Abstract
Transgenic Medicago truncatula plants were produced harboring chimeric gene constructs of the glutamine synthetase (GS) cDNA clones (MtGS1a or MtGS1b) fused in sense or antisense orientation to the nodule-specific leghemoglobin promoter Mtlb1. A series of transgenic plants were obtained showing a 2- to 4-fold alteration in nodule GS activity when compared with control plants. Western and northern analyses revealed that the increased or decreased levels of GS activity correlate with the amount of cytosolic GS polypeptides and transcripts present in the nodule extracts. An analysis of the isoenzyme composition showed that the increased or decreased levels of GS activity were attributable to major changes in the homo-octameric isoenzyme GS1a. Nodules of plants transformed with antisense GS constructs showed an increase in the levels of both asparagine synthetase (AS) polypeptides and transcripts when compared with untransformed control plants, whereas the sense GS transformants showed decreased AS transcript levels but polypeptide levels similar to control plants. The polypeptide abundance of other nitrogen metabolic enzymes NADH-glutamic acid synthase and aspartic acid amino-transferase as well as those of major carbon metabolic enzymes phosphoenolpyruvate carboxylase, carbonic anhydrase, and sucrose synthase were not affected by the GS-gene manipulations. Increased levels of AS polypeptides and transcripts were also transiently observed in nodules by inhibiting GS activity with phosphinothricin. Taken together, the results presented here suggest that GS activity negatively regulates the level of AS in root nodules of M. truncatula. The potential role of AS in assimilating ammonium when GS becomes limiting is discussed.
Nitrogen is the major nutrient limiting plant growth and crop yield, and thus many studies have been devoted to the mechanisms by which it is taken up and used by plants. Legumes obtain a significant fraction of their nitrogen from atmospheric N2 through symbiotic association with nitrogen fixing bacteria, termed rhizobia. The ammonium produced by nitrogen fixation is mainly released from the rhizobial bacteroids into the infected cells of the root nodules where it is assimilated into the organic pools by plant Gln synthetase (GS; EC 6.3.1.2). GS in conjunction with NADH-Glu synthase (NADH-GOGAT, EC 1.4.1.14) operates the Glu synthase cycle leading to the synthesis of Gln and Glu, which then serve as nitrogen donors for the biosynthesis of essentially all nitrogenous compounds. In temperate legumes, fixed nitrogen is exported from the nodules to the rest of the plant mainly as Asn, which is synthesized by the concerted action of two additional enzymes, Asp aminotransferase (AAT, EC 2.6.1.1) and Asn synthetase (AS; EC 6.3.5.4). The overall pathway of ammonium assimilation to Asn in nodules requires oxaloacetate as carbon skeleton, which is produced by metabolism of photosynthate (involving a nodule-enhanced Suc synthase [SUCS]) in conjunction with nodule CO2 fixation via the enzymes carbonic anhydrase (CA) and phosphoenolpyruvate carboxylase (PEPC). The metabolic activities of all of these enzymes are tightly linked as supported by their coordinated nodule-enhanced expression during root nodule development (for review, see Gordon et al., 2001). Biochemical and molecular analysis indicates that nodule-specific or nodule-enhanced forms of these isoenzymes are responsible for the majority of the increased enzymatic activities (Pathirana et al., 1997; Shi et al., 1997; Hohnjec et al., 1999; Trepp et al., 1999; Yoshioka et al., 1999; Galvez et al., 2000). Being the first enzyme of the pathway, GS is in a key position to regulate nitrogen assimilatory pathways in nodules.
In higher plants, GS is an octameric enzyme of 310 to 350 kD that occurs as a number of isoenzymes, located both in the plastids (GS2) and in the cytosol (GS1). These GS subunits are encoded by small multigene families of generally three to six members (Forde and Cullimore, 1989; McGrath and Coruzzi, 1991; Peterman and Goodman, 1991; Li et al., 1993). Individual GS genes are differentially regulated in different plant organs and under different physiological conditions (Lam et al., 1996). In all legumes studied to date, the expression of at least one GS1 gene is greatly enhanced during nodule development (Forde and Cullimore, 1989). The model legume Medicago truncatula contains the smallest GS gene family identified to date in a higher plant, comprising only three expressed genes, MtGS1a, MtGS1b (encoding cytosolic GS), and MtGS2 (encoding the plastid GS; Stanford et al., 1993; Melo et al., 2003). We have previously studied the expression and regulation of cytosolic GS in root nodules of M. truncatula (Carvalho et al., 1997; Carvalho et al., 2000a) and have shown that MtGS1a is highly up-regulated in the central infected cells of M. truncatula root nodules and accounts for the production of more than 90% of plant nodule GS activity.
A relevant question remaining to be answered is whether GS in nodules plays a regulatory role in controlling the flux through the nitrogen assimilatory pathway that could eventually lead to changes in plant productivity. There is evidence from studies using a GS-inhibitor producing bacterium that the activity of GS in nodules may control the yield of plants growing symbiotically on dinitrogen (Knight and Langston-Unkefer, 1988). Difficulties in repeating this experiment, though, have lead to skepticism of its results. The use of genetically modified plants deregulated for this particular step in the metabolic pathway provides an invaluable alternative tool to address this question (Harrison et al., 2000).
Recently, various approaches have been undertaken to modify ammonium assimilation in transgenic plants that have greatly contributed to the understanding of the precise roles and regulation of the enzymes involved (for review, see Hirel and Lea, 2001). Several attempts have been made to manipulate the GS levels by either overexpression or down-regulation of GS genes, most of them using constitutive promoters and heterologous transgenic systems. Not all of these genetic manipulations have been successful, particularly the anti-sense strategy has rarely been successful with GS (see Temple et al., 1994; Harrison et al., 2000; Hirel and Lea, 2001). However, one successful example is the reduction of GS using an anti-sense GS behind a phloem-specific promoter, which has revealed that GS in the phloem plays a major role in Pro production (Brugière et al., 1999). More recent work on plants overexpressing GS point to a relationship between GS levels and plant productivity although with mixed results. The over-expression of cytosolic GS in roots and nodules of Lotus japonicus appears to decrease plant productivity (Limami et al., 1999; Harrison et al., 2000), whereas in tobacco (Nicotiana tabacum) and poplar (Populus spp.), the overexpression of distinct isoforms of GS appears to stimulate growth (Gallardo et al., 1999; Migge et al., 2000; Fuentes et al., 2001; Oliveira et al., 2002). In this study, using an homologous transgenic system and a native promoter, we have succeeded in modulating GS activity, specifically in root nodules of M. truncatula, and we show that changes in GS activity negatively regulate the expression of AS.
RESULTS
Construction of Nodule-Specific Cytosolic GS-Sense/Antisense Chimeric Genes
To alter GS levels specifically in the central infected cells of root nodules, we selected a promoter able to drive a strong and nodule-specific expression. The M. truncatula gene Mtlb1 encoding leghemoglobin has been previously shown to be specifically and strongly expressed in root nodules of M. truncatula by northern analysis (Gallusci et al., 1991). A gene construct consisting of a 2.1-kb Mtlb1 promoter fragment driving β-glucuronidase and containing the 3′ nopaline synthase (nos) terminator (kindly provided by D. Barker) was introduced into the homologous transgenic system to analyze the cellular expression driven by this promoter fragment. Reporter gene expression driven by the Mtlb1 promoter was found to be very strong and exclusively located in the central infected cells of the nitrogen-fixing zone (Fig. 1). No β-glucuronidase (GUS) activity was observed in uninfected cells or in the peripheral tissues or vascular bundles. This promoter fragment was therefore chosen to specifically modulate cytosolic GS activity in the infected cells of M. truncatula root nodules. To achieve this aim, four constructs were made consisting of the 2.1-kb Mtlb1 promoter fragment fused to each of the M. truncatula cytosolic GS expressed cDNAs (MtGS1a and MtGS1b), in both sense and antisense orientation, followed by the 3′ nos terminator. The resulting constructs were cloned into the binary vector pBIN19 (Bevan, 1984) and were designated LbSA, LbSB, LbASA, and LbASB.
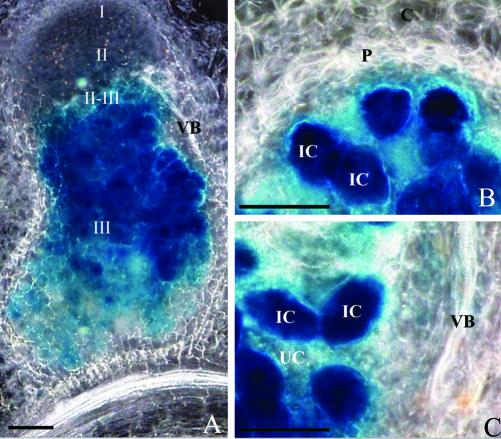
Figure 1.
Histochemical localization of GUS activity in longitudinal (A) and transversal (B and C) sections of root nodules of transgenic M. truncatula plants expressing an Mtlb1 2.1-kb promoter fragment-GUS fusion. Nodules were sectioned (80 μm thick), assayed for GUS activity, and photographed by dark-field microscopy. The blue precipitate indicates the location of GUS activity. I through III, Histologically defined nodule zones; IC, infected cells; UC, uninfected cells; P, parenchyma; C, cortex; VB, vascular bundle. Bars = 100 μm.
Selection of Transgenic Plants with Altered GS Activity in Root Nodules
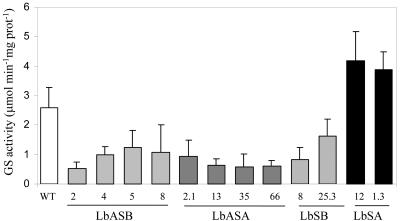
The binary constructs previously described were used to produce primary M. truncatula transformants by Agrobacterium tumefaciens-mediated transformation. We were able to regenerate 36 kanamycin-resistant transgenic M. truncatula plants. These T0 plants were propagated clonally as stem cuttings that when rooted were inoculated with Sinorhizobium meliloti strain 2011. GS activity was determined in root nodules of at least six independent T0 plants from each transformant line. From the 36 regenerated plants, 12 showed alterations in root nodule GS activity, when compared with untransformed control plants (Fig. 2). Four antisense GS1b (LbASB) and four antisense GS1a (LbASA) plants showed 2- to 4-fold reductions in GS activity. Two sense GS1a (LbSA) plants possessed a 2-fold increase in GS activity, whereas two sense GS1b (LbSB) plants showed the opposite effect, a reduction in GS activity. The GS activity levels were unaltered in both leaves and roots of the transgenic plants (data not shown). The transformants showed no visible phenotypic differences when compared with control plants. The number of transgene insertions in the genome of the plants that were showing alterations in activity was analyzed by Southern blot (data not shown), and analysis revealed one to three insertions in different transformants. The number of copies inserted in the genome could not be directly related to the GS activity levels in the nodules. Because all of the analyzed sense or antisense lines behaved in a qualitatively similar way, further studies used the two lines showing the greatest reduction (LbASB-2, antisense) and increase (LbSA-12, sense) in GS activity.
Figure 2.
Root nodule GS activity in transgenic plants. GS activity was determined in root nodules of at least six plants of each of the 12 independently transformed lines harboring constructs LbASB, LbASA, LbSB, LbSA, and compared with untransformed control plants (WT). The error bars represent the sd.
Transformants with Altered GS Activity Levels Exhibit Corresponding Alterations in Enzyme Transcripts and Polypeptides
To evaluate whether the alterations in enzyme activity could be correlated with their protein and transcript levels, nodule extracts from the selected transgenic plants were analyzed both by western and northern blots (Fig. 3, A and B). All analyses were performed in two independent plants from the selected lines to assess possible variations between individuals. The variations in enzyme activity could be proportionally correlated with the amounts of cytosolic GS polypeptides (GS1) detected by western-blot analysis of crude nodule extracts, whereas plastid GS (GS2) appeared not to be altered by these manipulations (Fig. 3A). Quantitation of multiple protein blots by image densitometry demonstrated a consistent reduction or increase in cytosolic GS protein content that was proportional to the levels of GS activity in root nodules, i.e. 3-fold reduction in transformant LbASB-2 and a 2-fold increase in transformant LbSA-12.
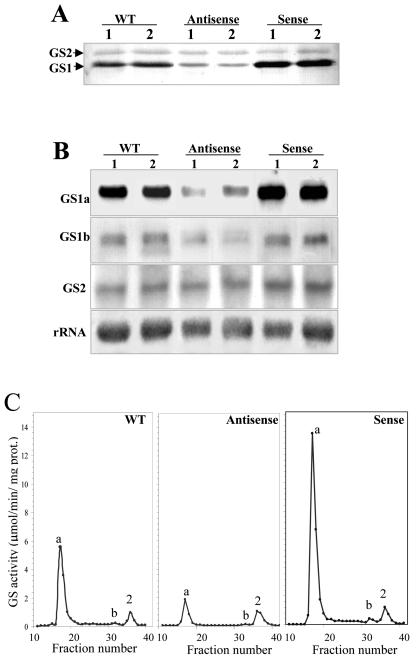
Figure 3.
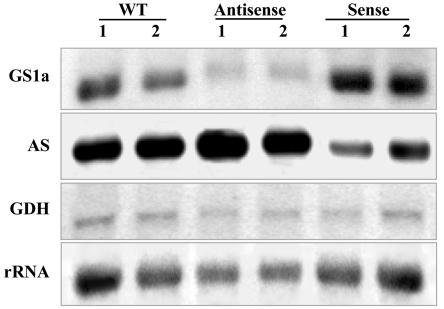
Comparison of GS polypeptides, transcripts, and isoenzyme levels in root nodules of two untransformed control plants (WT) and two independent plants of the transgenic lines LbASB-2 (Antisense) and LbSA-12 (Sense), showing the lowest and highest GS activity levels, respectively. A, Western-blot analysis of GS polypeptides. Equal amounts of proteins (10 μg) were loaded on the gel. GS1 and GS2 designate cytosolic and plastid GS, respectively. B, Analysis of mRNA abundance for the three M. truncatula GS genes. Total RNA (30 μg) was separated electrophoretically, blotted, and probed with specific GS probes for MtGS1a, MtGS1b, and MtGS2. The hybridization profile with a 25S rRNA gene probe is shown as an indicator of RNA loading. C, Elution profiles of GS activity following IEX-HPLC of soluble extracts from wild-type and transgenic nodules. The position of the homo-octameric GS1a (a), GS1b (b), and GS2 (2) isoenzymes are indicated.
To assess whether the alterations in GS enzyme activity and polypeptides were resulting from changes in the steady-state levels of the cytosolic transcripts and to determine which GS genes were being affected by the genetic manipulations, we analyzed the levels of each independent GS transcript by northern blots (Fig. 3B). Total nodule RNA from two independent transgenic plants from lines LbASB-2 and LbSA-12 and from the wild-type control plant were separated electrophoretically, blotted, and hybridized with gene-specific probes from the 3′-untranslated regions (UTRs) of MtGS1a, MtGS1b, and MtGS2. The hybridization signal was quantified, and the band intensities were standardized against the intensity of the 25S rRNA bands. MtGS1a was the most affected GS gene showing a remarkable reduction in transcripts in the antisense plants and a 2-fold increase in the GS overexpressing plants (Fig. 3B). Due to the very high homology between MtGS1a and MtGS1b coding sequences, the expression of MtGS1b mRNA in antisense orientation was capable of decreasing both MtGS1a and MtGS1b transcripts. Densitometry analysis suggests that the amount of MtGS1b transcripts remained unaltered in the sense plants and that MtGS2 transcript abundance was not greatly affected by either of the genetic manipulations (Fig. 3B).
The GS isoenzyme composition in the transgenic nodules was analyzed by ion-exchange HPLC (IEXHPLC; Fig. 3C). A comparison of the column elution profile of GS activity of nodule extracts from the transgenic plants revealed that homo-octameric GS1a was the major, if not the only, GS holoenzyme affected. Using the plastid GS2 as a reference, which remains unaltered in the transgenic plants, there is a 3-fold decrease in activity of the GS1a homo-octamer (elution at fraction 17) in antisense plants and a 2-fold increase in sense plants, when compared with the non-transformed plants. In all cases, the homo-octamer GS1b represents a minor proportion of GS activity.
Analysis of the Effects of the GS Manipulations on the Expression of Other Metabolic Enzymes
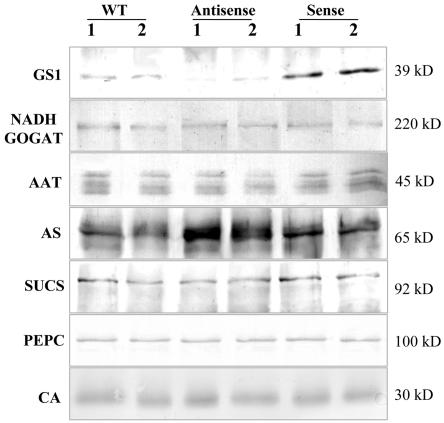
Using specific antibodies raised against the nodule-enhanced forms of nitrogen assimilatory enzymes from the closely related species alfalfa (Medicago sativa), we analyzed the levels of NADH-GOGAT, AAT, and AS by western blots (Fig. 4). The antibody raised against the alfalfa root nodule NADH-GOGAT (Anderson et al., 1989) recognized a single polypeptide with an estimated molecular mass of 220 kD in M. truncatula root nodule protein extracts, which was maintained at similar levels in sense and antisense GS plants as well as in the untransformed control plants. The antibody raised against the nodule-enhanced AAT-2 from alfalfa (Robinson et al., 1994) detected four polypeptides with approximate molecular masses of 45 kD, which probably correspond to different AAT M. truncatula isoenzymes. None of the polypeptides appears to be significantly altered in the nodules of the transgenic plants. The levels of AS were evaluated on western blots probed with an antibody raised against AS from alfalfa (Shi et al., 1997), which recognized a single polypeptide with an estimated molecular mass of 65 kD, whose abundance was found to be 2-fold increased in antisense plants and maintained at levels similar to the control in sense plants. To ensure that the differences detected were not due to differences in the amount of protein loaded on the gel, GS and AS polypeptide levels were analyzed in the same blot, together with SUCS. The membranes were sequentially incubated with AS, GS, and SUCS antibodies and developed with different color substrates. As can be observed in Figure 4, SUCS polypeptides of approximately 92 kD were maintained at about constant levels in anti-sense, sense, and control in contrast to the inverse changes in the abundance of the other two proteins. We also compared the levels of two other major carbon metabolic enzymes PEPC and CA in the GS-modulated transgenic plants and control untransformed plants by western blot using specific antibodies (Fig. 4). The antibodies detected single polypeptides in root nodules of M. truncatula of the expected molecular mass for each enzyme, 100 kD and 30 kD for PEPC and CA, respectively, but neither of the enzymes appear to be affected by the consistent alterations in GS activity.
Figure 4.
Western-blot analysis of polypeptide abundance for several key enzymes involved in carbon and nitrogen metabolism. Detection with AS, GS, and SUCS antibodies was performed in the same membrane for direct comparison. Equal amount of proteins were loaded in each gel but with varying quantities according to the abundance of the polypeptides in root nodules: 5 μg for GS, AS, and SUCS; 10 μg for PEPC and CA; and 20 μg for NADH-GOGAT. Plastid GS was not detected due to the low amount of protein loaded (5 μg). The approximate molecular masses of the polypeptides are indicated.
To assess whether the changes observed in the levels of AS polypeptides in the GS-modulated transgenic plants were the result of changes in the steady-state levels of AS transcripts, RNA blots were performed (Fig. 5). A M. truncatula cDNA corresponding to the nodule-enhanced AS of alfalfa (Shi et al., 1997) was used as probe. AS transcripts were consistently increased in plants down-regulated for GS and decreased in the overexpressing GS plants. Because GDH could represent an alternative route for ammonia assimilation in the transgenic lines, its transcript levels were also analyzed by northern blot. GDH transcript levels, when related to the rRNA, appeared not to be altered in the transgenic plants.
Figure 5.
Analysis of mRNA abundance for AS and Glu dehydrogenase (GDH) in sense and antisense GS transgenic plants. Total RNA from control and transgenic nodules (30 μg) was separated electrophoretically, blotted, and probed with specific probes for GS1a, AS, and GDH. The hybridization profile with a 25S rRNA gene probe is shown as an indicator of RNA loading.
Inhibiting GS Activity Using the Specific GS Inhibitor Phosphinothricin (PPT) Leads to Increased Expression of AS
To assess whether the increased levels of AS detected in the GS-antisense transgenic plants could also be observed by inhibiting total GS activity, we applied PPT to nodulated roots of wild-type plants and analyzed AS and GS content in nodules collected at different times after application of the herbicide. PPT is an analog of Glu and binds to the active site of GS irreversibly inactivating the enzyme (Manderscheid and Wild, 1986). At 24 h but not 8 h after PPT addition, the plants were beginning to show signs of chlorosis and stress.
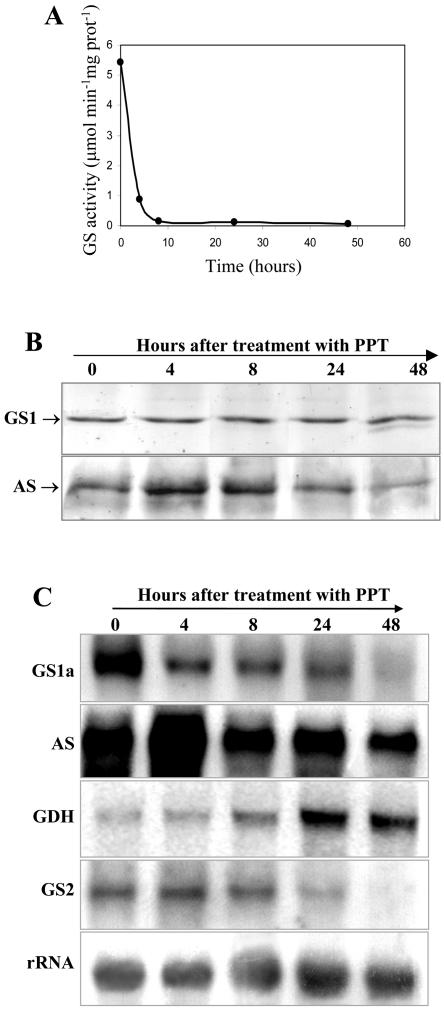
The inhibition of GS activity in root nodules was very effective; GS activity was totally lost after 8 h application of the inhibitor (Fig. 6A), and a similar inhibition of GS activity was observed in roots and in leaves (data not shown). Western-blot analysis revealed that the amount of GS protein was maintained, even though its activity was almost zero, suggesting that GS was inactivated but not yet degraded (Fig. 6B). Interestingly, 48 h after PPT application, a lower Mr GS polypeptide was detected, which probably results from a selective proteolysis of the enzyme. To directly compare the amount of GS and AS, the same membrane was subsequently hybridized with anti-AS antibodies. We could detect an increase in AS polypeptide content in root nodules after 4 and 8 h of PPT treatment, which then decreased after 24 and 48 h.
Figure 6.
Analysis of GS, AS, and GDH expression in nodules of PPT-treated plants. The analyses were performed in nodules collected at 0, 4, 8, 24, and 48 h after PPT treatment. A, Analysis of GS activity in the nodules of wild-type plants after application of PPT. B, Western-blot analysis of GS and AS polypeptides. Equal amounts of proteins (5 μg) were loaded on the gel. The same membrane was sequentially hybridized with AS and GS antibodies for direct comparison. Plastid GS was not detected due to the low amount of protein loaded on the gel. C, Northern-blot analysis of GS1a, AS, GDH, and GS2 transcripts. Equal amounts of total RNA (30 μg) were separated electrophoretically, blotted, and probed with gene-specific probes. The hybridization profile with a 25S rRNA gene probe is shown as an indicator of RNA loading.
This increase in AS content in the herbicide-treated nodules appears to occur at the transcript level, because it was also accompanied by an increase in the corresponding mRNA, which appears to be maximal after 4 h of incubation with the herbicide, then decreasing to normal levels (Fig. 6C). MtGS1a transcripts decreased in abundance in the course of the PPT treatment as well as the plastid GS mRNA (Fig. 6C). Interestingly, GDH transcripts showed a clear increase in abundance after 24 and 48 h. This effect was not only observed in nodules but also in roots and leaves (data not shown).
DISCUSSION
To evaluate whether GS in nodules could play a regulatory role in controlling the flux through the nitrogen assimilatory pathway, we have produced transgenic plants deregulated for this particular step in the metabolic pathway. Making use of the model legume M. truncatula in which we have previously characterized the GS genes (Carvalho et al., 1997, 2000a, 2000b; Melo et al., 2003), we have succeeded in both increasing and decreasing GS activity specifically in nodules, by expressing GS in sense/antisense orientation under the direction of a strong and nodule-specific promoter. The choice of the Mtlb1 leghemoglobin promoter relied on its nodule-specific expression that follows the same time course of expression as MtGS1a, the nodule-enhanced M. truncatula GS gene (Gallusci et al., 1991; Stanford et al., 1993). By analyzing GUS fusions to this promoter, it is clear that it is also highly specific for the infected cells of the nodule (Fig. 1), thus altering GS levels only in the cells in which ammonium is being produced by rhizobial nitrogen fixation.
Most of the previous attempts to manipulate GS levels by either overexpression or down-regulation of GS genes have used constitutive promoters and heterologous transgenic systems, sometimes leading to alterations in whole-plant metabolism and sometimes leading to lethality (see Temple et al., 1994; Harrison et al., 2000). These experiments have revealed much about the regulation of GS expression and its importance to the plant (Eckes et al., 1989; Hirel et al., 1992; Temple et al., 1993, 1996, 1998; Vincent et al., 1997; Limami et al., 1999; Gallardo et al., 1999; Migge et al., 2000; Fuentes et al., 2001; Ortega et al., 2001; Oliveira et al., 2002), but evaluating its role in regulating precise cellular pathways has not always been possible. One successful example, however, is the case of expressing an antisense GS using a phloem-specific promoter, which has revealed that GS in the phloem plays a major role in Pro production (Brugière et al., 1999). Very recently, Harrison et al. (2003) have used an antisense strategy to successfully lower GS activity in nodules of L. japonicus. The success of these strategies and of our strategy may rely first on targeting a single cell type and thus not altering nitrogen assimilation in other parts of the plant. A second important factor may be the use of promoters from the same plant family as the transgenic system thus ensuring that all transacting factors required for correct expression are available.
Our manipulations resulted in up to 2-fold increases and 4-fold decreases in GS activity specifically in nodules (Fig. 2). Furthermore, the increased or decreased activity could be directly attributable to changes in a single GS holoenzyme, homo-octameric GS1a, which makes up more than 90% of the nodule GS activity in normal plants (Fig. 3C) and is expressed in the same cells as the Mtlb1 promoter (Carvalho et al., 2000a). The observed modifications in GS activity in the transgenic plants were associated with proportional increases/reductions in transcripts and polypeptides (Fig. 3), suggesting that transcript regulation is a main control point of GS expression in root nodules of M. truncatula. Although a slight decrease in GS1b-specific mRNA was detected in the GS-antisense plants, the activity of the corresponding octameric isoenzyme is too low to significantly affect the overall GS activity (Fig. 3).
Analysis of these transgenic plants has revealed a remarkable negative regulation of AS by GS in root nodules. Thus when the level of GS is reduced, the expression of AS is increased and vice versa (Figs. 4 and 5). This regulation appears to be specific for AS because no changes were observed in the expression of five other key nitrogen and carbon assimilatory enzymes in nodules (NADH-GOGAT, AAT, CA, PEPC, and SUCS). The mechanism of AS regulation most likely occurs at the transcript level because the abundance of the AS mRNAs were affected, and in most cases, this was accompanied by associated changes in AS protein. However, when GS was increased by the sense strategy, little reduction was seen in AS protein suggesting that the observed lower level of AS mRNA could support the normal level of AS protein synthesis (Fig. 5). The inverse relationship between GS and AS was also seen, albeit transiently, when GS activity was inhibited by PPT (Fig. 6). In this experiment, the abundance of GS protein remained constant during the AS induction, suggesting that the regulation of AS is caused by changes in GS activity rather than in the levels of GS protein.
The finding that the levels of AS transcripts and polypeptides in the transgenic nodules were consistently increased when GS was reduced (Figs. 4 and 5) suggests that AS may compensate for the reduced GS ammonium assimilatory activity. It is known that AS can use ammonium instead of Gln as a nitrogen donor when the level of ammonium is high (Oaks and Ross, 1984; Lam et al., 1996), for example, the alfalfa nodule AS has only a 20-fold lower affinity for ammonium than for Gln (Shi et al., 1997). Consistent with our hypothesis, Harrison et al. (2003) have shown that a reduction in nodule GS in L. japonicus nodules leads to an increase in ammonium and Asn levels. However, to function in ammonium assimilation, AS activity also requires Asp. The fact that the increase in AS expression is not maintained when GS is completely inhibited by PPT (Fig. 6) suggests that maintaining partial GS activity is essential for maintaining the higher level of AS. Plant GS may thus be required to synthesize enough Gln to support Asp biosynthesis via NADH-GOGAT and AAT, in conjunction with oxaloacetate production via SUCS, CA, and PEPC. The levels of all of these enzymes were maintained in the GS-modulated plants (Fig. 5). The resulting pathway of dual ammonium assimilation via GS and AS would be energetically more favorable than ammonium assimilation through GS alone, involving an estimated 17% savings in energy requirements.
This potential energy savings, through partial assimilation of ammonium by plant AS, could explain the increase in plant productivity that has been observed in certain situations where GS levels were reduced either in genetic variants (Limami et al., 1999) or by the use of a GS inhibitor-producing bacterium (Knight and Langston-Unkefer, 1988). Moreover, if AS is already partially involved in nodule ammonium assimilation, this would offer an explanation of why overexpression of GS in root nodules of Lotus corniculatus leads to a decrease in Asn content and a decrease in biomass production (Harrison et al., 2000). However, in the most recent paper addressing this question (Harrison et al., 2003), only minor increases in biomass were observed following a specific reduction of nodule GS activity. Thus more studies are clearly required to address this point.
Finally it is noteworthy that in a situation where GS is completely inhibited and the associated increase in AS is transient (addition of PPT; Fig. 6), a remarkable increase in GDH expression was observed. This increase could be viewed as a desperate attempt by the plant to assimilate ammonium using the aminating activity of GDH. However, most evidence suggests that this mitochondrial enzyme usually operates in a deaminating direction and is induced in situations such as carbon starvation where the enzyme plays a role in amino acid degradation and production of organic acids for carbon recycling (Miflin and Habash, 2002). The experiment on PTT application may suggest that GDH is induced also in conditions of perceived nitrogen starvation (due here to lack of GS) where it may function to promote nitrogen rather than carbon recycling through amino acid degradation. Alternatively, the GDH induction may be a response to senescence, clear signs of which were evident at 24 h.
In conclusion, we have obtained transgenic M. truncatula plants with reduced and increased levels of GS in root nodules and have shown using these plants that GS negatively regulates AS expression in nodules. This conclusion was reinforced by studies on plants in which GS was chemically inhibited with PPT. This latter experiment also revealed a role of GS in negatively regulating the expression of GDH and that the effect of GS on AS and GDH expression appears to be due to its enzymatic activity rather than the level of its protein. Many questions remain to be answered concerning the mechanism of this effect and its consequence on the nodule nitrogen and carbon metabolic pathways in the plant. In particular, it needs to be established whether the nitrogen assimilatory pathways in the GS-reduced nodules are more energy efficient and whether this has a positive effect on biomass production. Clearly, these transgenic plants, which are altered in a single GS holoenzyme in a single cell type, will serve as a valuable tool for such investigations.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants of Medicago truncatula Gaertn. cv Jemalong (genotype H39) used for plant transformation were maintained in in vitro culture in an environmental cabinet at a temperature of 23°C by day and 19°C by night, with 13-h daylength and light intensity of 150 to 200 μmol m-2 s-1. After regeneration, the transgenic plants were grown aeroponically on ammonium nitrate as described by Lullien et al. (1987) under 13-h-light (23°C)/11-h-dark (19°C) cycles. For nodule induction, the plants were starved of nitrogen for 7 d before inoculation with Sinorhizobium meliloti strain RCR 2011 (GMI51). For the PPT experiment, 0.5 mm PPT was added to the growth medium of aeroponically grown nodulated wild-type plants (genotype H39). Plants were harvested just before PPT treatment and at 4, 8, 24, and 48 h after application of the herbicide. Nodules, roots, and leaves were separated and immediately frozen in liquid nitrogen.
Construction of M. truncatula Mtlb1 Gene Promoter-GS Fusions
Full-length GS cDNAs were isolated from plasmids pTrcGSa and pTrcGSb containing the complete coding sequences of MtGS1a and MtGS1b (accession numbers Y10267 and Y10268, respectively). These plasmids contain an NcoI site at the start codon that has been introduced by PCR (Carvalho et al., 1997). A 2.1-kb Mtlb1 leghemoglobin promoter fragment was isolated as a BamHI/NcoI fragment from a binary vector PLP100 containing a Mtlb1 promoter fragment-GUS fusion, which also contains a NcoI site at the GUS start codon. This plasmid was kindly provided by David Barker (Institut National de la Recherche Agronomique, Toulouse, France). The cloning strategy to create sense constructs required several intermediary steps. First, the 5′ end of each cDNA, up to amino acid 80 (BamHI site) was isolated as an NcoI/BamHI fragment and was subcloned into the NcoI/BamHI sites of pCRScript (Stratagene, La Jolla, CA). The Mtlb1 BamHI/NcoI promoter fragment was made blunt on the BamHI side and was inserted into NcoI/EcoRI (blunt) sites of each of the previous constructs, creating intermediary plasmids containing the promoter fused to the 5′ end of each GS cDNA. The rest of the construct, containing the remaining GS coding sequence containing the 3′-UTR from the bean (Phaseolus vulgaris) gln-α GS gene and the nopaline synthetase terminator, was built in another plasmid, to avoid repeated restriction sites. The 3′-UTR of the gln-α 1 cDNA was isolated from plasmid pR-2 (Gebhardt et al., 1986) and was inserted as a BamHI/HindIII fragment into pBluescript KS- (Stratagene). Subsequently, a BamHI/HindIII fragment containing the nos terminator (excised from plasmid pSLJ4K1) was introduced as a blunt fragment at the HindIII (blunt) site of the previous construct. A BamHI fragment containing the GS1a or GS1b cDNA lacking the 5′ end was then inserted into the BamHI sites of the previous constructs. From the resulting construct, a BamHI/XhoI (blunt) fragment (containing the GS cDNA+3′-UTR glnα 1+nosT) was isolated and inserted into BamHI/NotI (blunt) sites of the previously described intermediary vector containing the Mtlb1 promoter plus the 5′ end of each GS cDNA. An SalI fragment containing the 2.1-kb Mtlb1 promoter, full GS1a or GS1b coding sequences, gln-α 3′-UTR and the nos terminator was subsequently cloned into the SalI site of pBIN19 (Bevan, 1984) to generate plasmids LbSA and LbSB.
For the construction of the chimeric antisense GS genes, the nos terminator was introduced at the 5′ end of pBluescript KS- plasmids containing MtGS1a or MtGS1b full-length cDNAs. The previously described nopaline synthetase blunt fragment was introduced at the SmaI site of the pBluescript KS- GS cDNA clones. The Mtlb1 promoter was isolated from the previously described binary vector as an EcoRV/NcoI fragment, and the NcoI site was made blunt and inserted at the XhoI (Blunt) site of the constructs containing the GS-cDNAs fused to the nos terminator. A NotI (blunt)/SalI fragment containing the 2.1-kb Mtlb1 promoter, full GS1a or GS1b cDNAs in antisense orientation, and the nos terminator was subsequently cloned into SmaI/SalI sites of pBIN19 (Bevan, 1984) to generate plasmids LbASA and LbASB.
Transformation of M. truncatula and Regeneration of Transgenic Plants
The binary constructs LbSA, LbSB, LbASA, and LbASB were introduced into the Agrobacterium tumefaciens strain AGL1 and were used to transform leaf segments of M. truncatula Gaertn. cv Jemalong (genotype H39). Kanamycin-resistant plants were regenerated by somatic embryogenesis as described by Chabaud et al. (1996) and propagated in vitro. For further information, see http://medicago.toulouse.inra.fr/Mt/Protocol/Transformation.
GS Extraction and Assay
Plant material was homogenized at 0°C to 4°C in a mortal and pestle with 2 volumes of an extraction buffer containing 10 mm Tris-HCl, pH 7.5, 5 mm sodium Glu, 10 mm MgSO4, 1 mm dithiothreitol, 10% (v/v) glycerol, and 0.05% (v/v) Triton X-100. The homogenates were centrifuged at 13,000g for 20 min, at 4°C. The extracts were assayed for GS activity by the transferase assay (Cullimore and Sims, 1980).
Protein Extraction and Western Immunoblotting
Soluble protein concentration was measured by the Coomassie Blue dye-binding assay (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as a standard. Proteins were separated by 12.5% (w/v) SDS-PAGE and electroblotted onto nitrocellulose membranes. Immunodetection was performed with the polyclonal antibody raised against GS (Cullimore and Miflin, 1984) or with the antibodies raised against NADH-GOGAT (Anderson et al., 1989): AAT-2 (Robinson et al., 1994), AS (Shi et al., 1997), PEPC (Vidal et al., 1980), CA (Galvez et al., 2000), and SUCS (Ross and Davies, 1992). For NADH-GOGAT and PEPC, a 6% (w/v) acrylamide gel was used.
RNA Isolation and Northern-Blot Hybridization
Total RNA was isolated from nodules using TRIzol reagent (Invitrogen, Carlsbad, CA) as described by the manufacturer. RNA samples (30 μg per lane) were separated by electrophoresis in 1.5% (v/v) agarose/15% (v/v) formaldehyde gels and were transferred to positively charged nylon (Amersham Biosciences, Uppsala) membranes by capillary blotting. The DNA probes were prepared from plasmid inserts isolated from agarose gels and labeled with 32P by random priming (Sambrook et al., 1989). Hybridization was performed at 42°C in the presence of 50% (v/v) formamide and was washed to a final washing stringency of 0.2× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) and 0.1% (w/v) SDS at 65°C. Signal intensities were quantified using the densitometer function of ImageQuant (Molecular Dynamics, Sunnyvale, CA). As gene-specific probes for AS and GDH, we used the complete cDNA inserts from expressed sequence tag clones MtBB36C1 and MtBA49D5 from M. truncatula (Journet et al., 2002), homologous to the best characterized alfalfa (Medicago sativa) nodule-expressed genes. Hybridization to a tomato (Lycopersicon esculentum) 25S rRNA probe was used as a standard.
Analysis of GS Isoenzymes by IEX-HPLC
Soluble protein plant extracts were filtered through a 0.2-μm filter and then applied to a DEAE 5PW column (75-× 7.5-mm diameter, Hewlett Packard, Palo Alto, CA) pre-equilibrated in running buffer. Proteins were eluted at 0.5 mL min-1 with running buffer containing no salt (2 min), 0 to 0.1 m KCl (4 min), 0.1 to 0.4 m KCl (44 min), and 0.6 m KCl (10 min). Fractions of 0.5 mL were collected and assayed for GS activity.
Histochemical Localization of GUS Activity
Histochemical staining for GUS activity was performed according to Jefferson et al. (1987). Whole-plant fragments were fixed by vacuum infiltration for 5 min in an ice-cold solution of 0.5% (w/v) p-formaldehyde in 0.1 m phosphate buffer at pH 7.0, followed by incubation on ice for 1 h and two washes in phosphate buffer. The material was cut into 80-μm-thick sections using a vibratome (VT 1000, Leica, Wetzlar, Germany), and the sections were immersed in the GUS substrate solution containing 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucoronide cyclohexylammonium (Biosynth, Staad, Switzerland), 5 mm EDTA, 0.5 mm potassium ferrocyanide, 0.5 mm potassium ferricyanide, and 0.1 m potassium phosphate buffer, pH 7.0. The immersed tissues were incubated in the dark at 37°C for 1 to 16 h, depending on the intensity of the coloration. The sections were briefly cleared with NaOCl to improve the contrast between stained and nonreactive tissues. Samples were mounted in water between a slide and a coverslip and were observed by dark field microscopy using an light microscope (BX50, Olympus, Tokyo) and photographs were taken using an Olympus DP10 digital camera.
Acknowledgments
We gratefully acknowledge Dr. David Barker (Institut National de la Recherche Agronomique, Toulouse, France) for providing the Mtlb1 promoter-β-glucuronidase construct. We are also extremely grateful to the following persons for providing antibodies for this work: Dr. Carrol Vance (University of Minnesota, St. Paul) for antibodies against the alfalfa nodule-enhanced forms of NADH-GOGAT, AAT, and AS; Dr. Martin Crespi (Centre National de la Recherche Scientifique-Gif-sur Ivette, France) for antibodies against CA and PEPC; and Dr. Helge Kuster (University of Bielefeld, Germany) for antibodies against SUCS.
This work was part of the Biotechnology Research and Technological Development Shared Cost Project FIXNET supported by the European Union (grant no. CT 97-2319).
References
- Anderson MP, Vance CP, Heichel GH, Miller SS (1989) Purification and characterization of NADH-glutamate synthase from alfalfa root nodules. Plant Physiol 90: 351-358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan MW (1984) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12: 8711-8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière N, Dubois F, Limami A, Lelandais M, Roux Y, Sangwan RS, Hirel B (1999) Glutamine synthetase in the phloem plays a major role in controlling proline production. Plant Cell 11: 1995-2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho H, Lescure N, de Billy F, Chabaud M, Lima L, Salema R, Cullimore J (2000a) Cellular expression and regulation of the Medicago truncatula cytosolic glutamine synthetase genes in root nodules. Plant Mol Biol 42: 741-756 [DOI] [PubMed] [Google Scholar]
- Carvalho H, Lima L, Salema R, Cullimore J (2000b) Differential expression of the two cytosolic glutamine synthetase genes in various organs of Medicago truncatula. Plant Sci 159: 301-312 [DOI] [PubMed] [Google Scholar]
- Carvalho H, Sunkel C, Salema R, Cullimore J (1997) Heteromeric assembly of the cytosolic glutamine synthetase polypeptides of Medicago truncatula: complementation of a glnA Escherichia coli mutant with a plant domain-swapped enzyme. Plant Mol Biol 35: 623-632 [DOI] [PubMed] [Google Scholar]
- Chabaud M, Larsonneau C, Marmouget C, Huguet T (1996) Transformation of barrel medic (Medicago truncatula Gaertn.) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the MtENOD12 nodulin promoter fused to the gus reporter gene. Plant Cell Rep 15: 305-310 [DOI] [PubMed] [Google Scholar]
- Cullimore JV, Miflin B (1984) Immunological studies on glutamine synthetase using antisera raised to the plant forms of the enzyme from Phaseolus root nodules. J Exp Bot 153: 581-587 [Google Scholar]
- Cullimore JV, Sims AP (1980) An association between photorespiration and protein catabolism: studies with Chlamydomonas. Planta 150: 392-396 [DOI] [PubMed] [Google Scholar]
- Eckes P, Schmitt P, Daub W, Wengenmayer F (1989) Overproduction of alfalfa glutamine synthetase in transgenic tobacco plants. Mol Gen Genet 217: 263-268 [DOI] [PubMed] [Google Scholar]
- Forde BG, Cullimore JV (1989) Glutamine synthetase in higher plants. In BJ Miflin, ed, Oxford Surveys of Plant Molecular and Cell Biology, Vol 6. Oxford University Press, pp 247-296 [Google Scholar]
- Fuentes SI, Allen DJ, Ortiz-Lopez A, Hernandez G (2001) Over-expression of cytosolic glutamine synthetase increases photosynthesis and growth at low nitrogen concentrations. J Exp Bot 52: 1071-1081 [DOI] [PubMed] [Google Scholar]
- Gallardo F, Fu J, Canton FR, Garcia-Gutierrez A, Canovas FM, Kirby EG (1999) Expression of a conifer glutamine synthetase gene in transgenic poplar. Planta 210: 19-26 [DOI] [PubMed] [Google Scholar]
- Gallusci P, Dedieu A, Journet EP, Huguet T, Barker DG (1991) Synchronous expression of leghaemoglobin genes in Medicago truncatula during nitrogen fixing root nodules development and response to exogenously supplied nitrate. Plant Mol Biol 17: 335-349 [DOI] [PubMed] [Google Scholar]
- Galvez S, Hirsch AM, Wycoff KL, Hunt S, Layzell DB, Kondorosi A, Crespi M (2000) Oxygen regulation of a nodule-located carbonic anhydrase in alfalfa. Plant Physiol 124: 1059-1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Oliver JE, Forde BG, Saarelainen R, Miflin B (1986) Primary structure and differential expression of glutamine synthetase genes in nodules, roots and leaves of Phaseolus vulgaris L. EMBO J 5: 1429-1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon AJ, Lea P, Rosenberg C, Trinchant J (2001) Nodule formation and function. In PS Lea, JF Morot-Gaudry, eds, Plant Nitrogen. Springer-Verlag, Berlin, and Institut National de la Recherche Agronomique, Paris, pp 101-138
- Harrison J, Brugière N, Phillipson B, Ferrario-Mery S, Becker T, Limami A, Hirel B (2000) Manipulating the pathway of ammonia assimilation through genetic engineering and breeding: consequences on plant physiology and plant development. In MA Martins-Loução, SH Lips, eds, Nitrogen in a Sustainable Ecosystem: From the Cell to the Plant. Backhuys Publishers, Leiden, The Netherlands, pp 89-101
- Harrison J, Crescenzo MP, Hirel B (2003) Does lowering glutamine synthetase activity in nodules modify nitrogen metabolism and growth of Lotus japonicus L. Plant Physiol 133: 253-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel B, Lea PJ (2001) Ammonia assimilation. In PJ Lea, JF Morot-Gaudry, eds, Plant Nitrogen. Springer-Verlag, Berlin, and Institut National de la Recherche Agronomique, Paris, pp 79-99
- Hirel B, Marsolier MC, Hoarau A, Hoarau J, Brangeon J, Schafer R, Verma DP (1992) Forcing expression of a soybean root glutamine synthetase gene in tobacco leaves induces a native gene encoding cytosolic enzyme. Plant Mol Biol 20: 207-218 [DOI] [PubMed] [Google Scholar]
- Hohnjec N, Becker JD, Puhler A, Perlick AM, Kuster H (1999) Genomic organization and expression properties of the MtSucS1 gene, which encodes a nodule-enhanced sucrose synthase in the model legume Medicago truncatula. Mol Gen Genet 261: 514-522 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kananagh TA, Bevan MW (1987) β-Galactosidase as a sensitive and versatile gene fusion marker in plants. EMBO J 6: 3901-3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Journet EP, van Tuinen D, Gouzy J, Carreau V, Farmer MJ, Niebel A, Schiex T, Crespeau H, Jaillon O, Chatagnier O et al. (2002) Exploring root symbiotic programs in the model legume Medicago truncatula using EST analysis. Nucleic Acids Res 30: 5579-5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight TJ, Langston-Unkefer PT (1988) Enhancement of symbiotic dinitrogen fixation by a toxin-releasing plant pathogen. Science 241: 951-954 [DOI] [PubMed] [Google Scholar]
- Lam HM, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM (1996) The molecular genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol 47: 569-593 [DOI] [PubMed] [Google Scholar]
- Li M, Villemur R, Hussey P, Silflow CD, Grantt JS, Snustad P (1993) Differential expression of six glutamine synthetase genes in Zea mays. Plant Mol Biol 23: 401-407 [DOI] [PubMed] [Google Scholar]
- Limami A, Phillipson B, Ameziane R, Pernollet N, Jiang Q, Roy R, Deleens E, Chaumont-Bonnet M, Gresshoff PM, Hirel B (1999) Does root glutamine synthetase control plant biomass production in Lotus japonicus L.? Planta 209: 495-502 [DOI] [PubMed] [Google Scholar]
- Lullien V, Barker DG, de Lajudie P, Huguet T (1987) Plant gene expression in effective and ineffective root nodules of alfalfa (Medicago sativa). Plant Mol Biol 9: 469-478 [DOI] [PubMed] [Google Scholar]
- Manderscheid DR, Wild A (1986) Studies on the mechanism of inhibition by phosphinothricin of glutamine synthetase isolated from Triticum aestivum. J Plant Physiol 123: 135-142 [Google Scholar]
- McGrath RB, Coruzzi GM (1991) A gene network controlling glutamine and asparagine biosynthesis in plants. Plant J 1: 275-280 [DOI] [PubMed] [Google Scholar]
- Melo P, Lima L, Santos I, Carvalho H, Cullimore J (2003) Expression of plastid-located glutamine synthetase of Medicago truncatula: accumulation of the precursor in root nodules reveals an in vivo control at the level of protein import into plastids. Plant Physiol 132: 390-399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miflin BJ, Habash DZ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53: 979-987 [DOI] [PubMed] [Google Scholar]
- Migge A, Carrayol E, Hirel B, Becker T (2000) Leaf specific overexpression of plastidic glutamine synthetase stimulates the growth of transgenic tobacco seedlings. Planta 210: 252-260 [DOI] [PubMed] [Google Scholar]
- Oaks A, Ross DW (1984) Asparagine synthetase in Zea mays. Can J Bot 62: 68-73 [Google Scholar]
- Oliveira IC, Brears T, Knight TJ, Clark A, Coruzzi GM (2002) Overexpression of cytosolic glutamine synthetase: relation to nitrogen, light, and photorespiration. Plant Physiol 129: 1170-1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega JL, Temple SJ, Sengupta-Gopalan C (2001) Constitutive overexpression of cytosolic glutamine synthetase (GS1) gene in transgenic alfalfa demonstrates that GS1 may be regulated at the level of RNA stability and protein turnover. Plant Physiol 126: 109-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathirana MS, Samac DA, Roeven R, Yoshioka H, Vance CP, Grantt JS (1997) Analysis of phosphoenolpyruvate carboxylase gene structure and expression in alfalfa nodules. Plant J 12: 293-304 [DOI] [PubMed] [Google Scholar]
- Peterman TK, Goodman HM (1991) The glutamine synthetase gene family of Arabidopsis thaliana: light regulation and differential expression in leaves, roots and seeds. Mol Gen Genet 230: 145-154 [DOI] [PubMed] [Google Scholar]
- Robinson DL, Kahn ML, Vance CP (1994) Cellular localisation of nodule enhanced aspartate aminotransferase in Medicago sativa. Planta 92: 202-210 [Google Scholar]
- Ross HA, Davies HV (1992) Purification and characterisation of sucrose synthase from the cotyledons of Vicia faba L. Plant Physiol 100: 1008-1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Shi L, Twary SN, Yoshioka H, Gregerson RG, Miller SS, Samac DA, Grantt JS, Unkefer PJ, Vance CP (1997) Nitrogen assimilation in alfalfa: isolation and characterization of an asparagine synthetase gene showing enhanced expression in root nodules and dark-adapted leaves. Plant Cell 9: 1339-1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford AC, Larsen K, Barker DG, Cullimore JV (1993) Differential expression within the glutamine synthetase gene family of the model legume, Medicago truncatula. Plant Physiol 103: 73-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Bagga S, Sengupta-Gopalan C (1994) Can glutamine synthetase activity levels be modulated in transgenic plants by the use of recombinant DNA technology? Biochem Soc Trans 22: 915-920 [DOI] [PubMed] [Google Scholar]
- Temple SJ, Bagga S, Sengupta-Gopalan C (1998) Down-regulation of specific members of the glutamine synthetase gene family in alfalfa by antisense RNA technology. Plant Mol Biol 37: 535-547 [DOI] [PubMed] [Google Scholar]
- Temple SJ, Heard J, Kunjibettu S, Roche D, Sengupta-Gopalan C (1996) Total glutamine synthetase activity during soybean nodule development is controlled at the level of transcription and holoprotein turnover. Plant Physiol 112: 1723-1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple SJ, Knight TJ, Unkefer PJ, Sengupta-Gopalan C (1993) Modulation of glutamine synthetase gene expression in tobacco by the introduction of an alfalfa glutamine synthetase gene in sense and antisense orientation: molecular and biochemical analysis. Mol Gen Genet 36: 315-325 [DOI] [PubMed] [Google Scholar]
- Trepp GB, Temple SJ, Bucciarelli B, Shi LF, Vance CP (1999) Expression map for genes involved in nitrogen and carbon metabolism in alfalfa root nodules. Mol Plant-Microbe Interact 12: 526-535 [Google Scholar]
- Vidal J, Godbillon G, Gadal P (1980) Recovery of active, highly purified phosphoenolpyruvate carboxylase from specific immunoadsorbent column. FEBS Lett 118: 31-34 [Google Scholar]
- Vincent R, Fraisier V, Chaillou S, Limami MA, Deleens E, Phillipson B, Douat C, Boutin JP, Hirel B (1997) Overexpression of a soybean gene encoding cytosolic glutamine synthetase in shoots of transgenic Lotus corniculatus L. plants triggers changes in ammonium assimilation and plant development. Planta 201: 424-433 [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Gregerson RG, Samac DA, Hoevens KC, Trepp GB, Grantt JS, Vance CP (1999) Aspartate aminotransferase in alfalfa nodules: localization of mRNA during effective and ineffective nodule development and promoter analysis. Mol Plant-Microbe Interact 12: 263-274 [Google Scholar]