Abstract
Complete replacement, by biolistic plastid transformation, of the hexadecameric ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) of tobacco (Nicotiana tabacum) with the dimeric version from the bacterium, Rhodospirillum rubrum, resulted in fully autotrophic and reproductive tobacco plants that required high CO2 concentrations to grow (Whitney SM, Andrews TJ [2001] Proc Natl Acad Sci USA 98: 14738-14743). Growth and photosynthesis of these plants was compared with that of nontransformed tobacco and other controls where the rbcL gene for the large subunit of tobacco Rubisco was linked to the aadA selectable-marker gene, simulating the gene arrangement of the transformants with R. rubrum Rubisco. An arrangement of the rbcL and aadA genes that gave rise to an abundant monocistronic rbcL transcript and a one-fifth as abundant bicistronic rbcL-aadA transcript had Rubisco levels and photosynthetic properties similar to those of nontransformed tobacco. Direct linkage of the rbcL and aadA genes, resulting in exclusive production of a bicistronic mRNA transcript analogous to that of the transformants with R. rubrum Rubisco, reduced transcript abundance and tobacco Rubisco content. The analogous transcript with the R. rubrum rbcM gene substituted for rbcL was not only reduced in abundance, but was also translated less efficiently. The photosynthetic rates of the transformants and controls were measured at high CO2 concentrations, using a mass spectrometric method. The rates and their responses to atmospheric CO2 concentration mirrored the amounts and the kinetic properties of the Rubiscos present. The contents of total nitrogen, carbohydrates, and photosynthetic metabolites of the leaves were also consistent with the content and type of Rubisco.
All plants depend on the photosynthetic CO2-fixing enzyme, Rubisco, to supply them with combined carbon. Plants, algae, and many bacteria have the so-called Form I Rubisco, which has a complex quaternary structure composed of eight large (50-55 kD) subunits, which bear the active sites, and eight small (12-18 kD) subunits (Roy and Andrews, 2000; Spreitzer and Salvucci, 2002). Two tetramers of small subunits glue four dimers of large subunits together in a (L2)4(S4)2 arrangement (Andersson, 1996). However, some bacteria and dinoflagellates have Rubiscos without small subunits. In these Form II Rubiscos, the basic L2 dimer occurs singly, as in Rhodospirillum rubrum, or as higher oligomers (Tabita, 1999).
Recently, we replaced the Form I Rubisco of tobacco (Nicotiana tabacum) with the Form II dimer from R. rubrum by substituting the rbcL gene for the large subunit in the tobacco plastid genome with the rbcM gene from R. rubrum (Whitney and Andrews, 2001a). The replacement of Rubisco was complete. No tobacco small subunits were seen in the transformants, although the nuclear RbcS gene family was undisturbed. Not being able to assemble with the R. rubrum large subunits, the tobacco small subunits presumably were degraded on arrival in the chloroplasts. R. rubrum is a photosynthetic anaerobe and its Rubisco has a weaker affinity for CO2, and a much poorer ability to discriminate between CO2 and O2, than the higher-plant enzyme (Jordan and Ogren, 1981). Therefore, the transformants with R. rubrum Rubisco (termed tobacco- rubrum plants) required supplementation with CO2 to grow. Provided with CO2 at 70 times its concentration in air, the plants grew slowly, but normally, and were fully reproductive (Whitney and Andrews, 2001a).
The tobacco-rubrum plants had only approximately one-third as much R. rubrum Rubisco as the nontransformed controls had tobacco Rubisco (Whitney and Andrews, 2001a). Plastid transformation depends on use of the aadA spectinomycin-resistance gene to select transformants. Here, we show that the reduced Rubisco content was the result of reduced abundance of the bicistronic rbcM-aadA transcript produced by the transgenes, combined with a reduction in its translational efficiency, compared with the rbcL transcript in nontransformed plants.
The requirement of R. rubrum Rubisco for high concentrations of CO2 conferred on the transformed plants a requirement for CO2 concentrations so high that CO2 assimilation was barely detectable with conventional gas-exchange equipment based on infrared analyzers (Whitney and Andrews, 2001a). Here, we measure gas exchange with a membrane-inlet mass spectrometer capable of measuring CO2 uptake at much higher CO2 concentrations. We compared photosynthesis in tobacco-rubrum plants with that of nontransformed plants and the other controls with different arrangements of rbcL and aadA genes. We show that the rates of CO2 assimilation are entirely consistent with the contents and kinetic properties of the resident Rubisco, as are the leaves' contents of total nitrogen, carbohydrates, and photosynthetic metabolites.
RESULTS
Construction of Tobaccos With rbcL in Contexts Similar to That of rbcM in Tobacco-rubrum
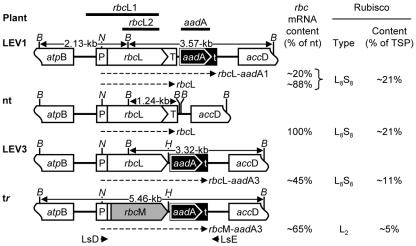
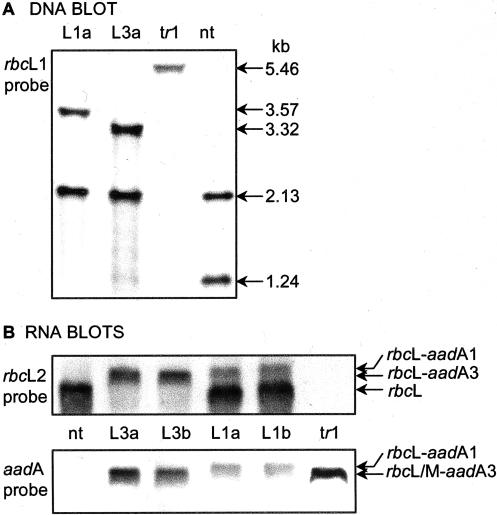
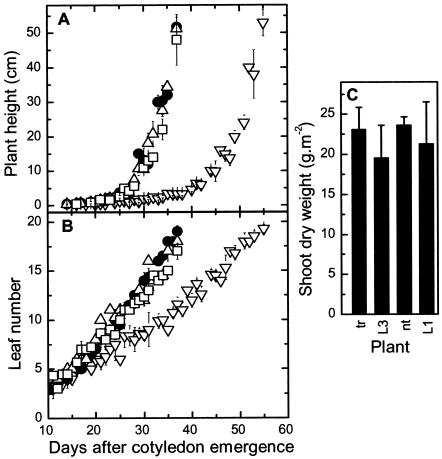
The antibiotic-resistance gene, aadA, required to select plastome transformants, was introduced to the tobacco plastome in two ways. In LEV1, a promoterless aadA gene was inserted immediately downstream of the rbcL terminator, giving rise to the possibility of two mRNA transcripts—a monocistronic one containing rbcL alone and a bicistronic one with rbcL and aadA. LEV3 was similar, except that the rbcL terminator was deleted so that only a bicistronic transcript was possible. LEV3 mimics the gene arrangement used in tobacco-rubrum (Fig. 1). Five and seven spectinomycin-resistant plantlets were obtained from 10 sets of leaves bombarded with the pLEV1 and pLEV3 transforming plasmids, respectively. After three rounds of regeneration on spectinomycin-containing medium, DNA blots showed that all of the plantlets were homoplasmic (Fig. 2). Two lines each of LEV1 (L1a and L1b) and LEV3 (L3a and L3b) transformants were grown to maturity in soil and their flowers were pollinated with wild-type pollen. In the CO2-enriched atmosphere required for growth of tobacco-rubrum plants, the LEV1 and LEV3 plants grew similarly to nontransformed tobacco and substantially faster than tobacco-rubrum plants. However, eventually, all of the plants reached a similar size and bore the same number of leaves (Fig. 3). The thickness of the leaves (0.4 ± 0.1 mm) and the density of stomata (1.0 ± 0.1 × 104 cm-2) were similar in all genotypes (data not shown).
Figure 1.
Organization of the transformed and nontransformed tobacco plastomes in the large single-copy region in the vicinity of rbcL. The LEV1 (Whitney et al., 1999) and LEV3 transformants have a promoter-less aadA gene inserted downstream of rbcL. For LEV1, the insertion point is downstream of the rbcL terminator sequence (T). For LEV3, the organization is similar, except that the rbcL terminator is deleted. The gene organization in tobacco-rubrum (tr) transformants is the same as for LEV3 transformants, except that the R. rubrum rbcM gene replaces rbcL (Whitney and Andrews, 2001a). The annealing positions of the probes, rbcL1, rbcL2, and aadA, and the primers, LsD and LsE, are shown. The positions of BamHI sites (B) and the sizes of the BamHI fragments that hybridize to the rbcL1 probe are indicated. The dashed arrows represent the various mono- and bicistronic rbc transcripts. H, HindIII; N, NcoI; P, rbcL promoter and 5′-untranslated region; t, rps16 terminator sequence; TSP, total soluble protein; nt; nontransformed.
Figure 2.
DNA and RNA blots of nucleic acids from leaves of transformed and nontransformed plants. A, Blot of BamHI-digested DNA from LEV1 (L1a), LEV3 (L3a), tobacco-rubrum (tr1), and nontransformed (nt) plants probed with the rbcL1 DNA fragment. B, Duplicate blots of total leaf RNA (10 μg) probed with the rbcL2 (top) and aadA (bottom) probes. RNA from independent LEV1 (L1a and L1b) and LEV3 (L3a and L3b) transformants is shown. Positions of the monocistronic rbcL (nt, L1), the bicistronic rbcL-aadA1 (L1), and rbcL/M-aadA3 (L3 and tr1 respectively) mRNAs are indicated.
Figure 3.
Comparison of plant growth in 2.5% (v/v) CO2 in air at 240 μmol quanta m-2 s-1. A, Plant height; B, number of leaves per plant (±sd) of T1-generation tobacco-rubrum (tr, ▿, n = 14), LEV3 (L3, □, n = 9), and LEV1 (L1, ▵, n = 9) transformants and nontransformed plants (nt, •, n = 6). C, Shoot dry weight measured 37 d (L1, L3, and nt) and 55 d (tr) after cotyledon emergence.
Abundance of Rubisco and rbc Transcripts
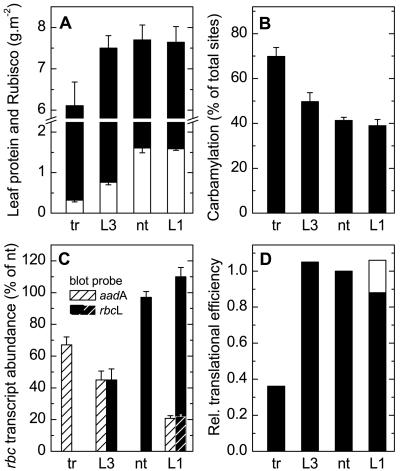
The Rubisco content of leaves of LEV1 plants was similar to that of the nontransformed controls, but, in LEV3 and tobacco-rubrum plants, it was reduced to 50% and 20%, respectively (Fig. 4A). Total soluble leaf protein was similar in LEV1, LEV3, and nontransformed plants, but was reduced in tobacco-rubrum plants, with most of the reduction attributable to the missing Rubisco (Fig. 4A). Under the high-CO2 growth conditions, Rubisco's carbamylation status inversely reflected its abundance (Fig. 4B).
Figure 4.
Soluble leaf protein, Rubisco, and rbcL/M transcript contents, and Rubisco carbamylation status in light-adapted leaves of transformants and controls. Measurements (±sd) were made on samples from leaf 5 of T2-generation tobacco-rubrum (tr, n = 4), LEV3 (L3, n = 3), and LEV1 (L1, n = 3) transformants and nontransformed plants (nt, n = 3; see “Materials and Methods”). A, Soluble leaf protein (white plus black), Rubisco content (white). B, Carbamylation status of active sites. C, The relative abundances of the bicistronic rbcL-aadA1, rbcL-aadA3, and rbcM-aadA3 transcripts in L1, L3, and tr plants, respectively, measured from RNA blots probed with the aadA probe (cross-hatched bars) and of the monocistronic rbcL transcript in nontransformed and L1 plants (black bars) and the bicistronic transcripts rbcL-aadA1 in L1 (white cross-hatched bars) and rbcL-aadA3 (black bars) in L3 plants probed with the rbcL probe. The mRNA abundance was normalized relative to that of the rbcL transcript in nontransformed plants. The relative responses to the rbcL and aadA probes was determined using the L3 plants, where the two probes detect the same mRNA species (Fig. 1). D, Relative translational efficiency of Rubisco mRNAs in tr, L3, and L1 transformants was obtained by dividing Rubisco content by transcript abundance and normalized relative to the result obtained with nontransformed leaves (see “Materials and Methods”). For L1 samples, the translational efficiency is calculated from the sum of the abundances of the rbcL and rbcL-aadA1 transcripts (black bar) and from the abundance of the rbcL transcript alone (black plus white bar).
Leaves of LEV1 and nontransformed plants had similar amounts of monocistronic rbcL transcripts (Figs. 2 and 4C). In addition, the LEV1 plants had a bicistronic rbcL-aadA transcript that was approximately 20% as abundant. The single bicistronic rbcL- aadA transcript of LEV3 leaves and rbcM-aadA transcript in tobacco-rubrum leaves (Fig. 2) was present at approximately 45% and 70%, respectively, of the level of the rbcL transcript in nontransformed leaves (Fig. 4C). Although the bicistronic rbcL-aadA transcript in LEV3 plants was less abundant, it was translated with an efficiency similar to that of the monocistronic rbcL transcript in LEV1 and nontransformed plants (Fig. 4D). By contrast, the translational efficiency of the bicistronic rbcM-aadA transcript in tobacco-rubrum plants was approximately one-third of that of the rbcL transcripts. Thus, the large reduction of Rubisco content of the tobacco-rubrum plants is a product of a modest reduction in message content and a larger reduction in its translational efficiency.
Abundance of rbcS mRNA Is Increased in Tobacco-rubrum Plants
RNA blotting showed that LEV1 and nontransformed plants produced similar amounts of mRNA from their nuclear rbcS gene family. LEV3 and tobacco-rubrum plants had 1.6- and 2.5-fold higher amounts, respectively (data not shown). Presumably, this is a response to the reduction in content, or total lack, of tobacco Rubisco.
CO2 Assimilation and Photosynthetic Metabolites
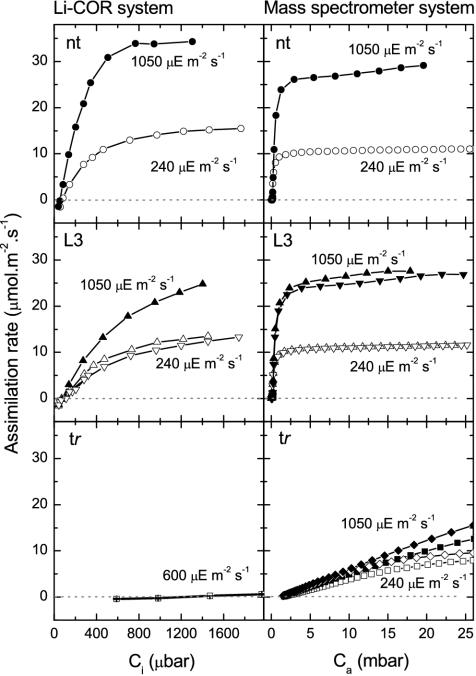
Where it was possible to compare them, there was generally reasonable agreement between CO2-assimilation measurements made with the infrared-analyzer and mass-spectrometer systems (Fig. 5). The membrane-inlet cuvette of the mass spectrometer system is unstirred and there are likely to be large boundary layer resistances adjacent to the leaf disc. These, or calibration imprecision, may be the cause of the slightly lower maximal assimilation rates measured with this system. At both levels of illumination, the reduced Rubisco content of the LEV3 plants compared with nontransformed plants was manifest as a reduction in slope of the curves at low CO2 pressures. However, at the much higher CO2 pressures under which the plants were grown, little difference in assimilation rate was apparent, indicating that both types of plants were limited by light-supported D-ribulose-1,5-bisphosphate (ribulose-P2) regeneration under these conditions. As reported previously (Whitney and Andrews, 2001a), tobacco- rubrum plants were massively impaired in CO2 assimilation at lower CO2 pressures. However, this impairment was substantially relieved at higher CO2 pressure so that, in the 25 mbar CO2 growth atmosphere, assimilation was only approximately 50% and 25% impaired at the higher and lower illumination, respectively (Fig. 5).
Figure 5.
Leaf CO2 assimilation rate as a function of intercellular (ci) or atmospheric (ca) CO2 partial pressure measured with a flow-through leaf chamber (Li-Cor, Lincoln, NE) and by mass spectrometry in a sealed unstirred cuvette. The fifth leaf (15 ± 0.5 cm in diameter) from nontransformed (nt; ○•, 17.6 μmol Rubisco sites m-2) and T2-generation tobacco-rubrum (tr; □▪, 5.2 μmol Rubisco sites m-2; ⋄ ♦, 6.2 μmol Rubisco sites m-2) and LEV3 (L3a; ▿ ▾, 6.9 μmol Rubisco sites m-2; L3b; ▵ ▴, 6.9 μmol Rubisco sites m-2) plants of similar physiological ages (24.6 ± 1.7 cm in height, bearing 15 leaves) grown at 450 μmol m-2 s-1 were measured at 25°C at the different light levels shown. The Li-Cor measurements for tobacco- rubrum plants reported previously (Whitney and Andrews, 2001a) are reproduced for comparison. Symbols are shown for every fifth (nt and L3) or 10th (tr) mass spectrometric measurement.
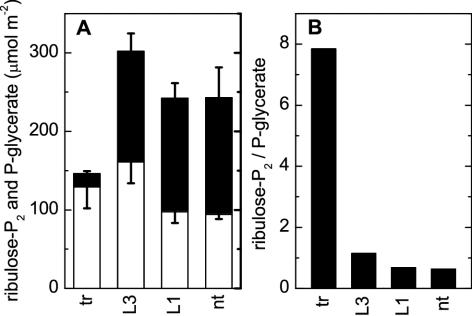
The reduced Rubisco content of the tobacco-rubrum plants caused only modest increases in the ribulose-P2 content of leaves sampled under the growth conditions—no larger than that seen in LEV3 leaves (Fig. 6). By contrast, the 3-phospho-D-glycerate (P-glycerate) content, which was similar in LEV1, LEV3, and nontransformed tobacco, was massively reduced in tobacco-rubrum leaves, leading to an approximately 10-fold increase in the P-glycerate: ribulose-P2 ratio (Fig. 6).
Figure 6.
Leaf contents (±sd) of ribulose-P2 and P-glycerate in T2-generation tobacco-rubrum (tr, n = 4), LEV3 (L3, n = 3), LEV1 (L1, n = 3), and nontransformed (nt, n = 3) plants grown at 450 μmol m-2 s-1 (see “Materials and Methods”). A, Ribulose-P2 (white) and P-glycerate (black) content; B, ribulose-P2:P-glycerate ratio.
Leaf Dry Matter and Its Components
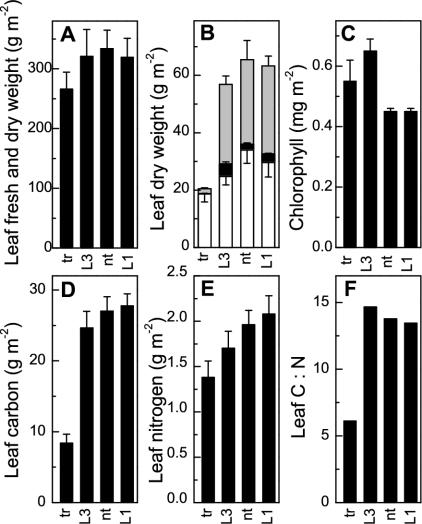
The photosynthetic impairment of the tobacco- rubrum plants was particularly obvious in their leaf dry matter. Although the fresh weight per unit area of the leaves was little diminished (<20%) compared with the other three genotypes, the dry-to-fresh weight ratio was reduced by approximately 60% (Fig. 7A). Most of this reduction was attributable to the near absence of starch and Glc from the tobacco- rubrum leaves (Fig. 7B). This massive reduction in storage carbohydrate caused an approximately 70% reduction in total leaf carbon (Fig. 7D), far outstripping more modest changes in total leaf nitrogen (Fig. 7E) and leading to a halving of the C:N ratio (Fig. 7F). Differences between the LEV1, LEV3, and nontransformed plants were small and consistent only in the case of total leaf N, which ranked approximately according to Rubisco content (Fig. 7E). Chlorophyll content was not reduced by reduction in or substitution of Rubisco (Fig. 7C), and the chlorophyll a:chlorophyll b ratio was 2.4 ± 0.1 in all cases (not shown), indicating that none of the genotypes suffered photoinhibition in the high-CO2 growth atmosphere.
Figure 7.
Structural and other components (±sd) of leaves of similar physiological age of T2-generation tobacco-rubrum (tr, n = 4), LEV3 (L3, n = 3), LEV1 (L1, n = 3), and nontransformed (nt, n = 3) plants (see “Materials and Methods”). A, Leaf fresh weight (total bar height) and its dry-weight component (white); B, leaf dry weight (total bar height) and its starch (gray, upward error bars) and Glc (black, downward error bars) components; C, chlorophyll; D through F, leaf carbon and nitrogen and their ratio.
DISCUSSION
Low Rubisco Content of Tobacco-rubrum Plants Is Caused Mostly by Impaired Translation
Previous 35S pulse-labeling experiments showed that little turnover occurred with R. rubrum Rubisco in tobacco-rubrum plants and tobacco Rubisco in the controls (Whitney and Andrews, 2001a, 2001b; Whitney et al., 2001); therefore, the content of Rubisco in both cases must be determined largely by the rate of synthesis. Thus, we sought to understand the reasons for the reduced Rubisco content of the tobacco- rubrum plants (20% of nontransformed plants in the present experiments; Fig. 4A) by comparing these bacterial-Rubisco transformants with transformants with analogous rbc gene arrangements that retained the tobacco rbcL gene (Fig. 1). Two reasons were uncovered. First, the bicistronic character of the rbcM-aadA mRNA transcript in tobacco-rubrum plants caused a modest (approximately 30%) reduction in its abundance compared with the monocistronic rbcL transcripts in the LEV1 and nontransformed controls. The analogous rbcL-aadA transcript in LEV3 plants was affected similarly (approximately 50% reduction; Fig. 4C). The longer mRNA molecules must be more unstable or transcribed less rapidly, or both. Second, the translational efficiency of the bicistronic rbcM-aadA transcript in tobacco-rubrum plants was severely impaired (approximately 65%). This impairment was not shared by the analogous rbcL-aadA transcript in LEV3 plants, therefore, its cause must lie in the intrinsic translatability of the rbcM message. The rbcM and rbcL genes had identical promoters, 5′-untranslated regions, and 5′-transcriptional control regions (the first 14 codons of the coding sequence; Kuroda and Maliga, 2001). The sequences 3′ to the rbc coding region were also identical in the rbcM-aadA and rbcL-aadA transcripts (Fig. 1). Therefore, the reduced translational efficiency must be caused by the rbcM coding sequence itself. Perhaps it causes the transcript to fold inappropriately, hindering binding of translation-activating factors or engagement with the ribosome. Alternatively, the GC-rich bacterial rbcM gene has a preference for some redundant codons used less frequently in the tobacco plastome (Shimada and Sugiura, 1991), and this may slow translation.
The unaltered translational efficiency of the bicistronic rbcL-aadA transcript in the LEV3 plants, compared with the monocistronic rbcL transcript in nontransformed or LEV1 plants (Fig. 4D), demonstrates that the tobacco chloroplast is unable to increase the translation rate of the bicistronic message to compensate for its lesser abundance. This contrasts with the situation in the Chlamydomonas reinhardtii chloroplast where reductions in transcript abundance have been shown to be sometimes compensated by increases in translational efficiency, making transcript abundance nonlimiting in the expression of some chloroplast-encoded proteins, including the large subunit of Rubisco (Hosler et al., 1989; Eberhard et al., 2002). This difference may reflect an underlying difference between higher plants and C. reinhardtii in the way translation of the rbcL message and other plastid transcripts are regulated.
Photosynthesis and Growth of Tobacco-rubrum Plants Are Consistent with the Content and Properties of R. rubrum Rubisco
Measurement of photosynthetic gas exchange with equipment based on a membrane-inlet mass spectrometer allowed measurements at much higher CO2 partial pressures than are accessible with conventional equipment based on sensitive infrared analyzers. These measurements confirmed earlier suspicions, based on measurements at low CO2 only (Whitney and Andrews, 2001a), that the photosynthetic characteristics of the tobacco-rubrum plants were in accord with their content of R. rubrum Rubisco and its kinetic properties (Fig. 5).
At high CO2, the 50% reduced tobacco Rubisco content of the LEV3 plants was still sufficient to support wild-type rates of CO2 assimilation that were limited solely by the rate of light-driven ribulose-P2 regeneration, regardless of the intensity of illumination (Fig. 5). This accords with the wild-type growth characteristics of these plants at 25 mbar CO2 pressure (Fig. 3).
By contrast, CO2 assimilation by the tobacco- rubrum plants clearly displayed the much higher Kcair (apparent Km for CO2 at air levels of O2) of the R. rubrum enzyme (Jordan and Ogren, 1981; Fig. 5). At 1,050 μmol m-2 s-1 illumination, CO2 assimilation was not CO2 saturated (i.e. not limited by ribulose-P2 regeneration) even at 25 mbar CO2. However, at 240 μmol m-2 s-1 illumination, the curve approached a plateau at higher CO2 pressures, indicating that assimilation had become limited by ribulose-P2 regeneration (Fig. 5). This is consistent with the smaller stimulation of CO2 assimilation at 25 mbar CO2 induced by increasing illumination from 240 to 1,050 μmol m-2 s-1 seen with tobacco-rubrum plants compared with LEV3 and nontransformed plants that are limited by ribulose-P2 regeneration at both light intensities. Boundary layer limitations to CO2 diffusion in the mass spectrometer cuvette preclude estimation of intercellular CO2 partial pressures (ci) from these measurements. Therefore, the kinetic properties of tobacco-rubrum Rubisco cannot be derived from these data. Nevertheless, the gross difference in the shape of the CO2-response curves of tobacco-rubrum leaves compared with LEV3 and nontransformed leaves readily reveals the presence of the bacterial Rubisco (Fig. 5).
The ribulose-P2 and P-glycerate contents of leaves sampled under 450 μmol m-2 s-1 illumination in the growth cabinet (an intensity midway between the two intensities used for gas-exchange measurements; Fig. 5) showed that photosynthesis in the tobacco- rubrum plants was limited by Rubisco activity under these conditions. Although the ribulose-P2 contents were not very different for all four genotypes, the severe reduction of P-glycerate content (and the large increase in ribulose-P2:P-glycerate ratio) testify to the Rubisco-activity limitation in the tobacco-rubrum leaves (Fig. 6). By contrast, the more balanced ribulose-P2 and P-glycerate contents (Fig. 6), coupled with diminished Rubisco carbamylation status (Fig. 4B), are consistent with ribulose-P2-regeneration limitation applying in LEV1, LEV3, and nontransformed plants under the same conditions.
Comparisons with Previous Manipulation of Rubisco by Nuclear Antisense
Before the advent of technology for manipulating the higher-plant plastome, genetic manipulation of Rubisco in plants was restricted to targeting the RbcS gene for the nuclear-encoded small subunit. Its expression was reduced by antisense genes in tobacco (Rodermel et al., 1988; Hudson et al., 1992), rice (Oryza sativa; Makino et al., 1997), and Flaveria bidentis (Furbank et al., 1996). Varying degrees of suppression were obtained, sometimes to very low Rubisco contents. The contents of sense RbcS mRNA, small subunits, Rubisco holoenzyme, and Rubisco activity were all reduced, leading to important insights into the mechanisms that coordinate the synthesis of the large and small subunits, and intergenomic integration in general (Rodermel et al., 1996; Rodermel, 1999). Such experiments also revealed the effects of varying the amount of Rubisco on the regulation of its activity and photosynthetic metabolism, and on leaf development (Quick et al., 1991; Hudson et al., 1992; von Caemmerer et al., 1994; Stitt and Schulze, 1994; Miller et al., 2000). Our present observations are fully consistent with the conclusions drawn from anti-RbcS studies and we extend them by demonstrating the consequences of changing the kinetic properties of Rubisco, rather than just its amount. Thus, the reductions in leaf CO2 assimilation (Fig. 5), P-glycerate:ribulose-P2 ratio (Fig. 6), Glc and starch contents, C:N ratio, dry weight:fresh weight ratio (Fig. 7), and the increase in Rubisco carbamylation status (Fig. 4) caused by the reduced Rubisco activity in our experiments all resemble similar perturbations resulting from the reduced Rubisco content of anti- RbcS plants. However, the gross changes in the CO2 response of assimilation in the tobacco-rubrum plants (Fig. 5) is an unprecedented consequence of the substitution of a Form II Rubisco.
The absence of detectable small subunits in the tobacco-rubrum plants (Whitney and Andrews, 2001a) is consistent with a model, derived partly from anti-RbcS studies, whereby the appropriate stoichiometry between small subunits and large subunits is maintained by rapid proteolytic degradation of excess unassembled small subunits (Rodermel, 1999). Our present observations demonstrate that transcriptional regulation of small subunit synthesis plays no part in this mechanism. Not only was the RbcS transcript undiminished when there was reduced (LEV3) or no (tobacco-rubrum) demand for small subunits, but its amount increased in apparent inverse proportion to the amount of tobacco Rubisco remaining (see “Results”). In the case of the very carbohydrate-deficient tobacco-rubrum plants, the increased RbcS mRNA content accords with ideas about the regulation of photosynthesis genes according to the demand for photosynthate (Koch, 1996). However, increased RbcS message in the LEV3 plants, whose photosynthesis was not Rubisco limited in the high- CO2 growth conditions and whose glut of leaf carbohydrates rivaled that of the controls, suggests a more direct response to low Rubisco content or some other more subtle perturbation of carbon metabolism caused by it.
Future Experiments
Although the tobacco-rubrum plants had sufficient Rubisco activity for their photosynthesis to encounter the ribulose-P2-regeneration limitation at low light intensity (Fig. 5), it would be interesting to investigate whether higher contents of R. rubrum Rubisco would be able to eliminate the growth impairment under the better illuminated conditions in the high- CO2 growth cabinet. The present experiments indicate that perhaps a doubling of the present content of bacterial Rubisco would be required to achieve this. This will probably necessitate improvement of the translatability of the rbcM mRNA. Experiments with a synthetic rbcM with a plastid-optimized codon preference are in progress.
MATERIALS AND METHODS
Construction and Transformation of Plasmid pLEV3
The transforming plasmid pLEV3 is equivalent to pRubLev14 (Whitney and Andrews, 2001a), except that the rbcM coding region is replaced with the coding region of the tobacco (Nicotiana tabacum) plastid rbcL gene (Fig. 1). A 1,869-bp product encompassing the rbcL promoter, 5′-untranslated region, and coding sequence was amplified from pLEV1 (Whitney et al., 1999) with primers TrbcLCla (Whitney and Andrews, 2001a) and TrbcLHind (5′-CAAGCTTTTACTTATCCAAAACGTCCAC-3′, HindIII site underlined, complement of rbcL terminator codon in bold) and cloned into pGEM-T Easy (Promega, Madison, WI) to give plasmid pTVErbcL. The plasmid was sequenced using BigDye terminator sequencing (Applied Biosystems, Foster City, CA) and the 1,595-bp rbcL NcoI-HindIII fragment was cloned into pRubLev14 to give pLEV3.
The plastid genome of tobacco cv Petit Havana [N,N] was transformed biolistically with plasmids pLEV1 and pLEV3, and spectinomycin-resistant plantlets were regenerated in tissue culture as described (Svab and Maliga, 1993). After three rounds of repeated tissue culture and regeneration, homoplasmic transformants were selected by DNA blotting of BamHI-digested leaf DNA using the rbcL1 probe (Fig. 1). Correct insertion into the plastid genome was confirmed by cloning and sequencing the 2,580- and 2,830-bp DNA products amplified by PCR from total DNA from LEV3 and LEV1 leaves, respectively, using the primers LsD (Whitney et al., 1999) and LsE (Whitney and Andrews, 2001a; Fig. 1).
Plant Growth
LEV1, LEV3, and tobacco-rubrum (tr1; Whitney and Andrews, 2001a) transformants and nontransformed control plants were germinated and grown in 5-liter pots of soil in an air-conditioned growth chamber at 25°C with 200 or 450 μmol quanta m-2 s-1 artificial illumination as previously described (Whitney et al., 1999). The atmosphere was supplemented with CO2 to 2.5% (v/v). Flowers of the transformants were hand pollinated with pollen from nontransformed plants at each generation. For growth analyses, the height of the plants and the number of leaves that they bore were recorded every 2 to 4 d until floral buds appeared, when the weights of the complete shoots were measured before and after oven drying at 80°C.
Leaf Sampling
All physiological, anatomical, and biochemical measurements were made on samples taken 8 h into the 14-h photoperiod from the fifth leaf below the apical meristem of T2-generation tobacco-rubrum, LEV3 and LEV1 transformants, and nontransformed plants of similar physiological age. At this stage, the plants were 15.5 ± 0.8 cm tall, they bore 12.5 ± 0.5 leaves, and the fifth leaf was 14 ± 0.5 cm in diameter. Because the tobacco-rubrum plants grew slower than the others, the former were chronologically older at the time they reached this stage. The plants were grown at 450 μmol quanta m-2 s-1 in 2.5% (v/v) CO2 as described above.
DNA and RNA Blotting
Total DNA was extracted from leaves, electrophoresed, and blotted as described previously (Whitney et al., 1999; Whitney and Andrews, 2001b). DNA blots were probed with an alkaline-phosphatase-conjugated (AlkPhos Direct labeling kit; AP Biotech, Sydney) 1,856-bp rbcL1 DNA probe (Whitney and Andrews, 2001a). Hybridizing fragments were visualized with AttoPhos reagent (Promega) using a Vistra FluorImager (AP Biotech).
Total RNA was isolated from the fifth leaf of T2-generation transformants and nontransformed plants (see above) with Tri-reagent (Sigma, St. Louis) according to the manufacturer's instructions. RNA was electrophoretically separated through formaldehyde-denaturing agarose gels (Sambrook et al., 2000) and was blotted onto Hybond N+ membranes (AP Biotech) as described (Kucharski and Maleska, 1998). Blots were hybridized with 32P-labeled DNA probes (Megaprime labeling kit; AP Biotech), the membranes were exposed to a storage phosphor screen (PhosphorImager 400S; Molecular Dynamics, Sunnyvale, CA), and densitometry was carried out with computer-generated images using ImageQuant software. Blots were hybridized with a 383-bp NcoI-XbaI rbcS DNA fragment from pJSTSSu (Whitney and Andrews, 2001b), an 814-bp aadA DNA probe (Whitney and Andrews, 2001a), or a 991-bp rbcL2 DNA fragment (Fig. 1) amplified from pLEV1 using the primers LSb (Whitney and Andrews, 2001a) and LSa (5′-CGAGCGTGTTGTTGGAGAAAAAGA-3′).
The abundances of rbcL- or rbcM-containing mRNA transcripts were measured in blots of gels loaded with 10 μg of total leaf RNA, using the rbcL2 and aadA DNA probes. Two RNA samples were extracted from each leaf sampled. The abundances of the monocistronic rbcL mRNA in nontransformed and LEV1 plants, and of the bicistronic rbcL-aadA1 and rbcL-aadA3 mRNAs in LEV1 and LEV3 leaf samples were measured using the rbcL2 probe. Similarly, the abundances of the bicistronic rbcM-aadA3, rbcL-aadA1, and rbcL-aadA3 mRNAs in the tobacco-rubrum, LEV1, and LEV3 leaf samples were measured in duplicate blots probed with the aadA probe (Fig. 2B).
Elemental and Anatomical Measurements
Total N and total C content of dried leaf material were measured using an elemental analyzer (model EA 1110; Carlo Erba Instruments, Milan).
Leaf discs (0.5 cm2) were snap frozen in liquid N2 and were immediately embedded onto a grooved aluminum stud using equal volumes of colloidal graphite (Agar Scientific, Essex, UK) and tissue-TEK embedding compound 4583 (Mile Scientific, Naperville, IL). While still frozen, and at approximately 13 Pa of water vapor pressure, leaf thickness and abaxial stomatal density were measured using a scanning electron microscope (model S-2250N; Hitachi, Tokyo) at an acceleration voltage of 27 kV.
Biochemical Analyses
Soluble leaf protein and the content and carbamylation status of Rubisco were measured as described (Whitney and Andrews, 2001a). Leaf ribulose-P2 and P-glycerate pool sizes were measured in leaf discs (2.4 cm2) rapidly frozen in liquid N2 under illumination in the growth chamber as described previously (He et al., 1997). Additional leaf discs (0.5 cm2) were assayed for starch and Glc (total starch and Glc assay kits; Megazyme, Boronia, Australia) as previously described (Atkin et al., 1999). Chlorophyll was measured after extraction with methanol (Porra et al., 1989).
Leaf Gas Exchange
Photosynthetic gas exchange by leaves in air was measured after transfer to the laboratory using a flow-through photosynthesis system (model LI-6400; Li-Cor) as described (Whitney et al., 1999). For mass spectrometric measurements at very high CO2 concentrations, a leaf punch (1 cm2) was taken from the same leaves and CO2 assimilation was measured simultaneously at 25°C in a closed cuvette (0.77 cm3 in volume) attached to a membrane-inlet mass spectrometer (Maxwell et al., 1998; Ruuska et al., 2000). The leaf discs were equilibrated with air containing 3.5% (v/v) CO2 and were illuminated at 240 or 1,050 μmol quanta m-2 s-1 for 10 min before sealing the cuvette and measuring the depletion of CO2.
Acknowledgments
We thank Stephanie McCaffery for assistance with the carbohydrate analyses and Heather Kane and Grant Pearce for reading the manuscript.
References
- Andersson I (1996) Large structures at high resolution: the 1.6Å crystal structure of spinach ribulose-1, 5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate. J Mol Biol 259: 160-174 [DOI] [PubMed] [Google Scholar]
- Atkin OK, Schortemeyer M, McFarlane N, Evans JR (1999) The response of fast- and slow-growing Acacia species to elevated atmospheric CO2: an analysis of the underlying components of relative growth rate. Oecologia 120: 544-554 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman FA (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149-160 [DOI] [PubMed] [Google Scholar]
- Furbank RT, Chitty JA, von Caemmerer S, Jenkins CLD (1996) Antisense RNA inhibition of RbcS gene expression reduces Rubisco level and photosynthesis in the C4 plant Flaveria bidentis. Plant Physiol 111: 725-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, von Caemmerer S, Hudson GS, Price GD, Badger MR, Andrews TJ (1997) Ribulose-1, 5-bisphosphate carboxylase/oxygenase activase deficiency delays senescence of ribulose-1, 5-bisphosphate carboxylase/oxygenase but progressively impairs its catalysis during tobacco leaf development. Plant Physiol 115: 1569-1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosler JP, Wurtz EA, Harris EH, Gillham NW, Boynton JE (1989) Relationship between gene dosage and gene expression in the chloroplast of Chlamydomonas reinhardtii. Plant Physiol 91: 648-655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson GS, Evans JR, von Caemmerer S, Arvidsson YBC, Andrews TJ (1992) Reduction of ribulose-1, 5-bisphosphate carboxylase/oxygenase content by antisense RNA reduces photosynthesis in transgenic tobacco plants. Plant Physiol 98: 294-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Ogren WL (1981) Species variation in the specificity of ribulose bisphosphate carboxylase/oxygenase. Nature 291: 513-515 [Google Scholar]
- Koch KE (1996) Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 509-540 [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleska R (1998) Arginine kinase is highly expressed in the compound eye of the honey bee, Apis mellifera. Gene 211: 343-349 [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2001) Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol 125: 430-436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino A, Shimada T, Takumi S, Kaneko K, Matsuoka M, Shimamoto K, Nakano H, Miyao-Tokutomi M, Mae T, Yamamoto N (1997) Does decrease in ribulose-1, 5-bisphosphate carboxylase by antisense RbcS lead to a higher N-use efficiency of photosynthesis under conditions of saturating CO2 and light in rice plants? Plant Physiol 114: 483-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Badger MR, Osmond CB (1998) A comparison of CO2 and O2 exchange patterns and the relationship with chlorophyll fluorescence during photosynthesis in C3 and CAM plants. Aust J Plant Physiol 25: 45-52 [Google Scholar]
- Miller A, Schlagnhaufer C, Spalding M, Rodermel S (2000) Carbohydrate regulation of leaf development: prolongation of leaf senescence in Rubisco antisense mutants of tobacco. Photosynth Res 63: 1-8 [DOI] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384-394 [Google Scholar]
- Quick WP, Schurr U, Scheibe R, Schulze E-D, Rodermel SR, Bogorad L, Stitt M (1991) Decreased ribulose-1, 5-bisphosphate carboxylaseoxygenase in transgenic tobacco transformed with “antisense” rbcS: impact on photosynthesis in ambient growth conditions. Planta 183: 542-554 [DOI] [PubMed] [Google Scholar]
- Rodermel S (1999) Subunit control of Rubisco biosynthesis: a relic of an endosymbiotic past? Photosynth Res 59: 105-123 [Google Scholar]
- Rodermel S, Haley J, Jiang CZ, Tsai CH, Bogorad L (1996) A mechanism for intergenomic integration: abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc Natl Acad Sci USA 93: 3881-3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel SR, Abbott MS, Bogorad L (1988) Nuclear-organelle interactions: nuclear antisense gene inhibits ribulose bisphosphate carboxylase enzyme levels in transformed tobacco plants. Cell 55: 673-681 [DOI] [PubMed] [Google Scholar]
- Roy H, Andrews TJ (2000) Rubisco: assembly and mechanism. In RC Leegood, TD Sharkey, S von Caemmerer, eds, Photosynthesis: Physiology and Metabolism. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 53-83
- Ruuska SA, Badger MR, Andrews TJ, von Caemmerer S (2000) Photosynthetic electron sinks in transgenic tobacco with reduced amounts of Rubisco: little evidence for significant Mehler reaction. J Exp Bot 51: 357-368 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (2000) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Shimada H, Sugiura M (1991) Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res 19: 983-995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreitzer RJ, Salvucci ME (2002) Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53: 449-475 [DOI] [PubMed] [Google Scholar]
- Stitt M, Schulze D (1994) Does Rubisco control the rate of photosynthesis and plant growth? An exercise in molecular ecophysiology. Plant Cell Environ 17: 465-487 [Google Scholar]
- Svab Z, Maliga P (1993) High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA 90: 913-917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita FR (1999) Microbial ribulose 1, 5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 60: 1-28 [Google Scholar]
- von Caemmerer S, Evans JR, Hudson GS, Andrews TJ (1994) The kinetics of ribulose-1, 5-bisphosphate carboxylase/oxygenase in vivo inferred from measurements of photosynthesis in leaves of transgenic tobacco. Planta 195: 88-97 [Google Scholar]
- Whitney SM, Andrews TJ (2001a) Plastome-encoded bacterial ribulose-1, 5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proc Natl Acad Sci USA 98: 14738-14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ (2001b) The gene for the ribulose-1, 5-bisphosphate carboxylase/oxygenase (Rubisco) small subunit relocated to the plastid genome of tobacco directs the synthesis of small subunits that assemble into Rubisco. Plant Cell 13: 193-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Baldet P, Hudson GS, Andrews TJ (2001) Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J 26: 535-547 [DOI] [PubMed] [Google Scholar]
- Whitney SM, von Caemmerer S, Hudson GS, Andrews TJ (1999) Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiol 121: 579-588 [DOI] [PMC free article] [PubMed] [Google Scholar]