ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol 5: 26-32 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA 2 domain protein. Plant Cell 10: 1043-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Ito M, Nishida I, Watanabe A (2000) Multiple signaling pathways in gene expression during sugar starvation: pharmacological analysis of din gene expression in suspension-cultured cells of Arabidopsis. Plant Physiol 124: 1139-1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P (2001) Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr Opin Plant Biol 4: 387-391 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117-1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280: 196-203 [DOI] [PubMed] [Google Scholar]
- Graham IA, Denby KJ, Leaver CJ (1994) Carbon catabolite repression regulates glyoxylate gene expression in cucumber cycle. Plant Cell 6: 761-772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford NG, Hardie DG (1998) SNF1-related protein kinases: global regulators of carbon metabolism in plants? Plant Mol Biol 37: 735-748 [DOI] [PubMed] [Google Scholar]
- Halford NG, Hey S, Jhurreea D, Laurie S, McKibbin RS, Paul M, Zhang Y (2003) Metabolic signalling and carbon partitioning: role of Snf1-related (SnRK1) protein kinase. J Exp Bot 54: 467-475 [DOI] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S (2000) The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant J 23: 577-585 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106-111 [DOI] [PubMed] [Google Scholar]
- Jang J-C, Sheen J (1994) Sugar sensing in higher plants. Plant Cell 6: 1665-1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J-C, Sheen J (1997) Sugar sensing in higher plants. Trends Plant Sci 2: 208-214 [Google Scholar]
- Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 387-405 [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 13: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Choi H, Im M, Kim SY (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343-357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE (1996) Carbohydrate modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509-540 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23: 587-596 [DOI] [PubMed] [Google Scholar]
- Leon P, Sheen J (2003) Sugar and hormone connections. Trends Plant Sci 8: 110-116 [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49: 199-222 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH (2000) A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol 41: 541-547 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH (2001) A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA 98: 4782-4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua NH (2003) AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev 17: 410-418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Alpi A, Perata P (2000) Glucose and disaccharide-sensing mechanisms modulate the expression of alpha-amylase in barley embryos. Plant Physiol 123: 939-948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W, Liu Y, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332-336 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Lynch TJ, Finkelstein RR (2001) Physical interactions between ABA response loci of Arabidopsis. Plant J 26: 627-635 [DOI] [PubMed] [Google Scholar]
- Quesada V, Ponce M, Micol J (2000) Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154: 421-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz M (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633-646 [DOI] [PubMed] [Google Scholar]
- Rolland F, Moore B, Sheen J (2002) Sugar sensing and signaling in plants. Plant Cell 14: S185-S205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421-433 [DOI] [PubMed] [Google Scholar]
- Rook F, Gerrits N, Kortstee A, Kampen Mv, Borrias M, Weisbeek P (1998) Sucrose-specific signalling represses translation of the Arabidopsis ATB2 bZIP transcription factor gene. Plant J 15: 253-263 [DOI] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) The complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41-48 [DOI] [PubMed] [Google Scholar]
- Sheen J, Zhou L, Jang JC (1999) Sugars as signaling molecules. Curr Opin Plant Biol 2: 410-418 [DOI] [PubMed] [Google Scholar]
- Signora L, De Smet I, Foyer CH, Zhang H (2001) ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J 24: 655-662 [DOI] [PubMed] [Google Scholar]
- Smeekens S (1998) Sugar regulation of gene expression in plants. Curr Opin Plant Biol 1: 230-234 [DOI] [PubMed] [Google Scholar]
- Smeekens S (2000) Sugar-induced signal transduction in plants. Annu Rev Plant Physiol Plant Mol Biol 51: 49-81 [DOI] [PubMed] [Google Scholar]
- Söderman EM, Brocard IM, Lynch TJ, Finkelstein RR (2000) Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol 124: 1752-1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Ecker JR (2000) Ethylene signaling: from mutants to molecules. Curr Opin Plant Biol 3: 353-360 [DOI] [PubMed] [Google Scholar]
- Takeda S, Mano S, Ohto M, Nakamura N (1994) Inhibitors of protein phosphatases 1 and 2A block the sugar-inducible gene expression in plants. Plant Physiol 106: 567-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku K, Loreti E, Vernieri P, Alpi A, Perata P, Yamaguchi J (2000) Glucose modulates the abscisic acid-inducible Rab16A gene in cereal embryos. Plant Mol Biol 42: 451-460 [DOI] [PubMed] [Google Scholar]
- Werner JE, Finkelstein RR (1995) Arabidopsis mutants with reduced response to NaCl and osmotic stress. Physiol Plant 93: 659-666 [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451-461 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439-473 [Google Scholar]
- Zhang H, Jennings AJ, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51: 51-59 [PubMed] [Google Scholar]
- Zhou L, Jang J-C, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294-10299 [DOI] [PMC free article] [PubMed] [Google Scholar]