Abstract
A project to systematically investigate respiratory supercomplexes in plant mitochondria was initiated. Mitochondrial fractions from Arabidopsis, potato (Solanum tuberosum), bean (Phaseolus vulgaris), and barley (Hordeum vulgare) were carefully treated with various concentrations of the nonionic detergents dodecylmaltoside, Triton X-100, or digitonin, and proteins were subsequently separated by (a) Blue-native polyacrylamide gel electrophoresis (PAGE), (b) two-dimensional Blue-native/sodium dodecyl sulfate-PAGE, and (c) two-dimensional Blue-native/Blue-native PAGE. Three high molecular mass complexes of 1,100, 1,500, and 3,000 kD are visible on one-dimensional Blue native gels, which were identified by separations on second gel dimensions and protein analyses by mass spectrometry. The 1,100-kD complex represents dimeric ATP synthase and is only stable under very low concentrations of detergents. In contrast, the 1,500-kD complex is stable at medium and even high concentrations of detergents and includes the complexes I and III2. Depending on the investigated organism, 50% to 90% of complex I forms part of this supercomplex if solubilized with digitonin. The 3,000-kD complex, which also includes the complexes I and III, is of low abundance and most likely has a III4I2 structure. The complexes IV, II, and the alternative oxidase were not part of supercomplexes under all conditions applied. Digitonin proved to be the ideal detergent for supercomplex stabilization and also allows optimal visualization of the complexes II and IV on Blue-native gels. Complex II unexpectedly was found to be composed of seven subunits, and complex IV is present in two different forms on the Blue-native gels, the larger of which comprises additional subunits including a 32-kD protein resembling COX VIb from other organisms. We speculate that supercomplex formation between the complexes I and III limits access of alternative oxidase to its substrate ubiquinol and possibly regulates alternative respiration. The data of this investigation are available at http://www.gartenbau.uni-hannover.de/genetik/braun/AMPP.
Structural basis for oxidative phosphorylation in mitochondria are five protein complexes termed NADH dehydrogenase (complex I), succinat dehydrogenase (complex II), cytochrome c reductase (complex III, which is a functional dimer), cytochrome c oxidase (complex IV), and ATP synthase (complex V). They were first characterized about 40 years ago by solubilizations of mitochondrial membrane proteins using detergents and differential precipitations or chromatographic separations. According to the popular “liquid state” model, the protein complexes of the respiratory chain are randomly arranged in the membrane and freely diffuse in lateral direction within the inner mitochondrial membrane (for review, see Rich, 1984). However, other results rather indicate an ordered association of these protein complexes forming larger structures. These so-called “supercomplexes” were first described for bacteria (Berry and Trumpower, 1985; Sone et al., 1987; Iwasaki et al., 1995; Niebisch and Bott, 2003). Later the existence of respiratory supercomplexes was also reported for yeast and mammalian mitochondria (Schägger and Pfeiffer, 2000).
In Brewer's yeast (Saccharomyces cerevisiae), which does not comprise complex I, three large mitochondrial complexes were identified by Blue-native gel electrophoresis after gentle protein solubilization using nonionic detergents: (a) dimeric ATP synthase, (b) a supercomplex containing dimeric complex III + one copy of complex IV, and (c) a supercomplex containing dimeric complex III + two copies of complex IV (Arnold et al., 1998; 1999; Cruciat et al., 2000; Schägger and Pfeiffer, 2000; Schägger, 2001a, 2002; Zhang et al., 2002). Dimeric ATP synthase from yeast includes three dimer-specific subunits, two of which are directly involved in dimer formation. Supercomplexes containing complexes III and IV were not only prepared by Blue-native gel electrophoresis but also by gel filtrations and co-immunoprecipitations (Cruciat et al., 2000). Their formation depends on the cardiolipin content of the inner mitochondrial membrane and also is influenced by growth conditions. Functional implications of complex III-complex IV associations were shown by ubiquinol-oxidase activity measurements in the presence of mild detergents (Schägger and Pfeiffer, 2000).
In mammalian mitochondria, five large complexes were found: (a) dimeric ATP synthase, (b) a supercomplex containing dimeric complex III + one copy of complex I, and (c-e) supercomplexes containing dimeric complex III + one copy of complex I + one to three copies of complex IV (Schägger and Pfeiffer, 2000, 2001; Schägger, 2001a, 2002). All of these super-complexes can be visualized on Blue-native gels after solubilization of mitochondrial proteins using digitonin. Solubilization using Triton X-100 additionally allow detection of a supercomplex consisting of dimeric complex III + monomeric complex I + four copies of complex IV. A high percentage of complex I forms part of supercomplexes, whereas dimeric complex III and monomeric complex IV also exist in singular form, because abundance of these protein complexes is significantly higher in comparison with complex I. NADH-cytochrome c activity measurements in dependence of various mild detergents have revealed functional importance of supercomplex formation between complexes III2 + I. The term “respirasome” was suggested for supercomplexes containing the complexes I, III2, and IV, which autonomously can carry out respiration in the presence of cytochrome c and ubiquinone (Schägger and Pfeiffer, 2000).
The supramolecular structure of the respiratory chain of plant mitochondria is unknown. The five protein complexes of oxidative phosphorylation are well characterized and structurally resemble their counterparts in fungi and mammals (Jänsch et al., 1996; Vedel et al., 1999; Heazlewood et al., 2003b; Sabar et al., 2003). Some plant-specific subunits of respiratory chain complexes were described, e.g. the subunits of the mitochondrial processing peptidase, which form an integral part of complex III in plant mitochondria (Braun et al., 1992a; Eriksson et al., 1994). Additionally, the electron transfer chain of plant mitochondria is very much branched due to the presence of several alternative oxidoreductases like a cyanide-insensitve terminal oxidase and rotenone-insensitive NAD(P)H dehydrogenases (for review, see Siedow and Umbach, 1995; Vanlerberghe and McIntosh, 1997; Mackenzie and McIntosh, 1999; Rasmusson et al., 1999).
Here, we describe a systematic investigation of supercomplexes in plant mitochondria. Using gentle protein solubilizations with nonionic detergents and Blue-native gel electrophoresis, three supercomplexes could be visualized: (a) dimeric ATP synthase, (b) a supercomplex formed by dimeric complex III and complex I, and (c) a supercomplex containing two copies of dimeric complex III and two copies of complex I. The complexes II and IV as well as the alternative oxidase (AOX) do not form part of super-complexes under all conditions applied. Furthermore, a larger and a smaller form of cytochrome c oxidase were found, which differ by at least two protein subunits, and a complex II is described, which has a very unusual subunit composition.
RESULTS
Identification of Respiratory Supercomplexes in Mitochondria from Arabidopsis
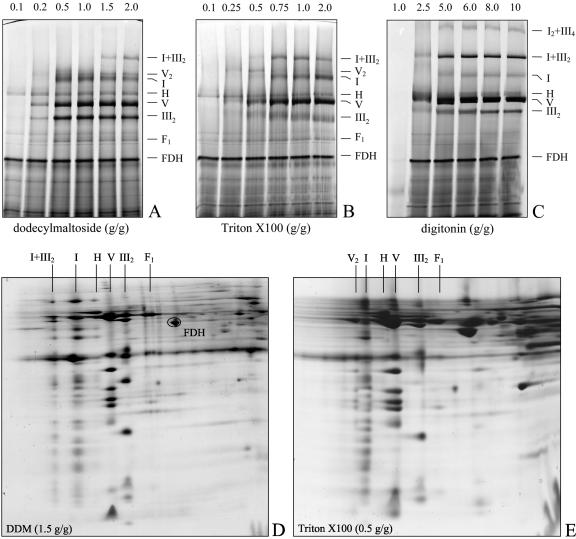
Blue-native gel electrophoresis was previously employed for the characterization of the respiratory chain of plant mitochondria (Jänsch et al., 1995, 1996; Brumme et al., 1998; Kügler et al., 1998; Karpowa and Newton, 1999; Ducos et al., 2001; Kruft et al., 2001; Mihr et al., 2001; Rasmusson and Agius, 2001; Werhahn and Braun, 2002; Bykova and Moller, 2003; Heazlewood et al., 2003a, 2003b, 2003c; Sabar et al., 2003). Nevertheless, respiratory supercomplexes were not described, most likely because very similar conditions for protein solubilization were chosen, which seem to have destabilizing effects on labile protein-protein interactions. In an attempt to systematically search for the occurrence of respiratory supercomplexes in plants, mitochondria from Arabidopsis were solubilized using varying concentrations of the nonionic detergents dodecylmaltoside, Triton X-100, and digitonin and analyzed by Blue-native PAGE (Fig. 1, A-C). Protein complexes were identified by their known subunit compositions upon analyses on second gel dimensions and by partial sequence analysis of selected proteins using mass spectrometry (Figs. 1, D and E, and 2; Table I).
Figure 1.
Resolution of mitochondrial protein complexes and supercomplexes by Blue-native PAGE. A through C, Solubilization of mitochondrial protein complexes from Arabidopsis using different detergents. Isolated mitochondria were treated with varying concentrations of dodecylmaltoside (A), Triton X-100 (B), or digitonin (C), and protein complexes were subsequently resolved by one-dimensional Blue-native PAGE. Detergent to protein ratios are given above the gels (in grams of detergent per gram protein), and the identity of protein complexes is given to the right of the gels. D and E, Two-dimensional resolution of mitochondrial protein complexes from Arabidopsis by Blue-native/SDS PAGE after solubilization with dodecylmaltoside (1.5 g per g protein) (D) and Triton X-100 (0.5 g per g protein) (E). Designations of the protein complexes are given above the gels. FDH, Formate dehydrogenase; F1, F1-part of the ATP synthase complex; III2, dimeric cytochrome c reductase; V, ATP synthase; H, HSP60 complex; I, NADH dehydrogenase; I+III2, supercomplex formed by complex I and dimeric complex III; IVa and VIb, large and small form of cytochrome c oxidase; V2, dimeric ATP synthase.
Figure 2.
Two-dimensional resolution of digitonin-solubilized mitochondrial protein complexes and supercomplexes by Blue-native/SDS-PAGE. Identities of protein complexes are given above the gel (for designations, see Fig. 1). Arrows indicate proteins identified by mass spectrometry (Table I).
Table I.
Identified subunits of mitochondrial protein complexes of Arabidopsis and bean
The numbers of the spots correspond to those given in Figures 2 and 5. Proteins were identified by electrospray tandem mass spectrometry (ESI), matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI), or Edman degradation (ED). Amino acid sequences given in italics represent N-terminal protein sequences determined by Edman degradation. Calculated molecular masses are given for all proteins with known N-terminal sequence, otherwise molecular masses are estimated on the basis of migration during SDS gel electrophoresis (see Figs. 2 and 5). Protein accessions correspond to the code of the Arabidopsis Genome Initiative.
| Spot | Identified Peptides | Identification Strategy | Protein | Molecular Mass | Arabidopsis Genome Initiative Accession No. |
|---|---|---|---|---|---|
| kD | |||||
| Arabidopsis | |||||
| 1 | WDPQISQVAGR | ESI | Subunit of complex I | ∼10 | At1g67350 |
| RDPYDDLLEDNYTPPSSSSSSSD | |||||
| 2 | SPNVAYLPGGQSMLFALNR | ESI | Prohibitin | 30.4 | At5g40770 |
| 3 | YEDISVLGQRPVEE | ESI | 8.0-kD subunit, complex III | 8.0 | At3g52730 |
| 4 | AVVYALSPFQQK | ESI | 8.2-kD subunit, complex III | 8.2 | At3g10860 |
| At5g05370 | |||||
| 5 | TFIDPPPTEEK | ESI | 6.7-kD subunit, complex III | 6.7 | At2g40765 |
| 6 | IPTAHYEFGANYYDPK | ESI | TOM40 | 34.1 | At3g20000 |
| IDSNGVASALLEER | |||||
| 7 | RLDDIDFPEGPFGTK | ESI | COX subunit Vb | ∼16 | At3g15640 |
| 8 | - | MALDI | Flavoprotein subunit, complex II | ∼65 | |
| 9 | DLVVDMTNFYNQYK | ESI | Iron sulphur subunit, complex II | ∼29 | At5g40650 |
| WNPDNPGKPELQDYKIDLK | |||||
| 10 | DLVVDMTNFYNQYK | ESI | Iron sulphur subunit, complex II | ∼29 | At5g40650 |
| At3g27380 | |||||
| 11 | AAEAVEEFGGILTSIK | ESI | Hyp. prot. (complex II) | 18.4 | At1g47420 |
| YAEYLDSFEPEEVYLK | MALDI | ||||
| SEDVSHMPEMDSXVLNAFK... | ED | ||||
| 12a | FMEWWER | MALDI | Hyp. prot. (complex II) | ∼15 | At1g08480 |
| LDTMAAQVK | ESI | ||||
| 12b | QGPNLNGLFGR | ESI | Cytochrome c | 12.4 | At4g10040 |
| At1g22840 | |||||
| 13 | STISGDIKTTQEEP | ED | Subunit 3 of complex II | 12 | At5g09600 |
| 14 | LVVDTTANQDPLVTK_ | ESI | Superoxide dismutase | 22.5 | At3g10920 |
| YASEVYEKESN_ | |||||
| 15 | QYIQEPATVEK | ESI | g subunit, complex V | ∼12 | At4g29480 |
| LASIPGRYETFWK | |||||
| 16 | LNQISILVQR | ESI | COX subunit II | 29.4 | y08501 |
| Bean | |||||
| 17 | YLEYHR | ESI | Subunit VIb of cytochrome c oxidase | ∼32 | Highly similar to At1g22450 |
| GDDAPE | |||||
| TPATPEE | |||||
| LETAPVDFR | |||||
| EATSEEAVVEK |
Solubilization of Arabidopsis mitochondria with 1 g dodecylmaltoside g-1 protein allows resolution of known singular complexes of the oxidative phosphorylation system (Fig. 1, A and D): complex I (1,000 kD), F0F1 ATP synthase (580 kD), complex III (480 kD), which always is dimeric for functional reasons, and the F1 part of ATP synthase (390 kD). Furthermore, the soluble HSP60 (750 kD) and formate dehydrogenase complexes (200 kD) are visible on the gel. Additionally, some low amount of dimeric ATP synthase can be seen at about 1,100 kD, which was overlooked on the Blue-native gels shown before by Kruft et al. (2001). In contrast to yeast and mammals, the amount of dimeric ATP synthase does not increase if mitochondrial proteins are solubilized with lower dodecylmaltoside concentrations (Fig. 1A). Usage of dodecylmaltoside to protein ratios >1 g per g allows visualization of a supercomplex of about 1,500 kD, which is composed of the complexes I and III and probably has the structure III2I. However, only a small proportion of total complex I forms part of this super-complex and an even smaller proportion of complex III, which is more abundant than complex I.
Solubilization of Arabidopsis mitochondria with Triton X-100 allows visualization of the same protein complexes and supercomplexes on Blue-native gels (Fig. 1, B and E). The amount of dimeric ATP synthase is highest between 0.25 and 0.5 g Triton X-100 g-1 protein, which is in line with observations reported for yeast (Arnold et al., 1998). The ratio of dimeric to monomeric ATP synthase is about 1 upon solubilization using 0.25 g Triton g-1 protein but decreases sharply upon solubilizations using higher amounts of detergent (Fig. 1B). About 50% of complex I forms part of the III2I supercomplex on Blue-native gels after protein solubilizations using 0.5 to 1.0 g per g Triton X-100 per g protein (Fig. 1B).
In general, higher detergent to protein ratios are necessary for protein solubilizations using digitonin, which is in accordance with results found for yeast and mammals. However, starting with a digitonin to protein ratio of 2.5 g per g, this detergent proved to be very suitable for supercomplex stabilization (Figs. 1C and 2). Under these conditions, about 80% of complex I forms part of the III2I supercomplex. Furthermore, a supercomplex of about 3,000 kD can be seen on Blue-native gels (Fig. 1C), which also is composed of subunits of the complexes III and I as found by two-dimensional Blue-native/SDS gel electrophoresis and silver staining (data not shown). This supercomplex most likely has a III4I2 structure, because the ratio of single complex I and complex III subunits is unchanged if compared with their ratio in the 1,500-kD III2I complex. Dimeric ATP synthase only is visible at very low digitonin to protein ratios (data not shown). The F1 part of the ATP synthase complex is not detectable on the Blue-native gels, indicating a stabilizing effect of the detergent on complex V (Fig. 1C). Furthermore, in contrast to dodecylmaltoside and Triton X-100, digitonin allows solubilization of three novel protein complexes of about 150, 220, and 300 kD. These protein complexes do not form visible bands on one-dimensional Blue-native gels, probably because the background on the gels is too high in this molecular mass range. However, these multisubunit complexes nicely are resolved on corresponding two-dimensional gels (Fig. 2). The subunit compositions of the 220- and 300-kD complexes very much resemble the one of cytochrome c oxidase from potato (Solanum tuberosum; Jänsch et al., 1996). In contrast, identity of the 150-kD complex was unclear on the basis of subunit composition.
Protein identifications by mass spectrometry allowed unambiguous identification of subunits of complexes I, III, and IV (Fig. 2; Table I). Furthermore the 1,000-kD prohibitin complex was identified, as was the preprotein translocase of the outer mitochondrial membrane, the so-called TOM complex, at 390 kD (Werhahn et al., 2003).
Dissection of Supercomplexes into Protein Complexes by Two-Dimensional Blue-Native/Blue-Native Gel Electrophoresis
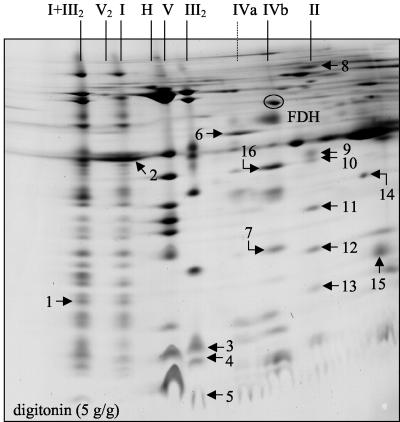
A novel two-dimensional Blue-native/Blue native gel electrophoresis method (Schägger and Pfeiffer, 2000) was employed to investigate whether the complexes I and III are the only components of the 1,500- and 3,000-kD supercomplexes. For this procedure, protein complexes and supercomplexes are separated by a first-dimension Blue-native-PAGE in the presence of digitonin. Afterward, protein supercomplexes are destabilized by incubation with dodecylmaltoside, which is followed by a second-dimension Blue-native-PAGE. On the resulting two-dimensional gels, supercomplexes are separated vertically into protein complexes, whereas singular protein complexes are located on a diagonal line. Two-dimensional Blue-native/Blue-native PAGE clearly revealed that the complexes I and III are the only constituents of the 1,500- and 3,000-kD supercomplexes in Arabidopsis (Fig. 3).
Figure 3.
Separation of supercomplexes into protein complexes by two-dimensional Blue-native/Blue-native gel electrophoresis. Identities of protein complexes are given above and to the left of the gel (for designations, see Fig. 1).
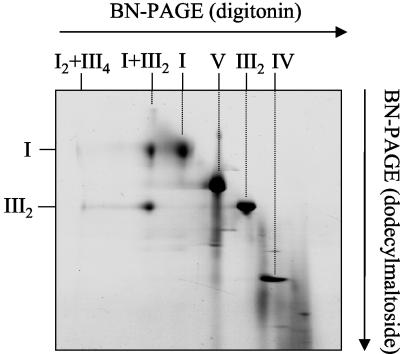
Characterization of Mitochondrial Supercomplexes in Potato, Bean (Phaseolus vulgaris), and Barley (Hordeum vulgare)
To investigate whether occurrence of the III2I and III4I2 supercomplexes and dimeric ATP synthase is a special characteristic of Arabidopsis or a general feature of plant mitochondria, the above described experiments were repeated with isolated organelles from potato, bean, and barley. All protein solubilizations were done with digitonin (5 g per g protein), which proved to be optimal for visualizations of mitochondrial protein complexes and supercomplexes in Arabidopsis on Blue-native gels. The III2I supercomplex is also present in potato, bean, and barley (Fig. 4). About 50% of complex I forms part of this supercomplex in bean and potato, whereas even 90% of complex I from barley is associated with dimeric complex III. Under the conditions applied, dimeric ATP synthase of all three plants only represents a very minor fraction of total ATP synthase complex. Furthermore, the large and the small form of cytochrome c oxidase are present in all plants investigated. However, the ratio of large to small forms varies considerably: In Arabidopsis and barley, the smaller form is very abundant, whereas in potato, the larger form is present in higher quantities, and in bean, both forms of complex IV are of equal abundance. The newly discovered 150-kD complex is also present in potato and bean but could not be clearly detected in barley under the conditions applied.
Figure 4.
Mitochondrial supercomplexes and protein complexes in Arabidopsis, potato, bean, and barley. Proteins were solubilized by 5 g per g digitonin and separated by two-dimensional Blue-native/SDS PAGE. Identities of protein complexes are given above the gels (for designations, see Fig. 1).
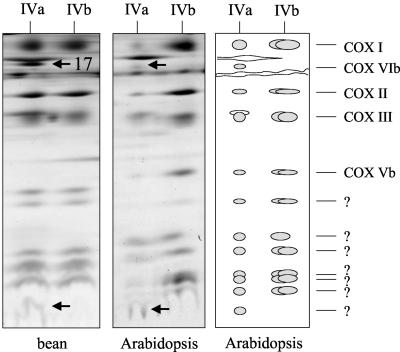
The Larger Form of Cytochrome c Oxidase Contains Additional Protein Subunits
Although Blue-native gel electrophoresis is not a suitable procedure for precise molecular mass determinations, the larger 300-kD form of the cytochrome c oxidase complex (IVa) probably cannot be considered to be a dimer of the 220-kD complex (IVb). Careful evaluation of the Blue-native gels in the region of the two forms of cytochrome c oxidase from Arabidopsis and bean revealed the presence of additional subunits in the larger form, which might explain the size difference between the two forms of this complex (Fig. 5). Data are especially clear for bean, because both forms of complex IV are equally abundant. A 32-kD protein and at least one very small subunit of <6 kD are unique to complex IVa. Proteins of comparable size are also present in Arabidopsis (Fig. 5) but are difficult to detect in potato and barley under the conditions applied. According to our interpretation of the Blue-native gels, the subunit composition of cytochrome c oxidase is as follows (see scheme in Fig. 5): Complex IVa includes 12 separable proteins, and complex IVb is composed of 10 proteins. Furthermore, complex IVb of Arabidopsis can be further subdivided into two complexes of very similar molecular masses that differ with respect to the presence of a 10-kD subunit.
Figure 5.
Large and small forms of cytochrome c oxidase in bean and Arabidopsis after two-dimensional resolution by Blue-native/SDS PAGE. Proteins only forming part of the larger form of this protein complex are indicated by arrows. Protein number 17 was identified by mass spectrometry (Table I). A scheme of the two-dimensional Blue-native/SDS gel of cytochrome c oxidase from Arabidopsis is given to the right.
The 32-kD subunit of complex IVa is homologous to the 10-kD COX VIb protein of heterotrophic eukaryotes (see Table I, protein 17), which is known to be easily detached from cytochrome c oxidase in yeast and mammals and which was shown to have regulatory functions on cytochrome c oxidase activity (LaMarche et al., 1992; Weishaupt and Kadenbach, 1992).
Complex II from Plant Mitochondria Contains Seven Subunits
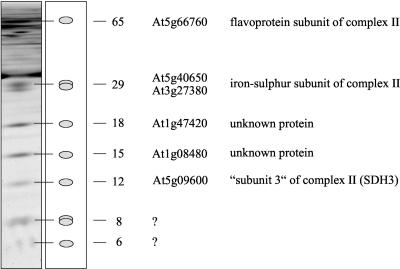
The newly discovered 150-kD complex of Arabidopsis comprises seven subunits of 65, 28, 18, 15, 12, 8, and 6 kD. In bean and potato, this complex has a very comparable subunit composition, except that the molecular masses of the three smallest subunits slightly vary (Fig. 4). To identify the 150-kD complex, subunits were subjected to analysis by mass spectrometry. Peptide sequences of five of the seven subunits allowed identification of corresponding genes of the Arabidopsis genome (Fig. 6; Table I). Surprisingly the 65-, 28-, and 12-kD proteins could be identified as being subunits of the succinate dehydrogenase complex (complex II) of the respiratory chain: the flavoprotein subunit (SDH1), the iron-sulfur subunit (SDH2), and the so-called subunit III (SDH3). Succinate dehydrogenase is well characterized for several bacteria, fungi, and mammals and is known to be a four-subunit complex comprising the above mentioned subunits and one additional subunit in the size range of 10 to 15 kD termed subunit IV or SDH4 (Lemire and Oyedotun, 2002; Yankovskaya et al., 2003). Although the subunits of this protein complex from plants were never biochemically characterized, counterparts of the SDH1-SDH4 proteins could be identified on the basis of sequence similarities of predicted proteins with known complex II subunits from mammals, fungi, algae, and protozoans (Daignan-Fornier et al., 1994; Burger et al., 1996; Figueroa et al., 2001; Figueroa et al., 2002). However, sequence identities for the SDH3 and SDH4 proteins, which constitute a hydrophobic membrane anchor of this protein complex, are very low (Burger et al., 1996). Most interestingly, complex II from plants seems to contain additional subunits of unknown function. The 18-kD protein corresponds to the putative At1g47420 protein of Arabidopsis, which was identified previously in the course of a proteomic approach to characterize novel mitochondrial proteins of this organisms (Kruft et al., 2001; protein No 4). Furthermore, the 15-kD protein represents the putative At1g08480 protein. However, presence of this protein within complex II is slightly uncertain, because one of the three identified peptides perfectly matches to Arabidopsis cytochrome c. Hence the corresponding protein spot contains more than one protein. The 8- and 6-kD subunits could not be identified by mass spectrometry; one of these subunits might represent the SDH4 protein.
Figure 6.
Subunit composition of complex II in Arabidopsis. Apparent molecular masses of the subunits and the accession numbers of the corresponding genes are given to the right of the gel.
Most likely all seven proteins of plant succinate dehydrogenase are single-copy subunits of the 150-kD complex, because the sum of their apparent molecular masses (153 kD) is very close to the apparent molecular mass of this protein complex on Blue-native gels (150 kD). Furthermore, probably no additional subunits form part of this complex.
The AOX Does Not Form Part of Respiratory Supercomplexes
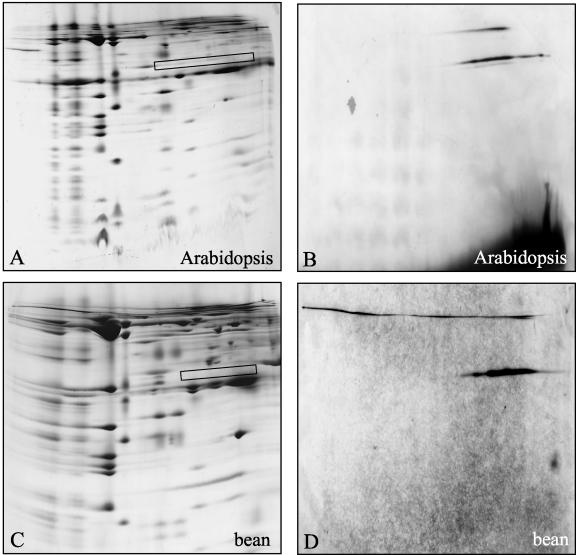
AOX represents a characteristic oxidoreductase of the respiratory chain of plant mitochondria. Its possible association with the complexes I to V after digitonin-solubilization of mitochondrial protein fractions was investigated by two-dimensional Blue-native/SDS gel electrophoresis and immunoblotting using a monoclonal antibody directed against AOX from Sauromatum guttatum (Elthon et al. 1989). On the two-dimensional gels, the antibody reacts with a protein of 32 kD in bean and Arabidopsis, which forms a smear on the first gel dimension between 30 and 300 kD (Fig. 7). Therefore, AOX seems not to form part of any supercomplexes but rather aggregates under the conditions applied. Also the rotenone-insensitive NAD(P)H dehydrogenases are not attached to respiratory protein complexes or super-complexes as investigated by similar experiments using antibodies directed against these proteins (data not shown).
Figure 7.
The AOX does not form part of mitochondrial supercomplexes. Mitochondrial proteins from Arabidopsis (A and B) and bean (C and D) were solubilized by 5% (w/v) digitonin and separated by two-dimensional Blue-native/SDS PAGE. Afterward, gels were either directly stained with Coomassie Blue (A and C) or electroblotted onto nitrocellulose membranes and immunostained with an antibody directed against AOX (B and D). The boxes on the Coomassie Blue gels indicate the position of the main immunosignals.
DISCUSSION
Supercomplexes in Plant Mitochondria
Respiratory protein complexes form supercomplexes in plant mitochondria. In the course of our investigations, V2, III2I, and III4I2 supercomplexes could be identified. Possibly further supercomplexes exist in vivo that are instable in the presence of detergents and Coomassie Blue. Digitonin proved to be the optimal compound for supercomplex solubilization, which is surprising because it specifically binds sterols, which are believed to be absent in inner mitochondrial membranes. Furthermore, digitonin also allows stable extraction of singular protein complexes that cannot be solubilized by dodecylmaltoside or Triton X-100. It therefore is a very suitable tool for plant mitochondrial research.
Between 50% and 90% of complex I from plants forms part of the III2I supercomplex on Blue-native gels upon digitonin solubilizations. In contrast, the same supercomplex from beef only contains 17% of complex I under identical conditions (Schägger, 2002). However, more than 50% of complex I from beef is part of even larger supercomplexes that include dimeric complex III and additionally one to four copies of complex IV. In Brewer's yeast, complex I-containing supercomplexes are absent due to the general absence of this protein complex, but also in this organism, the complexes IV and III are associated forming III2IV or III2IV2 supercomplexes (Schägger and Pfeiffer, 2000; Cruciat et al., 2000). In contrast, associations of complexes III and IV of plant mitochondria are not detectable on Blue-native gels under all conditions applied. Also the AOX and the rotenone-insensitive NAD(P)H dehydrogenases seem not to be present in supercomplexes. Furthermore, plant complex II only is present in singular form, which is in line with findings for yeast and mammals. Dimeric complex V from plants is present on Blue-native gels after solubilizations using low concentrations of detergents, but its abundance is lower than in other organisms (Arnold et al., 1998, 1999). Recently, a very stable dimeric ATP synthase complex was reported for mitochondria from Chlamydomonas reinhardtii (van Lis et al., 2003).
What is the functional role of supercomplexes in plant cells? In yeast, supercomplexes were reported to enhance activity rates of respiratory electron transport (Schägger and Pfeiffer, 2000). Furthermore, it is speculated that supercomplex formation increases the capacity of the inner mitochondrial membrane for protein insertion (Arnold et al., 1998). The protein content of this mitochondrial membrane, which is estimated to lie at about 70%, can only be realized if proteins are very efficiently packed. In plant mitochondria, the III2I supercomplex possibly has important consequences for the regulation of alternative respiration, because it might reduce access of AOX to its substrate ubiquinol. Because alternative respiration is known to increase under various stress conditions (Vanlerberghe and McIntosh, 1997), the occurrence of respiratory supercomplexes in Arabidopsis was investigated in mitochondria isolated from suspension cell cultures that were treated with antimycin A, a known inhibitor of complex III. However, our initial data reveal only small differences concerning respiratory supercomplexes in antimycin-treated and untreated cells, which are at the borderline of significance (data not shown). Therefore the role of supercomplexes in plant mitochondria has to be further investigated.
Respiratory Protein Complexes in Plant Mitochondria
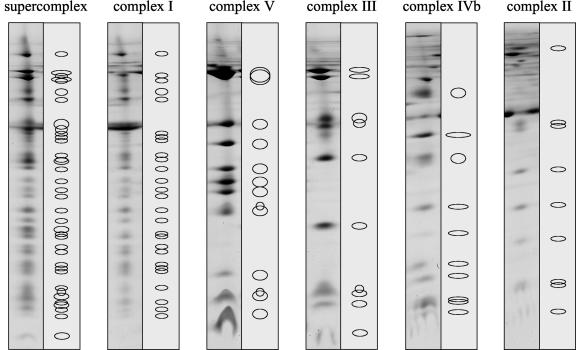
Recently, the subunit compositions of protein complexes of the oxidative phosphorylation system of Arabidopsis were studied intensively. Complex I from plants can be resolved into 27 to 30 different subunits on two-dimensional Blue-native/SDS gels (Fig. 8) but possibly comprises more than 40 proteins (Rasmusson et al., 1998). Heazlewood et al. (2003a) identified 30 subunits of Arabidopsis complex I after separation on two-dimensional gels by mass spectrometry. Several of the identified proteins have counterparts in fungi and mammals, but others seem to be unique to plants. Using a similar approach, Heazlewood et al. (2003c) identified 10 subunits of Arabidopsis complex V. Some further subunits remain to be characterized, because up to 13 proteins can be resolved on two-dimensional gels (Fig. 8). All 10 subunits of potato complex III were biochemically characterized (for review, see Braun and Schmitz, 1995) and counterparts for all 10 subunits are present in Arabidopsis protein databases at The Institute for Genomic Research or the Munich Information Center for Protein Sequences (β-MPP subunit, At3g02090; α-MPP subunit, At1g51980 and At3g16480; cytochrome b, Y08501; cytochrome c1, At5g40810 and At3g27240; 'Rieske FeS' protein, At5g13440 and At5g13430; counterpart to 14-kD subunit from potato, At4g32470 and At5g25450; counterpart to 7.8-kD subunit from potato, At2g01090 and At1g15120; counterpart to potato 8.0-kD subunit, At5g05370 and At3g10860; counterpart to potato 8.2-kD subunit, At3g52730; counterpart to potato 6.7-kD subunit, At2g40765).
Figure 8.
Subunit composition of respiratory protein complexes from Arabidopsis on two-dimensional Blue-native/SDS gels. Proteins were solubilized with digitonin. Sixty-seven different subunits of complexes I through V are separated: 27 subunits of complex I, seven of complex II, 10 of complex III, 10 of complex IV, and 13 subunits of complex V.
The least characterized respiratory protein complexes of plants are the complexes IV and II. Arabidopsis complex IV can be resolved in two different forms on Blue-native gels, which comprise 10 to 12 subunits (Figs. 5 and 8). The identity of the five largest subunits is known, whereas the identity of most smaller subunits remains to be established. The larger form of cytochrome c oxidase includes an additional 32-kD protein, which resembles the 10-kD subunit COX VIb of yeast and beef. This subunit is very hydrophilic, lacks membrane spanning helices, and is localized on the intermembrane-space side of cytochrome c oxidase (Tomizaki et al., 1999). Removal of this protein from complex IV was shown to activate cytochrome c oxidase of beef (Weishaupt and Kadenbach, 1992). Furthermore, COX VIb from beef was shown to be important for dimerization of cytochrome c oxidase (Tomizaki et al., 1999; Lee et al., 2001). Genes encoding COX VIb from plants were characterized previously in Arabidopsis and rice (Oryza sativa; Ohtsu et al., 1999, 2001). Interestingly, two forms of COX VIb proteins are encoded by these genes, which have molecular masses of 10 or 20 kD. Both forms of this protein very much resemble the 10-kD COX VIb from yeast and beef, but the 20-kD form has a long N-terminal extension. Curiously, both predicted forms of the plant COX VIb protein are much smaller than the 32-kD COX VIb found for Arabidopsis and bean on Blue-native gels. Also in rice, a 32-kD COX VIb protein was recently identified in the course of a mitochondrial proteome project (Heazlewood et al., 2003b). Consequently, the 32-kD COX VIb protein either is made by posttranslational modifications of smaller COX VIb proteins or is encoded by additional cox VIb genes, which so far were not discovered. Also, the functional role of this protein in plant mitochondria remains to be established. Possibly, it is important for dimerization of complex IV like in heterotrophic eukaryotes. However, dimeric cytochrome c oxidase was not detectable on our Blue-native gels under all conditions applied. Further investigations on cytochrome c oxidase of plant mitochondria are under way in our laboratory.
Surprisingly, complex II from Arabidopsis turned out to comprise seven subunits, which is three subunits more than the well-characterized succinat dehydrogenases from fungi, mammals, algae, protozoa, and several bacteria. Theoretically, these additional subunits could form a different protein complex, which accidentally runs at an identical position on Blue-native gels as a four-subunit complex II. However, this possibility is highly unlikely, because all seven proteins form an ideal line on Blue-native gels in three different plants: potato, bean, and Arabidopsis. Furthermore, the intensities of the protein spots on the two-dimensional gels is very much in the same range. Finally, the sum of the apparent molecular masses of the seven subunits corresponds to the apparent molecular mass of the protein complex on our native gels. As a general rule, respiratory protein complexes in mitochondria include several additional subunits if compared with their counterparts in prokaryotes. So far, complex II was the only exception, which seems not to be valid for plant mitochondria.
One of the newly described subunits of complex II from plants represents the hydrophilic At1g47420 protein, which was identified previously in the course of an Arabidopsis mitochondrial proteome project and which was reported to be one of the most abundant proteins on two-dimensional isoelectric focusing/SDS gels of mitochondrial fractions (Kruft et al., 2001). On the basis of sequence comparisons, the role of this protein is unclear. Highly similar proteins exist in barley (gi 18652408) and rice (CAD40922). Interestingly, the N-terminal presequence of 89 amino acids has some sequence identity with the N-terminal domain of the putative transcription factor APF1 of Arabidopsis (gi13507025). Also, the newly identified 15-kD subunit of complex II from Arabidopsis (At1g08480) does not exhibit significant sequence identity to previously characterized proteins. For the first time, to our knowledge, the plant SDH3 subunit was biochemically characterized. It comprises an exceptionally long mitochondrial targeting sequence of 105 amino acids as revealed by comparison of its N-terminal sequence and the amino acid sequence deduced from the corresponding gene. In summary, complex II from plants has unique features, which should be characterized by further biochemical and physiological investigations.
Further Protein Complexes in Plant Mitochondria
Besides the V2, III2I, and III4I2 supercomplexes and the respiratory complexes I to V, several additional protein complexes are visible on our Blue-native gels: the prohibitin complex at 1,000 kD, the HSP60 complex at 750 kD, the TOM complex at 390 kD, and a complex containing formate dehydrogenase at 200 kD (Fig. 2). Further plant mitochondrial protein complexes resolvable on Blue-native gels were not identified in the course of our study but were reported in earlier investigations, e.g. a Glu dehydrogenase complex (Heazlewood et al. 2003b). In contrast, some other protein complexes are known to be present in plant mitochondria but never were detected on Blue-native gels possibly due to their instability in the presence of detergents or Coomassie Blue, e.g. pyruvate dehydrogenase or the so-called AAA complexes. Recently, the occurrence of protein complexes comprising mitochondrial dehydrogenases of the citric acid cycle was reported on the basis of diffusion rate measurements of individual enzymes of this metabolic pathway (Haggie and Verkman, 2002). Most likely, these protein complexes are too unstable for biochemical preparations. Protein complexes and supercomplexes offer several physiological advantages in comparison with singular proteins, including substrate channeling, metabolic pathway regulation, and the realization of complicated biochemical reactions with reactive intermediates. Therefore the majority of mitochondrial proteins probably form part of protein complexes, and possibly most protein complexes are involved in the formation of even larger supermolecular structures, which remain to be discovered.
MATERIALS AND METHODS
Isolation of Mitochondria from Arabidopsis, Bean (Phaseolus vulgaris), Potato (Solanum tuberosum), and Barley (Hordeum vulgare)
Starting material for plant mitochondrial preparations were non-green Arabidopsis suspension cell cultures, potato tubers, 6-d-old etiolated barley seedlings, and 18-d-old etiolated bean seedlings. Arabidopsis cell lines were cultivated in the dark at 24°C to 26°C, 30% humidity, and gentle shaking (90 rpm) as described previously (Werhahn et al., 2001), and etiolated seedlings were grown at 24°C. All organelle preparations were carried out on the basis of filtration, differential centrifugation, and Percoll density centrifugation as outlined by Werhahn et al. (2001) for Arabidopsis; Focke et al. (2003) for barley; and Braun et al. (1992b) for potato and bean. Purified organelles were finally resuspended in a buffer containing 0.4 m mannitol, 1 mm EGTA, 0.2 mm phenylmethylsulfonyl fluoride (PMSF), and 10 mm Tricine/KOH, pH 7.2, at a protein concentration of 10 mg mL-1, divided into aliquots of 100 μL, and directly used for investigations (the amount of some supercomplexes was significantly reduced if mitochondrial fractions were frozen and stored before analyses).
Solubilization of Mitochondrial Proteins
Mitochondrial aliquots were centrifuged for 10 min at 14,300g, and sedimented organelles were resuspended in one of the following buffers (conditions adopted from Arnold et al., 1998; Schägger, 2001): (a) 100 μL of digitonin solubilization buffer (30 mm HEPES pH 7.4, 150 mm potassium acetate, 10% [v/v] glycerol, 2 mm PMSF, and [1-10 g per g protein] digitonin [Fluka, Buchs, Switzerland]); (b) 100 μL of dodecylmaltosid solubilization buffer (750 mm aminocaproic acid, 50 mm BisTris, pH 7.0, 0.5 mm EDTA, 1 mm PMSF, and docedylmaltoside [0.1-2 g per g protein; Roche, Mannheim, Germany]; and (c) 100 μL of Triton solubilization buffer (50 mm NaCl, 2 mm aminocaproic acid, 1 mm EDTA, 50 mm imidazole-HCl, pH 7.4, 10% glycerol, 5 mm PMSF, and Triton X-100 [0.1-2 g per g protein; Amersham-Pharmacia-Biotech Uppsala].
After incubation for 20 min on ice, samples were centrifuged at 18,000g for 30 min to remove insoluble material and were subsequently supplemented with 5 μL of Coomassie Blue solution (5% [w/v] Coomassie Blue in 750 mm aminocaproic acid). Dodecylmaltoside-solubilized samples were centrifuged immediately after resuspension of organelles in solubilization buffer and afterward were supplemented with 20 μL of Coomassie Blue solution. Coomassie Blue-treated protein samples were directly loaded onto Blue-native gels.
Two-Dimensional Blue-Native/SDS PAGE
One-dimensional Blue-native PAGE and two-dimensional Blue-native/SDS PAGE were carried out as described by Schägger (2001b). Gradient gels (4.5%-16% [w/v] acrylamide) were used for the Blue-native gel dimensions and two-step Tricine-SDS gels (10% and 16% [w/v] acrylamide) for second gel dimensions. The cathode buffer of Blue-native gel dimensions did not include detergent; only for electrophoresis of dodecylmaltoside-solubilized samples 0.03% of the detergent was added. Gels were either stained with Coomassie-colloidal (Neuhoff et al., 1985, 1990) or with silver (Heukeshoven and Dernick, 1986)
Two-Dimensional Blue-Native/Blue Native Gel Electrophoresis
Two-dimensional Blue-native/Blue-native PAGE was carried out as published by Schägger and Pfeiffer (2000). It proved to be important to stop first-dimension electrophoresis runs after 50% completion to avoid protein complexes and supercomplexes getting stuck in the gels. In contrast, it was important to extend the electrophoresis runs of second gel dimensions by factor two, because protein complexes stuck in gels were best resolved.
Protein Preparations for Mass Spectrometry
For mass spectrometry, gels were colloidal stained with Coomassie Blue (Neuhoff et al., 1990) and single proteins were cut out, transferred into an Eppendorf tube, and incubated with Milli-Q water for 10 min. Rebuffering was carried out by incubating the gel pieces for 15 min in acetonitrile and 0.1 m NH4HCO3, respectively. Subsequently, the proteins were dehydrated by acetonitrile and incubated with 20 μL of digestion solution (0.5 μg of trypsin [Promega, Madison, WI] in 20 μL of 50 mm NH4HCO3) overnight at 37°C. Peptide extraction was performed at 37°C as follows: Samples were supplemented with 20 μL of 50 mm NH4HCO3 and shaken for 15 min, and afterward, supernatants were taken and stored. Gel pieces were then shaken for 15 min in the presence of 20 μL of 5% (v/v) formic acid. Subsequently, the same volume of acetonitrile was added, and samples were shaken for another 15 min. Afterward, all supernatants were pooled and dried down to a volume of about 10 μL. Purification of the generated peptides was achieved using ZipTips (Millipore, Bedford, MA) according to the manufacturer's instructions.
Matrix-Assisted Laser Desorption Ionization/Time of Flight Mass Spectrometry
Determination of the molecular masses of Zip-Tip purified peptides was carried out by positive-ion matrix assisted laser desorption ionization/time of flight mass spectrometry using an Ultraflex instrument (Bruker, Newark, DE) equipped with delayed-extraction and a N2 laser (337 nm). For each sample, 1 μL of matrix solution (10 mg of α-cyano-4-hydroxycinnamic acid in 1 mL of 60% [v/v] acetonitrile/0.1% [v/v] formic acid) was placed on the Scout ion source and crystallized as a thin layer. One microliter of sample was given directly on the top of the thin matrix layer, and cocrystallization was carried out at room temperature. Spectra were recorded in reflection mode with an acceleration voltage of 25 kV and a reflection voltage of 26.3 kV. Monoisotopic masses from spectra were selected automatically and were used for protein identification with the help of MASCOT (Matrix Science, London).
Electrospray Ionization Tandem Mass Spectrometry
For peptide sequencing, 3 μL of Zip-Tip purified sample was filled into Au/Pd-coated nanospray glass capillaries (Protana, Odense, Denmark). The tip of the capillary was placed orthogonally in front of the entrance hole of a quadropole time-of-flight mass spectrometry instrument (Q-TOF II, Micromass, Watres, Milford, MA) equipped with a nanospray ion source. A capillary voltage between 750 and 1,000 V and a cone voltage of 30 V was applied. Two-fold charged peptides were chosen for collision-induced dissociation experiments, and the corresponding parent ions were selectively transmitted from the quadropole mass analyzer into the collision cell. Argon was used as collision gas, and the kinetic energy was set between 20 and 40 eV. The resulting daughter ions were separated by an orthogonal time-of-flight mass analyzer. Peptide sequencing and protein identification were carried out with the programs PeptideSequencing of the BioLynx software package (v3.5, Mircomass), Sonar of the Knexus software package (Proteo-metrics, Manitoba, Canada), and MASCOT (Matrix Science).
Acknowledgments
We are very grateful to Tom Elton for providing antibodies directed against the AOX and to Jean-Michel Grienenberger and Sergei Kushnir for encouraging the presented work. Furthermore, we thank Dagmar Lewejohann for the cultivation of Arabidopsis suspension cell cultures and for expert technical assistance.
This work was supported by the Fonds der Chemischen Industrie.
References
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H (1998) Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J 17: 7170-7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H (1999) ATP synthase of yeast mitochondria: isolation of subunit j and disruption of the ATP18 gene. J Biol Chem 274: 36-40 [DOI] [PubMed] [Google Scholar]
- Berry EA, Trumpower BL (1985) Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c-aa3 complexes. J Biol Chem 260: 2458-2467 [PubMed] [Google Scholar]
- Braun HP, Emmermann M, Kruft V, Schmitz UK (1992a) The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J 11: 3219-3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun HP, Emmermann M, Kruft V, Schmitz UK (1992b) Cytochrome c1 from potato: a protein with a presequence for targeting to the mitochondrial intermembrane space. Mol Gen Genet 231: 217-225 [DOI] [PubMed] [Google Scholar]
- Braun HP, Schmitz UK (1995) The bifunctional cytochrome c reductase/processing peptidase complex from plant mitochondria. J Bioenerg Biomembr 27: 423-436 [DOI] [PubMed] [Google Scholar]
- Brumme S, Kruft V, Schmitz UK, Braun HP (1998) New insights into the co-evolution of cytochrome c reductase and the mitochondrial processing peptidase. J Biol Chem 273: 13143-13149 [DOI] [PubMed] [Google Scholar]
- Burger G, Lang F, Reith M, Gray MW (1996) Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distinct eukaryotes. Proc Natl Acad Sci USA 93: 2328-2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykova NV, Moller IM (2003) Identification of 14 new phosphoproteines involved in important plant mitochondrial functions. FEBS Lett 540: 141-146 [DOI] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA (2000) The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem 275: 18093-18098 [DOI] [PubMed] [Google Scholar]
- Daignan-Fornier B, Valens M, Lemire BD, Bolotin-Fukuhara M (1994) Structure and regulation of SDH3, the yeast gene encoding the cytochrome b560 subunit of respiratory complex II. J Biol Chem 269: 15469-15472 [PubMed] [Google Scholar]
- Ducos E, Touzet P, Boutry M (2001) The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes. Plant J 26: 171-180 [DOI] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89: 1311-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson AC, Sjöling S, Glaser E (1994) The ubiquinol cytochrome c oxidoreductase complex of spinach leaf mitochondria is involved in both respiration and protein processing. Biochim Biophys Acta 1186: 221-231 [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Jordana X (2001) Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol Biol 46: 241-250 [DOI] [PubMed] [Google Scholar]
- Figueroa P, Léon G, Elorza A, Holuigue L, Araya A, Jordana X (2002) The four subunits of the mitochondrial respiratory complex II are encoded by the multiple nuclear genes and targeted to mitochondria in Arabidopsis thaliana. Plant Mol Biol 50: 725-734 [DOI] [PubMed] [Google Scholar]
- Focke M, Gieringer E, Schwan S, Jänsch L, Binder S, Braun HP (2003) Fatty acid biosynthesis in mitochondrial from grasses: Malouyl-CoA is generated by a mitochondrial-localized acetyl-CoA carboxylase. Plant Physiol (in press) [DOI] [PMC free article] [PubMed]
- Haggie PM, Verkman AS (2002) Diffusion of tricarboxylic acid cycle enzymes in the mitochondrial matrix: evidence for restricted mobility of a multienzyme complex. J Biol Chem 277: 40782-40788 [DOI] [PubMed] [Google Scholar]
- Heazlewood JA, Howell KA, Millar AH (2003a) Mitochondrial complex I from Arabidopsis and rice: orthologs of mammalian and yeast components coupled to plant-specific subunits. Biochim Biophys Acta 1604: 159-169 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Howell KA, Whelan J, Millar AH (2003b) Towards an analysis of the rice mitochondrial proteome. Plant Physiol 132: 230-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Whelan J, Millar AH (2003c) The products of the mitochondrial ORF25 and ORFB genes are FO components of the plant F1FO ATP synthase. FEBS Lett 540: 201-205 [DOI] [PubMed] [Google Scholar]
- Heukeshoven J, Dernick R (1986) Silver staining of proteins. In Electrophoresis Forum '86, B.J. Radula, ed. Elektrophoresis Forum '86. In B.J. Radola, ed, Technische Universität München. pp 22-27
- Iwasaki T, Matsuura K, Oshima T (1995) Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7: I. The archael terminal oxidase supercomplex is a functional fusion of respiratory complexes III and IV with no c-type cytochromes. J Biol Chem 270: 30881-30892 [DOI] [PubMed] [Google Scholar]
- Jänsch L, Kruft V, Schmitz UK, Braun HP (1995) Cytochrome c reductase from potato does not comprise three core proteins but contains an additional low molecular weight subunit. Eur J Biochem 228: 878-885 [PubMed] [Google Scholar]
- Jänsch L, Kruft V, Schmitz UK, Braun HP (1996) New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J 9: 357-368 [DOI] [PubMed] [Google Scholar]
- Karpowa OV, Newton KJ (1999) A partially assembled complex I in NAD4-deficient mitochondria of maize. Plant J 17: 511-521 [Google Scholar]
- Kruft V, Eubel H, Werhahn W, Jänsch L, Braun HP (2001) Proteomic approach to identify novel mitochondrial functions in Arabidopsis thaliana. Plant Physiol 127: 1694-1710 [PMC free article] [PubMed] [Google Scholar]
- Kügler M, Brumme S, Jänsch L, Werhahn W, Schmitz UK, Braun HP (1998) Characterization of plant mitochondria by blue native polyacrylamide gel electrophoresis (BN-PAGE). In IM Moller, P Gardeström, K Glimelius, E Glaser, eds, Plant Mitochondria: From Gene to Function. Blackhuys Publishers, Leiden, The Netherlands, pp 273-276
- LaMarche AE, Abata MI, Chan SH, Trumpower BL (1992) Isolation and characterization of COX12, the nuclear gene for a previously unrecognised subunit of Saccharomyces cerevisiae cytochrome c oxidase. J Biol Chem 267: 22473-22480 [PubMed] [Google Scholar]
- Lee SJ, Yamashita E, Abe T, Fukumoto Y, Tsukihara T, Shinzawa IK, Ueda H, Yoshikawa S (2001) Intermonomer interactions in dimer of bovine heart cytochrome c oxidase. Acta Crystallogr D Biol Crystallogr 57: 941-947 [DOI] [PubMed] [Google Scholar]
- Lemire BL, Oyedotun KS (2002) The Saccharomyces cerevisiae mitochondrial succinate:ubiquinone oxidoreductase. Biochim Biophys Acta 1553: 102-116 [DOI] [PubMed] [Google Scholar]
- Mackenzie S, McIntosh L (1999) Higher plant mitochondria. Plant Cell 11: 571-585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihr C, Baumgärtner M, Dieterich JH, Schmitz UK, Braun HP (2001) Proteomic approach for investigation of cytoplasmic male sterility (CMS) in Brassica. J Plant Physiol 158: 787-794 [Google Scholar]
- Neuhoff V, Stamm R, Eibl H (1985) Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6: 427-448 [Google Scholar]
- Neuhoff V, Stamm R, Pardowitz I, Arold N, Ehrhardt W, Taube D (1990) Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11: 101-117 [DOI] [PubMed] [Google Scholar]
- Niebisch A, Bott M (2003) Purification of a cytochrome bc1-aa3 supercomplex with quinol oxidase activity from Corynebacterium glutamicum: identification of a fourth subunit of cytochrome aa3 oxidase and mutational analysis of diheme cytochrome c1. J Biol Chem 278: 4339-4346 [DOI] [PubMed] [Google Scholar]
- Ohtsu K, Hamanaka S, Yamazaki K, Nakazono M, Hirai A (1999) Characterization of a cDNA encoding a novel subunit for cytochrome c oxidase (COX6b) from rice. Breed Sci 49: 211-215 [Google Scholar]
- Ohtsu K, Nakazono M, Tsutsumi N, Hirai A (2001) Characterization and expression of the genes for cytochrome c oxidase subunit VIb (COX6b) from rice and Arabidopsis thaliana. Gene 264: 233-239 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Agius SC (2001) Rotenone-insensitive NAD(P)H dehydrogenases in plants: immunodetection and distribution of native proteins in mitochondria. Plant Physiol Biochem 39: 1057-1066 [Google Scholar]
- Rasmusson AG, Heiser VV, Zabaleta E, Brennicke A, Grohmann L (1998) Physiological, biochemical and molecular aspects of mitochondrial complex I in plants. Biochim Biophys Acta 1364: 101-111 [DOI] [PubMed] [Google Scholar]
- Rasmusson AG, Svensson AS, Knoop V, Grohmann L, Brennicke A (1999) Homologues of yeast and bacterial rotenone-insensitive NADH dehydrogenases in higher eukaryotes: two enzymes are present in potato mitochondria. Plant J 20: 79-87 [DOI] [PubMed] [Google Scholar]
- Rich PR (1984) Electron and proton transfer through quinones and cytochrome bc complexes. Biochim Biophys Acta 768: 53-79 [DOI] [PubMed] [Google Scholar]
- Sabar M, Gagliardi D, Balk J, Leaver CJ (2003) ORFB is a subunit of F(1)F(O)-ATP synthase: insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep 4: 1-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H (2001a) Respiratory chain supercomplexes. International Union of Biochemistry and Molecular Biology (IUBMB) Life 52: 119-128 [DOI] [PubMed] [Google Scholar]
- Schägger H (2001b) Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol 65: 231-244 [DOI] [PubMed] [Google Scholar]
- Schägger H (2002) Respiratory supercomplexes of mitochondria and bacteria. Biochim Biophys Acta 1555: 154-159 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777-1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2001) The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J Biol Chem 276: 37861-37867 [DOI] [PubMed] [Google Scholar]
- Siedow JN, Umbach AL (1995) Plant mitochondrial electron transfer and molecular biology. Plant Cell 7: 821-831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone N, Sekimachi M, Kutoh E (1987) Identification and properties of a quinol oxidase supercomplex composed of a bc1 complex and cytochrome oxidase in the thermophilic bacterium PS3. J Biol Chem 262: 15386-15391 [PubMed] [Google Scholar]
- Tomizaki T, Yamashita E, Yamaguchi H, Aoyama H, Tsukihara T, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S (1999) Structure analysis of bovine heart cytochrome c oxidase at 2.8 Å resolution. Acta Crystallogr 55: 31-45 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703-734 [DOI] [PubMed] [Google Scholar]
- van Lis R, Atteia A, Mendoza-Hernandez G, Gonzalez-Halphen D (2003) Identification of novel mitochondrial protein components of Chlamydomonas reinhardtii: a proteomic approach. Plant Physiol 132: 318-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedel F, Lalanne E, Sabar M, Chétrit P, de Paepe R (1999) The mitochondrial respiratory chain and ATP synthase complexes: composition, structure and mutational studies. Plant Physiol Biochem 37: 629-643 [Google Scholar]
- Weishaupt A, Kadenbach B (1992) Selective removal of subunit VIb increases the activity of cytochrome c oxidase. Biochemistry 46: 11477-11481 [DOI] [PubMed] [Google Scholar]
- Werhahn W, Braun HP (2002) Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis 23: 640-646 [DOI] [PubMed] [Google Scholar]
- Werhahn W, Jänsch L, Braun HP (2003) Identification of novel subunits of the TOM complex from Arabidopsis thaliana. Plant Physiol Biochem 41: 407-416 [Google Scholar]
- Werhahn W, Niemeyer A, Jänsch L, Kruft V, Schmitz UK, Braun HP (2001) Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis thaliana: identification of multiple forms of TOM20. Plant Physiol 125: 943-954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S (2003) Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299: 700-704 [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W (2002) Gluing the respiratory chain together: Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem 277: 43553-43556 [DOI] [PubMed] [Google Scholar]