Abstract
Two variants of creeping bentgrass (Agrostis stolonifera cv palustris), developed using tissue culture, have been used to determine the roles of chloroplast-localized small heat shock proteins (CP-sHSPs) in heat tolerance. Results from previous research indicate that the heat-tolerant variant expressed two additional CP-sHSP isoforms not expressed in the heat-sensitive variant, that accumulation of the additional CP-sHSP isoforms was genetically linked to thermotolerance, and that the presence of the additional isoforms in the heat-tolerant variant provided greater protection to photosystem II during heat stress. To determine the basis of the differential expression, we isolated the genes encoding the CP-sHSPs from both variants and characterized their structure and expression. Two genes, ApHsp26.2 and ApHsp26.7a, were isolated from the heat-tolerant variant, and three genes, ApHsp26.2m, ApHsp26.8, and ApHsp26.7b, were isolated from the heat-sensitive variant. The sequence of ApHsp26.2m from the heat-sensitive variant was identical to ApHsp26.2, except for a point mutation that generated a premature stop codon. Therefore, the protein product of ApHsp26.2m did not accumulate in the heat-sensitive line. Mass spectrometry analysis confirmed that ApHsp26.2 encoded for the CP-sHSP isoforms unique to the heat-tolerant variant. An identical mutation was detected in one of the three parental lines used to develop the creeping bentgrass variants. This suggests that ApHsp26.2m was inherited from this parent and did not arise from a mutation that occurred during tissue culture. The presence of two isoforms encoded by the same gene might be due to differential processing of the N-terminal amino acids during or after import into the chloroplast.
When plants are exposed to heat stress, they respond by synthesizing a set of protective proteins, the heat shock proteins (HSPs) (Howarth, 1991). Synthesis of HSPs results in acquired thermotolerance, which allows plants previously subjected to moderately high temperatures to survive subsequent exposure to lethal temperatures (Yarwood, 1961; Lin et al., 1984; Lindquist, 1986). HSPs generally act as molecular chaperones that prevent protein misfolding and aggregation (Vierling, 1991). There are two major groups of HSPs in plants: the high-molecular mass HSPs ranging from 60 to 100 kD and small HSPs (sHSPs) ranging from 15 to 30 kD (Vierling, 1991). The sHSPs are much more abundant in higher plants than in other organisms (Vierling, 1991). They are classified into five gene families based on sequence homology and intracellular localization. Classes I and II are localized to the cytosol, and the other three classes are localized to the chloroplast, endoplasmic reticulum, and mitochondria (Vierling, 1991; Waters et al., 1996).
Studies using transgenic plants have shown that enhanced heat tolerance in higher plants is correlated with HSP101 expression in Arabidopsis (Hong and Vierling, 2000; Queitsch et al., 2000) and HSP18.1 expression in carrot (Daucus carota; Malik et al., 1999). Recent studies indicate that chloroplast-localized small HSPs (CP-sHSPs), with an apparent molecular mass in the range of approximately 21 to 30 kD, also play an important role in heat tolerance, especially in the protection of PSII, the most heat labile component of photosynthesis (Heckathorn et al., 1998). CP-sHSPs are nuclear encoded, translated in the cytoplasm, and transported into the chloroplast (Vierling, 1991). They are characterized by a highly conserved Met-rich region of approximately 28 amino acids. Oxidation of the Met residues appears to influence the oligomerization state of the CP-sHSPs during heat stress (Gustavsson et al., 1999). Besides the Met-rich region, there are two additional conserved regions at the C terminus of CP-sHSP, consensus regions I and II, that are present in all the sHSPs. Consensus regions I and II are involved in oligomerization of sHSPs (Vierling, 1991; Waters et al., 1996).
In tomato (Lycopersicon esculentum) cultivars (Preczewski et al., 2000) and ecotypes of Chenopodium album (Downs et al., 1999) that differ in heat tolerance, there is a positive correlation between the ability of the photosynthetic system to withstand heat stress and the amount of CP-sHSP accumulation. Heat-tolerant species such as Ferocactus tended to accumulate more CP-sHSP per unit protein than heat-sensitive species such as pea (Pisum sativum; Downs et al., 1998). In addition, experiments conducted in vitro demonstrated that immunodepletion of CP-sHSPs from purified thylakoids reduced the level of PSII electron transport at high temperatures, whereas addition of CP-sHSPs to purified thylakoids protected PSII activity at high temperature (Heckathorn et al., 1998). Constitutive expression of a tobacco (Nicotiana tabacum) CP-sHSP gene in tobacco plants improved PSII thermotolerance and its ability to withstand photooxidative stress (Miyao-Tokutomi et al., 1998). Overexpression of Arabidopsis CP-sHSP only enhanced heat tolerance in high, but not low, light conditions in Arabidopsis (Harndahl et al., 1999).
We have been using two variants of Penncross, a cultivar of creeping bentgrass (Agrostis stolonifera cv palustris Huds.; Hanson, 1959) as a model to study the role of CP-sHSPs in heat tolerance (Park et al., 1996, 1997; Luthe et al., 2000). Penncross seeds are produced by random crossing of three bentgrass parental lines (Hanson, 1959). Callus lines initiated from these seeds were used to select and regenerate heat-tolerant and -sensitive bentgrass lines (Luthe et al., 2000). The heat-tolerant variant selected bentgrass (SB) was regenerated from callus that survived selection at 40°C for 1 week. The heat-sensitive variant nonselected bentgrass (NSB) was regenerated from callus that was not subjected to heat stress (Luthe et al., 2000). A study of the heat shock response in SB and NSB indicated that SB synthesized two additional isoforms of the CP-sHSP family that did not accumulate in NSB (Park et al., 1996). The production of the additional CP-sHSP isoforms was genetically correlated with improved heat tolerance and was the first demonstration of a genetic link between HSP polymorphisms and heat tolerance in plants (Park et al., 1996). Mutations altering CP-sHSP expression in these lines may have occurred during callus induction and/or heat selection. Alternatively, heat treatment may have selected for cells containing the most heat-tolerant alleles of the CP-sHSP genes present in the Penncross parental lines.
In addition to expressing the additional CP-sHSP isoforms, SB recovered from heat stress faster and was able to resume normal levels of protein synthesis approximately 2 h earlier than NSB (Park et al., 1997). Recently, in vivo physiological studies have shown that PSII is protected to a greater extent in SB than in NSB during high temperature stress (Heckathorn et al., 2002), and this may account for its higher thermotolerance. Components of the PSII and oxygen-evolving complex, such as D1, OEC33, OEC23, and OEC16, were protected from damage during heat shock in SB but not in NSB (Heckathorn et al., 2002). Protection of PSII and the OEC may result from increased association of the CP-sHSPs, either directly or indirectly with the thylakoids in SB (Heckathorn et al., 2002).
To further elucidate the mechanism of heat tolerance in these two variants and to determine why there is differential expression of the CP-sHSP isoforms, we isolated and sequenced the CP-sHSP gene family members in both NSB and SB and characterized their expression and probable inheritance.
RESULTS
Isolation of CP-sHSP Genes
PCR-based genomic walking was used to isolate two genes (ApHsp26.2 and ApHsp26.7a) from SB and three genes (ApHsp26.2m, ApHsp26.7b, and ApHsp26.8) from NSB. The cDNA sequences of these ApHsp26 family members shared more than 88% similarity, whereas the 5′-untranslated region (UTR), 3′-UTR, and intron regions were less conserved. The deduced amino acid sequences of ApHsp26.7a and b differed by five nucleotides, two of which caused changes in amino acids. ApHsp26.7a has Ser instead of Arg at position 30 and Tyr instead of Ala at position 80. Because the sequence of the promoter regions and UTRs are highly conserved between ApHsp26.7a and ApHsp26.7b (data not shown), they are probably the same gene with single-nucleotide polymorphisms.
When the nucleotide sequence for ApHsp26.2 isolated from SB was compared with the corresponding gene in NSB (ApHsp26.2m), two mutations in ApHsp26.2m were identified. A T was substituted for a C at codon 68, which converts the codon (CAG) for Gln to a stop codon (TAG). There also was an insertion 43 nucleotides downstream of the first mutation. However, this should have no effect on protein expression because it was downstream from the premature stop codon. The promoter regions of ApHsp26.2, ApHsp26.2m, and ApHsp26.8 contain a homopurine stretch, which may be a nuclear matrix attachment site, near -450, but ApHsp26.7 did not (Nover, 1989). There are identical heat shock elements at -190 and TATA box at -25 in all five genes (data not shown).
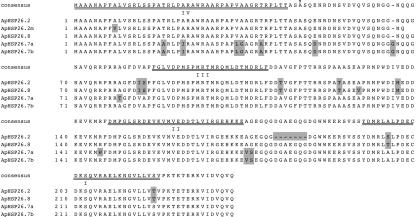
The deduced amino acid sequences indicated that all of the putative ApHsp26 proteins contained a chloroplast transit peptide, the Met-rich region, and consensus regions I and II (Fig. 1). Among these genes, the deduced amino acid sequences of ApHsp26.2 and ApHsp26.8 shared the highest sequence similarity (97.9%). The major difference between ApHsp26.2 and the other proteins is the absence of a seven-amino acid block (DGAEGQG) between consensus regions I and II that is present in the others (Fig. 1). The deduced molecular masses and isoelectric points of the ApHsp26 isoforms after removal of the chloroplast transit sequence are shown in Table I. The protein encoded by ApHsp26.2 was smaller and more basic than the other isoforms.
Figure 1.
Deduced amino acid sequence alignment of ApHsp26.2, ApHsp26.2m, ApHsp26.8, ApHsp26.7a, and ApHsp26.7b. Sequences that differ from the consensus are shaded with gray. Consensus region I (I), consensus region I (II), Met-rich region III (III), and chloroplast transit peptide IV (IV) are double underlined. The arrow marks the N-terminal amino acid for the lower spot in regions 1 and 3 in Figure 5.
Table I.
Deduced mass and pl values for the mature forms of ApHSP26s
| SB
|
NSB
|
|||
|---|---|---|---|---|
| Property Variant | ApHSP26.2 | ApHSP26.7a | ApHSP26.8 | ApHSP26.7b |
| Mass (D) | 21,603.2 | 22,226.8 | 22,259.8 | 22,166.7 |
| pl | 5.00 | 4.77 | 4.86 | 4.85 |
Reverse Transcriptase (RT)-PCR Analysis of ApHsp26s Expression
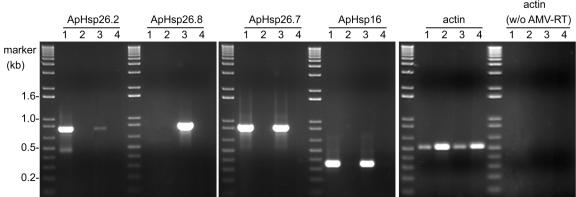
Because the DNA sequence similarity among ApHsp26 family members was higher than 88%, RTPCR analysis was used to determine the transcriptional level of each gene. One common upstream primer was designed from the 5′-UTR, and three downstream gene-specific primers were designed from the 3′-UTRs, the most variable region of each gene. Results from RT-PCR indicate that transcripts of the correct size (795, 815, and 863 bp for ApHsp26.2, ApHsp26.8, and ApHsp26.7, respectively) accumulated during heat shock and not at the control temperature (Fig. 2). The transcript corresponding to ApHsp26.2 appeared to be less abundant in NSB than SB. ApHsp26.8 could not be amplified from SB genome, and this probably accounts for the absence of its transcript in SB. Heat shock resulted in the nearly equal accumulation of transcripts for ApHsp26.7 and ApHsp16 (GenBank accession no. AF007762) in both variants, which indicated that the amount or quality of RNA used for RT-PCR did not account for the difference in ApHsp26.2 expression between SB and NSB. Primers for actin were used as a positive control, and its transcript was present in all samples, but it appeared to be less abundant in the heat-shocked plants. When RT-PCR was conducted in the absence of avian myeloblastosis virus-RT, there was no product verifying that there was no genomic DNA contamination of the RNA samples (Fig. 2).
Figure 2.
Analysis of mRNA level of ApHsp26.2, ApHsp26.8, ApHsp26.7a/b, ApHsp16, and actin by RT-PCR. The RNA was isolated from SB heat shocked for 4 h at 40°C (1), SB control (2), NSB heat shocked for 4 h at 40°C (3), and NSB control (4).
ApHsp26.2m Was Inherited and Did Not Arise during Tissue Culture
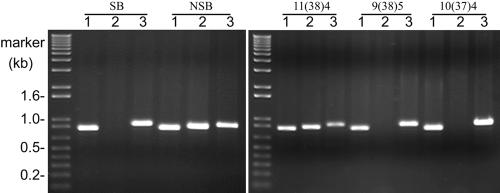
Seeds of the cultivar Penncross are produced by random crossing of three bentgrass parental lines: 9(38)5, 10(37)4, and 11(38)4 (Hanson, 1959). Callus lines initiated from these seeds were used to select and regenerate SB and NSB (Luthe et al., 2000). Mutations altering CP-sHSP expression might have occurred during callus formation and heat selection (Kaeppler et al., 2000). Alternatively, the mutations in ApHsp26.2 may have been inherited from the Penncross parents. To differentiate between these possibilities, either ApHsp26.2 or ApHsp26.2m was amplified from all three parental lines (Fig. 3). Sequence data indicated that the parental lines 11(38)4 and 10(37)4 contained the functional ApHsp26.2, whereas 9(38)5 contained ApHsp26.2m (data not shown). ApHsp26.7a/b was amplified from all three parental lines, whereas ApHsp26.8 was only amplified from 11(38)4 (Fig. 3). Hence, 11(38)4 had three functional CP-sHSP genes, 10(37)4 had two, and 9(38)5 had only one. As a consequence, the mutations that prevent NSB from accumulating the product of ApHsp26.2 were probably inherited from 9(38)5 and did not occur during tissue culture.
Figure 3.
Genomic PCR analysis of SB, NSB, and the three Penncross parental lines [11(38)4, 9(38)5, and 10(37)4]. The primer sets used to amplify ApHsp26.2 or ApHsp26.2m (1), gene ApHsp26.8 (2), and gene ApHsp26.7a or ApHsp26.7b (3) were the same as those used for RT-PCR analysis.
Results from immunoblot analysis showed that the smaller and more basic isoforms absent in NSB also were absent in 9(38)5 (Fig. 4). This provides additional evidence that suggests that ApHsp26.2 encodes at least one of the unique isoforms present only in SB.
Figure 4.
Immunoblot analysis of CP-sHSPs present in the three parental lines of Penncross. Total proteins were extracted from leaves of the parental lines 11(38)4, 9(38)5, and 10(37)4 that were heat shocked at 40°C for 4 h. Proteins were separated by two-dimensional gel electrophoresis, blotted, and probed with an antibody to the Met-rich region of the CP-sHSPs. The white box indicates the position of CP-sHSP that is absent in 9(38)5.
Protein Identification by Matrix-Assisted Laser-Desorption Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) and N-Terminal Sequencing
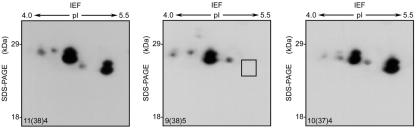
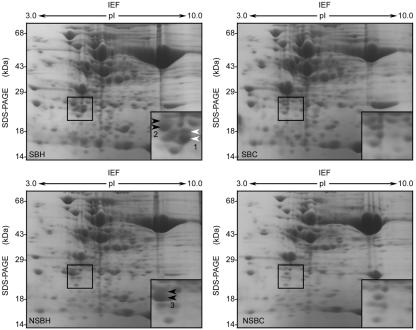
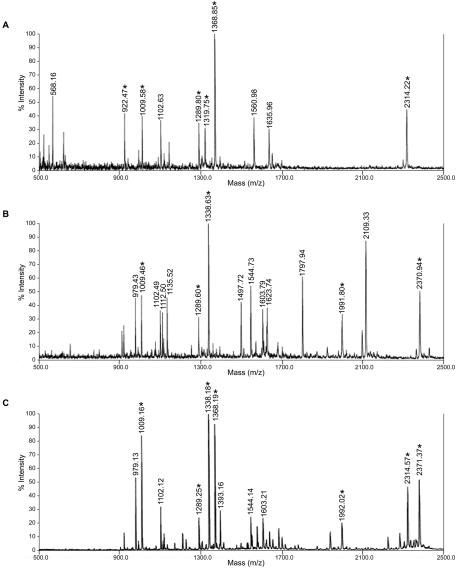
Two CP-sHSPs genes (ApHsp26.2 and ApHsp26.7a) are expressed in SB. Although these two genes share 89.8% similarity in their deduced amino acid sequences, the predicted tryptic peptide fingerprints are different and, only six of 15 peptides have the same masses (Table II). In NSB, the predicted tryptic peptide fingerprints of ApHSP26.8 and ApHSP26.7b also are different (Table II). These differences allowed us to use MALDI-TOF MS to predict the gene that encoded each CP-sHSP isoform. Heat-shocked and control samples of SB and NSB were separated by two-dimensional gel electrophoresis and stained with Coomassie Brilliant Blue (Fig. 5). The stained two-dimensional gels, immunoblots (Fig. 4), and in vivo labeling analysis (Park et al., 1996) indicated that each isoform appeared as a doublet comprised of proteins with similar isoelectric points but slightly different molecular masses. Both members of each doublet were excised and analyzed by MALDI-TOF MS. The doublet spots from each isoform generated the same spectrum, so only one spectra is shown in Figure 6.
Table II.
Predicted peptide mass fingerprints of the mature forms of ApHSP26s
| Property Variant | Mass | ApHSP26.2 Peptide Sequences | Mass | ApHSP26.7a Peptide Sequences |
|---|---|---|---|---|
| D | D | |||
| SB | 2,314.0766a | DNSVDVQVSQNGGNQQGNAVQR | 2,371.0981 | DNSVDVQVSQNGGGNQQGNAVQR |
| 1,936.9295 | AGFDISPFGLVDPMSPMRb | 1,991.8365 | EVSEGQGDGAEGQGDGWWK | |
| 1,368.6794 | LFDDTVGFPTTR | 1,936.9295 | TGFDVAPFGLVDPMSPMR | |
| 1,319.5651 | EAGEGQGDGWWK | 1,338.6688 | LFDDAVGFPTTR | |
| 1,289.6769 | VMVEDDTLVIR | 1,289.6769 | VMVEDDTLVIR | |
| 1,279.5333 | MPWDIMEDDK | 1,247.5612 | MPWDIVEDDK | |
| 1,039.6510 | NGVLLVTVPK | 1,239.5649 | MWFDMPGLSR | |
| 1,019.4714 | LSLPDECDK | 1,025.6353 | NGVLLVSVPK | |
| 1,009.4441 | QMLDTMDR | 1,009.4441 | QMLDTMDR | |
| 986.4901 | SPATASEAPR | 1,003.4764 | LALPDECDK | |
| 944.4142 | SVSSYDMR | 956.4795 | SPAAASEAPR | |
| 922.4451 | FDMPGLSR | 944.4142 | SVSSYDMR | |
| 862.4013 | ASASQENR | 848.3857 | ASASQDNR | |
| 800.4512 | VIDVQVQ | 800.4512 | VIDVQVQ | |
|
635.2995
|
TETER
|
635.2995
|
TETER
|
|
| Mass | ApHSP26.8 Peptide Sequences | Mass | ApHSP26.7b Peptide Sequences | |
| D | D | |||
| NSB | 2,314.0766 | DNSVDVQVSQNGGNQQGNAV QR | 2,371.0981 | DNSVDVQVSQNGGGNQQGNAVQR |
| 1,936.9295 | AGFDISPFGLVDPMSPMR | 1,991.8365 | EVSEGQGDGAEGQGDGWWK | |
| 1,933.7947 | EAGEGQGDGAEGQGDGWWK | 1,906.9189 | AGFDVAPFGLVDPMSPMR | |
| 1,368.6794 | LFDDTVGFPTTR | 1,338.6688 | LFDDAVGFPTTR | |
| 1,289.6769 | VMVEDDTLVIR | 1,289.6769 | VMVEDDTLVIR | |
| 1,279.5333 | MPWDIMEDDK | 1,247.5612 | MPWDIVEDDK | |
| 1,039.6510 | NGVLLVTVPK | 1,025.6353 | NGVLLVSVPK | |
| 1,033.4870 | LTLPDECDK | 1,009.4441 | QMLDTMDR | |
| 1,014.5214 | SPATASEVPR | 1,003.4764 | LALPDECDK | |
| 1,009.4441 | QMLDTMDR | 956.4795 | SPAAASEAPR | |
| 944.4142 | SVSSYDMR | 944.4142 | SVSSYDMR | |
| 922.4451 | FDMPGLSR | 922.4451 | FDMPGLSR | |
| 862.4013 | ASASQENR | 848.3857 | ASASQDNR | |
| 800.4512 | VIDVQVQ | 800.4512 | VIDVQVQ | |
| 635.2995 | TETER | 635.2995 | TETER |
a Mono-isotopic masses higher than 500 D are listed, all Cys were in the reduced form, and Met were not oxidized.
b In a given grass variant, tryptic peptides with masses common to both ApHSP26s are in bold.
Figure 5.
Two-dimensional gel electrophoresis of total protein isolated from heat-shocked (at 37°C for 12 h) SB (SBH), heat-shocked NSB (NSBH), control SB (SBC), and control NSB (NSBC) leaf tissues. Proteins were stained with Coomassie Brilliant Blue. Six protein spots from three regions (1-3) were excised from the gel for protein identification. Insets, Magnified sections of the gels. Black arrows, CP-sHSPs isoforms present in both SB and NSB. White arrows, CP-sHSP isoforms present only in SB.
Figure 6.
The MALDI-TOF MS spectra of CP-sHSP isoforms from region 1 (A), region 2 (B), and region 3 of the two-dimensional gels shown in Figure 5. Asterisks indicate peaks matching the peptide finger prints of ApHSP265.
When the spectra of the SB isoforms (Regions 1 and 2) were compared with the predicted tryptic peptide fingerprints of ApHsp26.2 and ApHSP26.7a (Table II), six peptides from Region 1 matched ApHsp26.2, and five peptides from Region 2 matched ApHsp26.7a (Table III). The key difference between ApHsp26.2 and ApHsp26.7a was the presence of the peptide (EAGEGQGDWWK) with a mass of 1,319.75. As predicted from the derived amino acid sequences (Fig. 1), this peptide is missing the seven-amino acid sequence (DGAEGQG) that is present in the corresponding tryptic peptide (EVSEGQGDGAEGQG- DGWWK) present in the other isoforms. This larger tryptic fragment with a mass of 1,991.80 was present in ApHsp26.7a. This indicated that the smaller isoform, unique to SB and the parental lines 10(37)4 and 11(38)4, probably is encoded by ApHsp26.2 and the larger isoform in Region 2 by ApHsp26.7a. When spectra of the isoforms in NSB (Region 3) were compared with the predicted tryptic peptide fingerprints of ApHsp26.8 and ApHsp26.7b, four trypsin-digested peptides matched ApHsp26.8, and five peptides matched ApHsp26.7b (Table III). The peptide (EAGEGQGDWWK) with a mass of 1,319.75 that distinguishes ApHsp26.2 from the other isoforms was not present in the spectra of the peptides from NSB. Therefore, the major isoform in NSB appeared to contain a mixture of proteins encoded by ApHsp26.8 and ApHsp26.7b.
Table III.
Summary of MALDI-TOF MS data from the proteins in regions 1, 2, and 3 inFigure 5
| Region | Observed Massa | Peptide Sequence Matched | Protein Identified | Sequence Coverage of Mature Protein |
|---|---|---|---|---|
| D | % | |||
| 1 | 2,314.22 | DNSVDVQVSQNGGNQQGNAVQR | ||
| 1,368.85 | LFDDTVGFPTTR | |||
| 1,319.75 | EAGEGQGDGWWK | ApHSP26.2 | 38 | |
| 1,289.80 | VMVEDDTLVIR | |||
| 1,009.58 | QMLDTMDR | |||
| 922.47 | FDMPGLSR | |||
| 2 | 2,370.94 | DNSVDVQVSQNGGGNQQGNAVQR | ||
| 1,991.80 | EVSEGQGDGAEGQGDGWWK | |||
| 1,338.63 | LFDDAVGFPTTR | ApHSP26.7a | 37 | |
| 1,289.60 | VMVEDDTLVIR | |||
| 1,009.46 | QMLDTMDR | |||
| 3 | 2,371.37 | DNSVDVQVSQNGGGNQQGNAVQR | ||
| 1,992.02 | EVSEGQGDGAEGQGDGWWK | |||
| 1,338.18 | LFDDAVGFPTTR | ApHSP26.7b | 37 | |
| 1,289.25 | VMVEDDTLVIR | |||
| 1,009.16 | QMLDTMDR | |||
| 2,314.57 | DNSVDVQVSQNGGNQQGNAV QR | ApHSP26.8 | 27 | |
| 1,368.19 | LFDDTVGFPTTR |
a None were significantly different from the deduced mass values.
N-terminal sequencing of the CP-sHSPs was conducted to further characterize the CP-sHSP isoforms, especially the doublets within each region (Fig. 5) because they generated the same peptide mass fingerprints. However, only the lower spots from Regions 1 and 3 gave reliable N-terminal sequences. The N-terminal sequence obtained for the lower doublet band in Region 1 was ASQENR, which was identical to the deduced amino acid sequence beginning at position 47 of ApHsp26.2 (Fig. 1). The N-terminal sequence obtained for the lower spot in Region 3 was ASQ(D/E)NRD, which appears to be a mixture of N-terminal sequences from ApHSP26.8 (ASQENRD) and ApHSP26.7b (ASQDNRD). The observed transit peptide cleavage site differs by two amino acids from the predicted site (Fig. 1). Because the upper and lower spots from each region gave the same peptide mass fingerprint, the upper spots may have been cleaved at the predicted site. The amino acids at the predicted cleavage site are not charged, so the cleavage variants would have the same pI but slightly different masses, as observed on two-dimensional gels.
DISCUSSION
Heat tolerance in the bentgrass variant SB is genetically linked to the ability to synthesize additional CP-sHSP isoforms that do not accumulate in NSB (Park et al., 1996; Luthe et al., 2000). Results from physiological studies suggest that PSII activity is more robust during heat stress in SB than in NSB (Heckathorn et al., 2002). This appears to be due to protection of proteins in the oxygen evolving complex by either a direct or indirect association of the CP-sHSPs with thylakoids (Heckathorn et al., 2002). This study focused on determining why the additional CP-sHSP isoforms accumulate in SB but not NSB. It is possible that the gene encoding the additional isoforms is present in genome of SB but not NSB or that mutations in the promoter region prevent its expression in NSB or enhance its expression SB. Alternatively, expression could be regulated transcriptionally or posttranscriptionally. Translational or posttranslational factors also may regulate its accumulation (Zimmerman et al., 1989). To determine why there was differential expression, five CP-sHSP genes were isolated from SB and NSB. We cannot be certain that all CP-sHSP genes were isolated. Primers from the conserved Met-rich and consensus regions I and II were used to amplify the CP-sHSP genes. It is possible, but unlikely, that there are CP-sHSP genes with reduced homology in these regions that were not amplified. ApHsp26.2 and ApHsp26.7a were isolated from SB and ApHsp26.2m, ApHsp26.8, and ApHsp26.7b were isolated from NSB. The nucleotide sequence of ApHsp26.2 and ApHsp26.2m are the same except for two point mutations. The first mutation resulted in a premature stop codon at position 68, which results in a truncated protein. If the encoded protein was expressed and imported into the chloroplast, it would have an apparent molecular mass of only 2 kD. Because the antibody used in this study specifically recognized the Met-rich region in the middle of the CP-sHSP, the truncated protein, even if it accumulated, would not have been detected on the immunoblots. These data provided indirect evidence that ApHsp26.2 coded for the CP-sHSP isoform in SB and indicate that its failure to accumulate in NSB was due to the mutations in the corresponding gene.
By sequencing the bentgrass CP-sHSP genes, we were able to compare the deduced amino acid sequences and properties of their products. Identity among the deduced amino acid sequences was 89.8% or greater. The major difference among the proteins was the absence of a seven-amino acid block (DGAEGQG) in ApHsp26.2 (Fig. 1). Deletion of this block of amino acids and removal of the transit sequence results in a protein with a predicted molecular mass of 21.60 kD and pI of 5.00 (Table I). Hence, the protein encoded by ApHsp26.2 is predicted to be smaller and more basic than the processed forms of ApHsp26.8, ApHsp26.7a, and ApHsp26.7b (Table I).
Transcriptional analysis of ApHsp26.2, ApHsp26.8 and ApHsp26.7 using RT-PCR with gene-specific primers indicated that accumulation of ApHsp26.2 mRNA was greater in SB than NSB during heat shock. We believe that these results accurately reflect the relative difference in ApHsp26.2 mRNA abundance because ApHsp26.7 and ApHsp16 were transcribed in equal amounts in both variants. In addition, actin mRNA was detected in all samples. A comparison of approximately 800 bp in the 5′ promoter regions of ApHsp26.2 and ApHsp26.2m revealed only two nucleotide differences at positions -621 and -655 that were upstream from the homopurine stretch (data not shown). Because there were no differences in the region containing the heat shock elements, it is unlikely that differences in the promoter sequences accounted for the lower amount of ApHsp26.2m mRNA. The premature stop codon in ApHsp26.2m could result in transcript destabilization due to incomplete translation of the mRNA. It has been reported in yeast (Saccharomyces cerevisiae) and mammalian systems that a premature stop codon can trigger nonsense-mediated mRNA decay (Leeds et al., 1991; Belgrader et al., 1993; Zhang et al., 1997; Maquat and Carmichael, 2001). This is a mechanism to prevent the accumulation of abnormal or potentially harmful protein products caused by premature stop codons or frameshift mutations. A similar mechanism might also exist in plants. Our results differ from those reported for tobacco (Lee et al., 1998), in which transcripts from the mutated and functional CP-sHSP genes were present at the same level.
Differential accumulation of ApHsp26.2 mRNA in SB and NSB suggests that ApHsp26.2 codes for the unique isoforms found in SB. In addition, protein properties extrapolated from the deduced amino acid sequence suggest that ApHsp26.2 encodes these unique isoforms. To determine if this was the case, MALDI-TOF MS was used to assign each of the ApHsp26 gene products to the isoform spots on two-dimensional gels. Among the CP-sHSPs isolated from creeping bentgrass, only ApHSP26.8 has a similar peptide mass fingerprint to ApHSP26.2 (Table II), but we could not amplify ApHSP26.8 from the SB genome. In addition, only AsHps26.2 has the peptide (EAGEGQGDWWK) that lacks the seven-amino acid sequence (DGAEGQG) predicted to be in the corresponding tryptic peptide present in the other CP-sHSP isoforms. These data suggest that the spectrum from region 1 was generated from the trypsin digestion of the ApHSP26.2 mature protein product. Results from MALDI-TOF MS also suggested that ApHsp26.7a encodes the other CP-sHSP isoforms in SB. In NSB, the CP-sHSPs appeared to be a mixture of proteins encoded by ApHsp26.8 and ApHsp26.7b. “Comigration” of the CP-sHSPs encoded by these two genes is feasible because they have similar molecular masses and isoelectric points (Table I).
When this study was initiated, we postulated that mutations generated in SB or NSB either during tissue culture and/or heat selection altered their heat tolerance. To determine the source of the mutation, genomic PCR was used to amplify ApHsp26.2, ApHsp26.8, and ApHsp26.7 from each of the parental lines. Mutations in ApHsp26.2m from the parental line 9(38)5 were identical to those in NSB, and parental lines 11(38)4 and 10(37)4 had functional ApHsp26.2. In a preliminary experiment, we noticed that dry matter accumulation in 9(38)5 after high temperature stress was lower than that of the other two parental lines. It is possible that heat selection of callus initiated from Penncross seeds eliminated the callus derived from the most heat-sensitive parent. Three other heat-tolerant variants, derived from Penncross in a different series of selections, also synthesized the additional ApHsp26 isoforms, which supported this hypothesis (Luthe et al., 2000). Creeping bentgrass was introduced to the United States from the cool, humid regions of Europe (Hanson and Juska, 1969). As a consequence, the differences in CP-sHSP expression among the parental lines of Penncross likely reflect adaptation to their environment. The accumulation of mutations in CP-sHSP genes could result from the lack of selection pressure when bentgrass is continuously cultivated in temperate regions. This supports results from other research (Downs et al., 1998) that suggests a relationship between thermo-adaptation and CP-sHSP expression.
It still is not clear how the ApHsp26.2 isoforms increase the heat tolerance of SB. It seems unlikely that the seven-amino acid block (DGAEGQG) missing in this isoform could significantly alter its function. However, the results from limited phylogenetic analysis indicate that this sequence or a modified version was present in the monocot but not the dicot species examined (D.S. Luthe, unpublished data). If there are no qualitative differences between the ApHsp26 isoforms, perhaps quantitative differences in total CP-sHSP levels are more important in conferring increased heat tolerance. Densitometry of immunoblots (data not shown) indicated that the relative amount of CP-sHSP per milligram of protein applied to the gel was approximately 2-fold higher in SB than in NSB. Because it has been shown that differences in CP-sHSP levels are positively correlated with thermotolerance (Downs et al., 1998; Preczewski et al., 2000; Knight and Ackerly, 2001), perhaps overall CP-sHSP abundance is the major factor contributing to heat tolerance.
MATERIALS AND METHODS
Plant Growth Conditions
Two variants of creeping bentgrass (Agrostis stolonifera cv palustris), SB and NSB, along with three parental lines of Penncross [designated 9(38)5, 10(37)4, and 11(38)4] were grown in a lighted growth chamber (approximately 150 μmol s -1 m-2) on a 22°C/18°C 14-h day/night cycle. The Penncross parental lines were obtained from Dr. David Huff (Pennsylvania State University, State College). Heat shock treatments were carried out at 40°C in a growth chamber. Alternatively, excised leaf segments were incubated in a water bath at 40°C in sterilized incubation buffer (1% [w/v] Suc, 1 mm K-PO4 [pH 6.0], and 0.02% [v/v] Tween 20).
Two-Dimensional Gel Electrophoresis and Immunoblot Analyses
Samples for two-dimensional gel electrophoresis were prepared using the phenol extraction method as previously described (Park et al., 1996). Protein concentration was measured using bovine serum albumin as standard (Ghosh et al., 1988). For MS analysis, 5 mg of protein was separated first by isoelectric focusing (pH 3-10) as previously described (Park et al., 1996) and further separated by 10% to 15% (w/v) gradient SDS-PAGE. For western-blot analysis, 0.2 mg of protein was separated on a Ready IPG strip (pH 4-7, Bio-Rad Laboratories, Hercules, CA), then further separated by 10% to 15% (w/v) gradient SDS-PAGE.
Proteins were blotted from two-dimensional gels onto a nitrocellulose membrane using a semidry electroblotter (Owl Separation Systems, Portsmouth, NH). The CP-sHSPs were detected by chemiluminescence using the ECL system (Amersham, Piscataway, NJ) according to the manufacturer's instructions. The primary antibody, Abmet (generously provided by Dr. Scott Heckathorn, Syracuse University, Syracuse, NY), which specifically recognizes the Met-rich region of CP-sHSPs (Downs et al., 1998), was used at a dilution of 1:4,000 (v/v).
Isolation of CP-sHSP Genes by PCR-Based Genomic Walking
An adaptor-ligated genomic library was constructed by ligating DraI- or EcoRV-digested genomic DNA with adaptors from the Marathon cDNA amplification kit (CLONTECH Laboratories, Palo Alto, CA). DNA (200 ng) from the adaptor-ligated genomic library was used as template for a 25-μL PCR reaction. Primers designed to amplify possible CP-sHSP gene family members were based on the sequence of gene ApHsp26 (GenBank accession no. AF019144) previously isolated in our laboratory. Primer GSP5-1 (5′GTCGTCGAACAGCCGGTCCATCGTG) and the nested primer GSP5-2 (5′CAGCCGGTCCATCGTGTCCAGCATC) were designed from the Met-rich region to amplify the 5′-flanking regions. The 3′-flanking regions were amplified using primer GSP3-1 (5′GAACGCCCCCTTCGCTCTCGTAA), designed from the transit peptide region, and the nested primer GSP3-2 (5′ GACGGGTGGTGGAAG), designed from a region between consensus regions I and II. For the first round of the PCR, primer AP1 (Marathon cDNA amplification kit) was used with GSP5-1 or GSP3-1. The second round of the PCR was conducted on the undiluted product from the first PCR with primer AP2 (Marathon cDNA amplification kit) and GSP5-2 or GSP3-2. The PCR products were purified using the Gene Clean Kit (BIO101, Vista, CA) and cloned into the pGEM-T Easy vector (Promega, Madison, WI). All sequencing was done using an ABI PRISM 310 autosequencer (PE Applied Biosystems, Warrington, UK). The GenBank accession numbers are AY153758 (ApHsp26.2), AY153759 (ApHsp26.2m), AY153760 (ApHsp26.8), AY153761 (ApHsp26.7a), and AY153762 (ApHsp26.7b).
DNA and Protein Sequence Analysis
Sequence editing and alignment were conducted using the DNASTAR package (DNASTAR Inc., Madison, WI). The transit peptide was predicted using ChloroP (Emanuelsson et al., 1999). The peptide mass fingerprints, theoretical mass, and pI were predicted using PeptideMass (http://us.expasy.org/tools/peptide-mass.html).
RNA Isolation and RT-PCR Analysis
Intact plants were heat shocked at 40°C in a growth chamber for 4 h. Total RNA was isolated (Bowden and Lord, 1979) from heat-shocked and control plants that were maintained at 22°C. The RT-PCR reactions were performed using the Enhanced Avian RT-PCR kit (Sigma, St. Louis). Specific primers were designed from the 5′- and 3′-UTRs of ApHsp26s. Primers RT-uni (5′GCGATCTCCGAGCACTTCTCTG) and RT-a (5′TCACGGGACAGAAACAAAGATGA) were used to amplify gene ApHsp26.2 and ApHsp26.2m specifically. Primers RT-uni and RT-b (5′ACGGGACACAAGCTCACTGAAA) were used for gene ApHsp26.8. Primers RT-uni and RT-c (5′GAGCACTCATTCGCAACACAAC) were used for gene ApHsp26.7a and ApHsp26.7b. Primers actin-5 (5′ GAGAAGATGACCCAGATCATGTTTG) and actin-3 (5′ TCCTAATATCCACGTCGCACTTCAT) were used to amplify the actin gene. Primers HSP16-5 (5′ACCCCTTCTCCCTCGACCTCTG) and HSP16-3 (5′AGCCCGGCCTTCACCTCCTC) were used to amplify gene ApHsp16 (GenBank accession no. AF007762).
Protein Identification by MALDI-TOF MS
The SB and NSB plants were subjected to heat shock at 37°C in a lighted growth chamber for 12 h to maximize the synthesis and accumulation of HSPs. Total protein fraction was isolated from heat-shocked and control leaves as previously described (Park et al., 1996) and separated by two-dimensional gel electrophoresis. The gel was stained with Coomassie Brilliant Blue R-250, and spots corresponding to CP-sHSP isoforms were excised. Trypsin was used for in-gel digestion of the CP-sHSP proteins (Li et al., 1997). The digested peptides were eluted from the gel (Li et al., 1997) and desalted using C18 reverse-phase ZipTips (Millipore Corp, Bedford, MA). Desalted peptides (0.5 μL) were loaded on a 100-well target plate and overlaid with 0.5 μL of matrix solution (8 mg mL-1 α-cyano-4-hydroxycinnamic acid in 50% [v/v] acetonitrile and 0.3% [v/v] trifluoroacetic acid). A Voyager-Elite MALDI-TOF mass spectrometer with a nitrogen laser (337 nm; PerSeptive Biosystems, Foster City, CA) was used in the linear, delay extraction, and positive ion mode. The spectrometer was calibrated externally with calibration mixture 1 (PerSeptive Biosystems). N-terminal amino acid sequences were determined in the Department of Biochemistry at the University of Mississippi, Mississippi Medical Center (Jackson).
This work was supported by the Mississippi Agricultural and Forestry Experiment Station and by the National Science Foundation (Award no. IBN-0114632 to D.S.L.). This article was approved for publication as No. 10242 of the Mississippi Agricultural and Forestry Experiment Station.
References
- Belgrader P, Cheng J, Maquat LE (1993) Evidence to implicate translation by ribosomes in the mechanism by which nonsense codons reduce the nuclear level of human triosephosphate isomerase mRNA. Proc Natl Acad Sci USA 90: 482-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden L, Lord JM (1979) Isolation and cell-free translation of total messenger RNA from germinating castor bean endosperm. Plant Physiol 63: 769-7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs CA, Heckathorn SA, Bryan JK, Coleman JS (1998) The methionine-rich low-molecular-weight chloroplast heat-shock protein: evolutionary conservation and accumulation in relation to thermotolerance. Am J Bot 85: 175-183 [PubMed] [Google Scholar]
- Downs CA, Ryan SL, Heckathorn SA (1999) The chloroplast small heat-shock protein: evidence for a general role in protecting photosystem II against oxidative stress and photoinhibition. J Plant Physiol 155: 488-496 [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978-984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Gepstein S, Heikkila JJ, Dumbroff EB (1988) Use of a scanning densitometer or an ELISA plate reader for measurement of nanogram amounts of protein in crude extracts from biological tissues. Anal Biochem 169: 227-233 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Harndahl U, Emanuelsson A, Roepstorff P, Sundby C (1999) Methionine sulfoxidation of the chloroplast small heat shock protein and conformational changes in the oligomer. Protein Sci 8: 2506-2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AA (1959) Grass Varieties in the United States. U.S. Department of Agriculture, Washington, DC
- Hanson AA, Juska FV (1969) Turfgrass Science. American Society of Agronomy, Inc., Madison, WI
- Harndahl U, Hall RB, Osteryoung KW, Vierling E, Bornman JF, Sundby C (1999) The chloroplast small heat shock protein undergoes oxidation-dependent conformational changes and may protect plants from oxidative stress. Cell Stress Chap 4: 129-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckathorn SA, Downs CA, Sharkey TD, Coleman JS (1998) The small, methionine-rich chloroplast heat-shock protein protects photosystem II electron transport during heat stress. Plant Physiol 116: 439-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckathorn SA, Ryan SL, Baylis JA, Wang D, Hamilton EW, 3rd, Cundiff L, Luthe DS (2002) In vivo evidence from an Agrostis stolonifera selection genotype that chloroplast small heat-shock proteins can protect photo-system II during heat stress. Funct Plant Biol 29: 933-944 [DOI] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392-4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth CJ (1991) Molecular responses of plants to an increased incidence of heat shock. Plant Cell Environ 14: 831-841 [Google Scholar]
- Kaeppler SM, Kaeppler HF, Rhee Y (2000) Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol 43: 179-188 [DOI] [PubMed] [Google Scholar]
- Knight CA, Ackerly DD (2001) Correlated evolution of chloroplast heat shock protein expression in closely related plant species. Am J Bot 88: 411-418 [PubMed] [Google Scholar]
- Lee BH, Tanaka Y, Iwasaki T, Yamamoto N, Kayano T, Miyao M (1998) Evolutionary origin of two genes for chloroplast small heat shock protein of tobacco. Plant Mol Biol 37: 1035-1043 [DOI] [PubMed] [Google Scholar]
- Leeds P, Peltz SW, Jacobson A, Culbertson MR (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev 5: 2303-2314 [DOI] [PubMed] [Google Scholar]
- Li G, Waltham M, Anderson NL, Unsworth E, Treston A, Weinstein JN (1997) Rapid mass spectrometric identification of proteins from two-dimension polyacrylamide gels after in gel proteolytic digestion. Electrophoresis 18: 391-402 [DOI] [PubMed] [Google Scholar]
- Lin CY, Roberts JK, Key JL (1984) Acquisition of thermotolerance in soybean seedlings. Plant Physiol 74: 152-160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S (1986) The heat-shock response. Annu Rev Biochem 55: 1151-1191 [DOI] [PubMed] [Google Scholar]
- Luthe DS, Krans JV, Park S-Y, Wang D (2000) The presence and role of heat shock proteins in creeping bentgrass. In RE Wilkinson, ed, Plant Environment Interactions, Ed 2. Marcel Dekker, Inc., New York, pp 283-319
- Malik MK, Slovin JP, Hwang CH, Zimmerman JL (1999) Modified expression of a carrot small heat shock protein gene hsp17.7 results in increased or decreased thermotolerance. Plant J 20: 89-99 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Carmichael GG (2001) Quality control of mRNA function. Cell 104: 173-176 [DOI] [PubMed] [Google Scholar]
- Miyao-Tokutomi M, Lee BH, Mizusawa N, Yamamoto N (1998) Active oxygen and photoinhibition of photosystem II. In G Garab, ed, Photosynthesis Mechanisms and Effects, Vol 3. Kluwer Academic Press, Dordrecht, The Netherlands, pp 2097-2102 [Google Scholar]
- Nover L (1989) Control of gene expression. In L Nover, D Neumann, K-D Scharf, eds, Heat Shock and Other Stress Response Systems of Plants, Vol 16. Springer-Verlag, Berlin, pp 30-42 [PubMed] [Google Scholar]
- Park S-Y, Chang K-C, Shivaji R, Luthe DS (1997) Recovery from heat shock in heat tolerant and nontolerant variants of creeping bentgrass. Plant Physiol 115: 229-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-Y, Shivaji R, Krans JV, Luthe DS (1996) Heat shock response in heat tolerant and non-tolerant variants of Agrostis palustris Huds. Plant Physiol 111: 515-524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preczewski P, Heckathorn SA, Downs CA, Coleman JS (2000) Photosynthetic thermotolerance is positively and quantitatively correlated with production of specific heat-shock proteins among nine genotypes of tomato. Photosynthetica 38: 127-134 [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579-620 [Google Scholar]
- Waters ER, Lee GJ, Vierling E (1996) Evolution, structure and function of the small heat shock proteins in plants. J Exp Bot 47: 325-338 [Google Scholar]
- Yarwood CE (1961) Acquired tolerance of leaves to heat. Science 134: 941-942 [DOI] [PubMed] [Google Scholar]
- Zhang S, Welch EM, Hogan K, Brown AH, Peltz SW, Jacobson A (1997) Polysome-associated mRNAs are substrates for the nonsense-mediated mRNA decay pathway in Saccharomyces cerevisiae. RNA 3: 234-244 [PMC free article] [PubMed] [Google Scholar]
- Zimmerman JL, Apuya N, Darwish K, O'Carroll C (1989) Novel regulation of heat shock genes during carrot somatic embryo development. Plant Cell 1: 1137-1146 [DOI] [PMC free article] [PubMed] [Google Scholar]