Abstract
Highly conserved mismatch repair (MMR) systems promote genomic stability by correcting DNA replication errors, antagonizing homeologous recombination, and responding to various DNA lesions. Arabidopsis and other plants encode a suite of MMR protein orthologs, including MSH2, the constant component of various specialized eukaryotic mismatch recognition heterodimers. To study MMR roles in plant genomic stability, we used Arabidopsis AtMSH2::TDNA mutant SALK_002708 and AtMSH2 RNA-interference (RNAi) lines. AtMSH2::TDNA and RNAi lines show normal growth, development, and fertility. To analyze AtMSH2 effects on germ line DNA fidelity, we measured insertion-deletion mutation of dinucleotide-repeat sequences (microsatellite instability) at nine loci in 16 or more progeny of two to four different wild-type or AtMSH2-deficient plants. Scoring 992 total alleles revealed 23 (2.3%) unique and 51 (5.1%) total repeat length shifts ([+2], [-2], [+4], or [-4] bp). For the six longest repeat loci, the corresponding frequencies were 22/608 and 50/608. Two of four AtMSH2-RNAi plants showed similar microsatellite instability. In wild-type progeny, only one unique repeat length allele was found in 576 alleles tested. This endogenous microsatellite instability, shown for the first time in MMR-defective plants, is similar to that seen in MMR-defective yeast and mice, indicating that plants also use MMR to promote germ line fidelity. We used a frameshifted reporter transgene, (G)7GUS, to measure insertion-deletion reversion as blue-staining β-glucuronidase-positive leaf spots. Reversion rates increased only 5-fold in AtMSH2::TDNA plants, considerably less than increases in MSH2-deficient yeast or mammalian cells for similar mononucleotide repeats. Thus, MMR-dependent error correction may be less stringent in differentiated leaf cells than in plant equivalents of germ line tissue.
Highly conserved protein systems are used by most organisms to preserve DNA integrity in the face of replication errors, attack from exogenous or endogenous mutagens, and spontaneous events such as deamination or depurination. Several challenges to genomic stability are unique to plant physiology and life forms. Unable to move, plants must cope with (sometimes obligate) exposure to environmental mutagens such as solar UV-B light or heavy metals. Oxygen-producing metabolism subjects cells to the mutational hazards of reactive oxygen species. Perhaps most important, plants lack a true reserved germ line; their gametes are derived from cells that have undergone many somatic divisions, with the potential for mutation fixation at each DNA replication. Although protective responses, such as production of UV-filtering flavonoids, may attenuate DNA damage, environmental challenges to the genome cannot be eliminated. Thus, plant genome maintenance systems at least as rigorous as those found in other organisms would seem essential. In fact, Arabidopsis orthologs of most gene products implicated in maintenance of genomic stability in other eukaryotes have been identified (for review, see Hays, 2002). We focus here on the multiprotein DNA mismatch repair (MMR) system.
Although DNA replicative polymerases copy template DNA with striking fidelity, incorrect bases are incorporated into nascent DNA at rates of 10-6 to 10-7 per base pair replicated. Insertions or deletions of nucleotides (potential frame shift mutations) may be more frequent where nucleotide-repeat sequences can give rise to slip-mispairing (for review, see Kunkel and Bebenek, 2000). The MMR system has evolved to correct a large portion of these errors, further reducing the error rate to 10-9 to 10-10. Repair entails recognition of the mismatch, identification of the nascent strand for excision of DNA surrounding the mismatch, and DNA resynthesis, notably by a replicative polymerase, to fill the excision gap. The importance of such a system is evidenced by its high evolutionary conservation: All eukaryotes and most eubacteria examined have retained genes encoding homologous MMR proteins.
Mismatched bases also arise during recombination. MMR-mediated correction of occasional mismatched heteroduplexes formed during homologous recombination results in gene conversion. MMR also antagonizes homeologous recombination between diverged but similar sequences, apparently in response to mismatches in recombinational intermediates (Chambers et al., 1996). Within a species, this may prevent chromosomal rearrangement, by aborting recombination between duplicated genes. Antagonism of recombination between dissimilar sequences also presents a genetic barrier to interspecies crosses (Matic et al., 1995). The recent demonstration (Dong et al., 2002) that a wheat (Triticum aestivum) MMR homolog (MSH7; see below) is linked to a mutation (ph2a) known to increase recombination frequency in wide crosses is interestingly consistent with this observation, although direct involvement of the gene product has yet to be proven.
Seven homologs of the prototypic prokaryotic MutS protein (MSH) have been identified in eukaryotes, at least three of which (MSH2, MSH3, and MSH6) have been firmly implicated in mismatch correction (for review, see Kolodner and Marsischky, 1999). Mismatch recognition, the responsibility of MutS homodimers in bacteria, is accomplished by MutSα (MSH2·MSH6 heterodimer) in the case of base-base mispairs or single extrahelical nucleotides, or by MutSβ (MSH2·MSH3 heterodimer) for larger extrahelical loopouts. Similarly, the bacterial MutL homodimer, thought to couple mismatch recognition to identification and excision of the nascent strand, is replaced by MutL-homolog (MLH1·PMS2) heterodimers for most post-replication error correction. Escherichia coli and some other bacteria identify nascent strands by their transitory non-methylated d(GATC) sites, which their MutH proteins nick when stimulated by mismatch-bound MutS and MutL. Many bacteria and all eukaryotes lack GATC methylation and MutH homologs; by one hypothesis, they use instead the 3′ ends of nascent DNA or 5′ ends of Okazaki fragments as a basis for strand identification.
A seventh MSH has been identified in Arabidopsis (Culligan and Hays, 2000) and other plants (Horwath et al., 2002) but not thus far in animals. AtMSH7 is most similar to AtMSH6 and also forms heterodimers in vitro with AtMSH2 (designated MutSγ), but the heterodimers exhibit somewhat different affinities for the range of mismatches. AtMSH2-AtMSH6 and AtMSH2-AtMSH7 heterodimers may perform overlapping as well as unique roles in base-mismatch recognition in plants (Culligan and Hays, 2000). Despite the apparent need for rigorous genome maintenance and the presence of clear orthologs of MMR proteins, a recent study found somewhat higher somatic mutation rates in leaves than have been observed in other organisms (Kovalchuk et al., 2000). This puts into question the role of MMR in planta.
Microsatellites, simple repeats of one or a few nucleotides, are found throughout eukaryotic genomes. Microsatellite instability, manifested as repeat length polymorphisms, is a hallmark of MMR deficiency and is used clinically to assess MMR proficiency in mammalian tumors. Instability is thought to arise during replication, when transient melting and out-of-frame re-annealing of nascent and template DNA strands in repeat regions cause extrahelical loopouts that escape proofreading by replicative polymerases. These are corrected efficiently by MMR in wild-type cells, but in MMR-deficient cells, rates of insertion-deletion mutations, especially at longer repeat sequences, increase dramatically—as much as 4 orders of magnitude in long mononucleotide runs (Tran et al., 1997).
To investigate the role of MMR in plant genomic stability, we analyzed effects of deficiency in the essential MMR protein AtMSH2, the constant component in MutSα, MutSβ, and MutSγ. Insertion-deletion mutations in endogenous repeat sequences in a minority of the cells of an organism are difficult to detect in a background of normal sequences. We have circumvented this problem in two ways. First, we constructed frame-shift reporter transgenes by inserting out-of-frame repeat sequences in the uidA (GUS) gene and scoring revertant cells as blue spots—positive staining for β-glucuronidase (GUS)—in transgenic AtMSH2-deficient and -proficient plants. Second, the sequences at several endogenous microsatellite loci of multiple progeny from AtMSH2-defective plants, some of which might be expected to be homozygous or heterozygous for insertion-deletion mutations that occurred in the parent, were compared with microsatellite sequences in progeny of wild-type plants. We used these assays to demonstrate microsatellite instability in plants in which AtMSH2 was disrupted by a T-DNA insertion or a transgene that caused RNA interference (RNAi) of AtMSH2 expression. The effect of MSH2 deficiency on generation and transmission of altered repeat length alleles appears similar to that seen in other higher eukaryotes, clearly implicating MMR in maintaining plant genomic stability. However, the MMR-deficient phenotype scored in leaf tissues appears less pronounced.
RESULTS
Identification of an AtMSH2::TDNA Plant
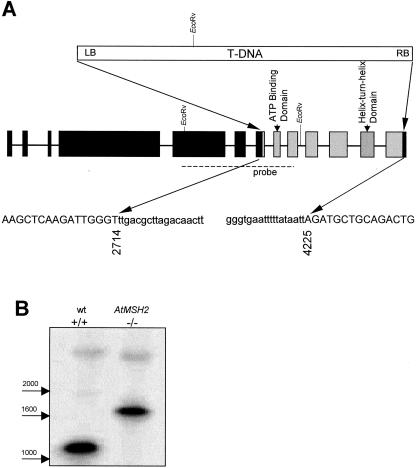
We identified two putative AtMSH2 insertion-mutations, SALK_002707 and SALK_002708, in the Salk Institute T-DNA insertion library database (http://signal.salk.edu/cgi-bin/tdnaexpress) by a BLAST search. PCR screening of plants from each line with pairs of primers respectively specific for AtMSH2 or T-DNA revealed in SALK_002708 a T-DNA insertion beginning in AtMSH2 exon 7. The DNA sequence of the PCR products showed the T-DNA left border beginning after bp 2,714, the T-DNA right border region followed by the final 37 bp of the coding region, and deletion of 1,510 bp of AtMSH2 between the two junctions (Fig. 1). Besides interrupting the coding sequences, the T-DNA insertion caused deletion of two highly conserved MSH2 regions essential for function, the ATP-binding domain and the helix-turn-helix domain (Alani et al., 1997).
Figure 1.
Structure of T-DNA insertion in At-MSH2. A, Sequences of PCR products generated with gene-specific and T-DNA-specific primers were used to deduce the structure of the disrupted AtMSH2 in the line SALK_002708. A single insertion of pROK2 T-DNA at positions 2,714 and 4,225 caused deletion of exons 8 to 12 and portions of exons 7 and 13 (gray boxes) in this line. Sequences of junction regions are below. Capital letters indicate AtMSH2 and lowercase letters indicate the insertion, beginning 2 bp downstream of the left border at the exon 7 junction and preceded by approximately 150 bp of rearranged sequence following the right border at the exon 13 junction. B, DNA blot of EcoRV-digested wild-type and T-DNA insertion homozygotes probed with a radiolabeled At-MSH2 fragment (dashed line).
Progeny of all eight T3 generation SALK_002708 plants tested were found to be homozygous for the T-DNA insertion. DNA-blot (Southern) analysis confirmed the presence of the predicted 1.8-kb AtMSH2::TDNA fragment (Fig. 1). No morphological abnormalities of AtMSH2::TDNA plants were apparent, and seed sets and germination rates were not significantly different from those of wild-type plants (data not shown).
Analysis of Repeat-Sequence Insertion-Deletion Mutation with Frame-Shifted GUS Transgenes
To quantitatively analyze insertion-deletion mutations of specific repeat sequences (microsatellites)—ultimately in a variety of genetic backgrounds—we constructed a series of GUS transgene alleles containing out-of-frame mono- or dinucleotide repeats and introduced them into Arabidopsis. A similar approach was used by Kovalchuk et al. (2000), who measured base-substitution reversion of nonsense codons in a series of GUS-transgene alleles by histochemical detection of GUS+-revertant (blue) spots in whole plants. Previously, a number of investigators had observed highly elevated rates of frame-shift reversion of reporter alleles in MMR-deficient E. coli (Cupples et al., 1990), yeast (Strand et al., 1993), and mammalian cells (Parsons et al., 1993), consistent with instability of endogenous microsatellite sequences in MMR-deficient human tumors (Loeb, 1994).
We inserted frameshifting (G)7, (G)10, (G)13, or (AC)17 runs near the 5′ end of the GUS coding sequence. When a GUS control allele (containing an in-frame (G)12 run), was transformed into Arabidopsis, 35 of 36 lines (progeny of independent transformation events) stained completely blue, demonstrating that the amino acids encoded by the repeated nucleotides did not significantly decrease GUS activity. To eliminate ambiguities that might be caused by T-DNA (GUS) insertions at multiple loci, we identified transformed lines whose T2 progeny segregated 3:1 for antibiotic resistance. Plants from these putative single-locus lines harvested after 2 weeks, then stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronide for detection of GUS activity and decolorized, were generally white; GUS activity was seen primarily in spots varying from single cells to 1 mm in diameter, and more rarely seen in sectors.
Initially, approximately 35 plants—corresponding to a total of roughly 2.4 × 108 cell divisions (see “Materials and Methods”)—of each of the independent single-locus transformed lines were analyzed for each GUS allele. GUS activity was detected by staining in plants from all six (G)13GUS lines, but no GUS activity was detected in five of 11 (G)7GUS lines, one of six (G)10GUS lines, or one of 17 (AC)17GUS lines. We did not determine whether GUS-negative lines contained inactive copies of GUS or transgenes whose location caused expression or reversion levels to be below the limit of detection. The frequency of spots varied more than 10-fold among independent lines transformed with (G)7GUS or (G)10GUS, and varied more than 2 orders of magnitude among independent lines carrying (G)13GUS or (AC)17GUS. Kovalchuk et al. (2000) similarly observed a wide range of base-substitution reversion rates for the same allele in independent transformants. Nonetheless, there was a trend of increasing spot frequency with repeat length; the average numbers of spots/plant for all plants carrying the same allele were 0.6, 1.2, 11.2, and 30.5 for (G)7GUS, (G)10GUS, (G)13GUS, and (AC)17GUS lines, respectively.
We used Southern blotting of single-locus GUS-transformed lines to identify single-locus single-copy lines for further study. To confirm our assumption that increased mutation rates would be detectable by an increase in the frequency of GUS+ spots, we subjected two different single-copy (G)7GUS lines to UV-C light (1,200 J m-2), a mutagen shown to increase frameshifts at repeat sequences in bacteria (Cupples et al., 1990). In each line, UV-C light increased GUS+ spot frequencies approximately 5-fold (data not shown). For comparison, strong doses of UV-C light stimulated (+1) and (-1) frameshift reversions at (G)6 runs in the lacZ gene by factors of five to 20 (Cupples et al., 1990). Thus the observed GUS+ spots most likely correspond to (-1) deletions and (+2) insertions at the (G)7 runs.
We chose a single-copy (G)7GUS line ((G)7GUS-1) selfed to homozygosity for further studies for two reasons. First, the typical reversion frequency of this line—one spot per plant—was high enough to generate statistically significant data using a reasonable number of plants, but could be accurately scored. In contrast, the typically high spot numbers in plants with longer-repeat alleles (more than 50 per plant in six of 17 (AC)17 GUS lines) made quantitative scoring problematic. Second, line (G)7GUS-1 exhibited a stable mutation rate, approximately 0.9 spots per plant, in three successive generations (Table I).
Table I.
Insertion-deletion reversion of (G)7GUS transgenes
| Generations after Transformation with (G)7GUS | Genotype (n)a | GUS+ Revertant Spots per Plantb |
|---|---|---|
| T2 | AtMSH2 +/+ (103) | 1.0 (±0.3) |
| T3 | AtMSH2 +/+ (87) | 1.2 (±0.2) |
| T4 | AtMSH2 +/+ (98) | 0.9 (±0.2) |
| T5c | AtMSH2 +/+ (9) | 1.2 (±1.0) |
| AtMSH2 +/− (27) | 0.9 (±0.6) | |
| AtMSH2 −/− (17) | 5.7 (±0.8) | |
| T6d | AtMSH2 +/+ (92) | 0.8 (±0.2) |
| AtMSH2 −/− (114) | 5.2 (±0.9) |
a Number of plants analyzed.
b Means (95% CI) for 2-week-old plants.
c F2 progeny segregating for AtMSH2::TDNA.
d F3 progeny homozygous for wild-type or mutant AtMSH2 allele.
Reversion Mutation of Transgene Repeat Sequences in AtMSH2-Defective Plants
In comparing GUS reversion rates, we wanted to minimize any background variations, such as in GUS expression, that might arise during propagation of MMR-deficient and -proficient lines. Therefore we chose to analyze segregating F2 progeny of single F1 parents heterozygous or hemizygous for AtMSH2::TDNA and for (single-copy) (G)7GUS-1 alleles, respectively. First, we crossed Arabidopsis line (G)7GUS-1 with an AtMSH2-/- plant. A resultant F1 plant was selfed, yielding F2 progeny whose AtMSH2 genotypes were determined by diagnostic PCR of DNA extracted from single cotyledons excised before GUS staining. The (G)7GUS-1 genotype was similarly confirmed by diagnostic PCR with GUS-specific primers, and only those progeny shown to carry the (G)7GUS-1 allele were scored for blue spots. Both AtMSH2+/+ and AtMSH2+/- F2 progeny showed approximately one spot per plant, similar to the rate for the original (G)7GUS-1 parental line (Table I). However, F2 AtMSH2-/- progeny exhibited approximately five times as many blue spots per plant. To confirm these results, some of the F2 plants homozygous for both the (G)7GUS-1 allele and either AtMSH2+/+ or AtMSH2-/- were selfed to produce F3 progeny. The rate of reversion in F3 (G)7GUS-1/AtMSH2+/+ plants was similar to the original parental G(7)GUS-1 line (Table I), indicating that segregating the T-DNA insertion away had restored wild-type function of AtMSH2. However, F3 (G)7GUS-1/AtMSH2-/- progeny reverted at approximately 5-fold the wild-type rate, similar to AtMSH2-/- F2 progeny.
Instability of Endogenous Microsatellite Sequences in MMR-Defective Plants
We considered it essential to analyze effects of plant MMR deficiency on endogenous repeat sequences for several reasons. Microsatellite instability in a variety of MMR-deficient mammalian cells and whole animals has been extensively studied, providing a basis for comparisons with observations in plants. Because a large number of Arabidopsis and other plant microsatellite sequences have been identified, it was possible to choose a variety of repeat units and repeat lengths, whereas only the (G)7GUS allele was convenient for extensive studies. Furthermore, because detection of expansions/contractions of microsatellite sequences does not depend on gene expression, one potential source of variation among sublines—epigenetic effects on gene transcription, which might affect detectability of GUS+ revertant and/or transcription-coupled repair of endogenous DNA damage—was eliminated, although chromosomal location might affect mutation rates by other mechanisms. Finally, the appearance (or not) of microsatellite length polymorphisms in the (multiple) progeny of a single parent would reflect events in the equivalent of a plant germ line: the floral tissues, the meristematic tissues that give rise to them, and the ancestry of the latter. In contrast, the total of GUS+ revertant (frameshifted) spots scored in whole plants is dominated by events in differentiated leaf cells, which might be subject to less rigorous genomic stability maintenance than the quasi-germ line cells.
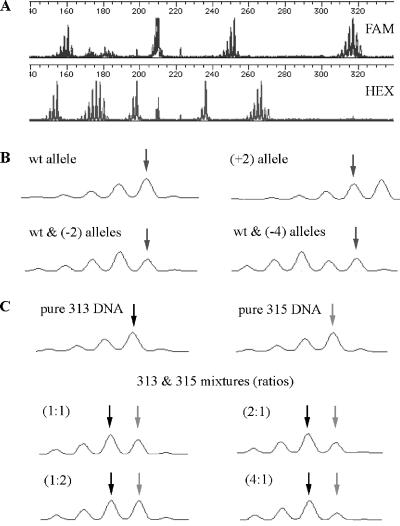
We chose for analysis high multiple repeats of dinucleotide units—to increase the likelihood of slippage and to enhance resolution respectively—that were scattered throughout the genome. Of 10 such microsatellite loci originally identified, nine could be efficiently PCR-amplified using primers that yielded 154- to 318-bp products, most of which could be resolved by capillary electrophoresis. By choosing primers labeled with different fluorescent dyes where product sizes were close, we were able to analyze mutations (if any) at nine different loci (18 alleles) in each electrophoresis run (Fig. 2A). The electropherograms of PCR products templated with individual progeny DNA clearly indicated shifts in repeat lengths (insertions or deletions) of one or more 2-bp units at either one or both alleles at each locus (Fig. 2B). By mixing DNA from two plants, each known to be homozygous for a different repeat length at a particular locus, we reconstructed these patterns and confirmed our ability to deduce genotypes from electropherograms (Fig. 2C). When mixed at a ratio of≥4:1, the minority DNA product was undetectable, strengthening our conclusion that alleles measured in progeny originated in the parent and were transmitted through the germ line. Novel alleles that arose after the 8- or 16-cell stage of progeny development would represent much less than 20% (4:1 mix) of total alleles present and would thus be undetectable in our measurements.
Figure 2.
PCR amplification of DNA containing each of nine different microsatellites from each progeny plant with one of each locus-specific primer pairs, analysis of product mixtures by capillary electrophoresis, detection of fluorescent products, and generation of electropherograms were as described under “Materials and Methods.” A, Electropherograms showing all PCR products; one of each primer pair was labeled with FAM or HEX fluorescent dye. B, Representative electropherogram patterns for locus NGA1107. Apparent absolute size of primary wild-type (wt) product band is 317 bp. Shown also are products where both alleles showed an increase of one repeat unit, increasing apparent product length to 319 bp (+2), where one allele maintained wild-type length and one showed a loss of one repeat [wt & (-2)], and where one allele remained wild type and one showed a shift of two repeats, decreasing apparent length to 313 bp [wt & (-4)]. Arrows indicate position of 317-bp (wild-type) product. C, Patterns for reconstruction mixtures. DNA from plants homozygous (pure) for alleles at locus NGA1107 encoding 313-bp (black arrow) or 315-bp (gray arrow) products or mixtures of the two at indicated ratios (concentrations of DNAs measured before PCR by staining with PicoGreen [Molecular Probes, Eugene, OR]) were analyzed as described above. Note that the primary criterion for scoring a pattern as reflecting a mixture of two different length products—second peak from right greater than a significantly large first peak—is met only for 1:1 and 2:1 mixtures.
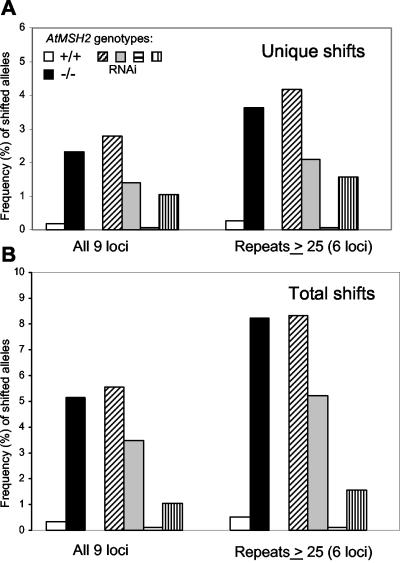
Table II shows analyses of progeny of two wild-type and three AtMSH2-/- plants: a total of 576 alleles from AtMSH2+/+ plants and 992 alleles from AtMSH2-/- plants. We detected only one unique repeat length change (both altered in one plant) in the wild-type plants, but we detected 23 unique length changes and a total of 51 length changes in the AtMSH2-/- progeny. As expected, most shifts occurred in higher repeat length loci: only once among all three loci of 20 repeat units or less, but at least once in all loci with 25 or more repeats. Where a single shift, e.g. (+2), is seen in multiple progeny from the same parent, we cannot distinguish among a single shift early in the lineage of a parental plant that gave rise to a clone of parental cells with the same new allele, multiple independent events that produced that allele, or an intermediate situation. In all cases, several progeny might show the same shift. Thus, Table II shows both the frequency of unique repeat length shifts observed among all targets tested for each locus (typically two alleles for each of 16 plants), expected to be at most 9.4% or 12.6% (three or four unique shifts in 32 targets), and total length shifts observed, which could in principle be 100%, if parental lengths in both alleles were altered very early in development and all progeny were derived from cells with altered-length alleles. Figure 3 shows that for all loci in AtMSH2::TDNA plants, the mean frequency of unique repeat length shifts per allele was 2.3%, compared with 0.2% for the single repeat length shift in AtMSH2+ plants. When we considered the subset of loci with 25 or more repeat lengths, the frequencies were 3.6% versus 0.3%. In the case of two loci, NGA6 and NGA1107, the different lengths observed in the progeny of AtMSH2-/--1 (Table II) were present at predicted Mendelian ratios for an already-heterozygous parent, as if in each case, a (+2) length shift had occurred at both loci early in the development of the parent plant or had occurred in its parent. A similar result was observed at locus NGA151 in the progeny of AtMSH2-/--2 (Table II). These preexisting shifts were not included in the tabulated data, but no such patterns were observed in the AtMSH2+ plants, where MMR is expected to correct microsatellite slippage.
Table II.
Effects of AtMSH2 disruption on stability of nucleotide-repeat sequence (microsatellite) alleles
Frequencies of unique and total repeat-length shifts at indicated loci in progeny of indicated plants.
| Locus
|
AtMSH2::TDNA−/−
|
AtMSH2+/+
|
|||
|---|---|---|---|---|---|
| 1a | 2b | 3b | 1c | 2d | |
| AtEAT | 1.6e (1.6) | 0 | 0 | 0 | 0 |
| (AT)11 | |||||
| T3F17 | 0 | 0 | 0 | 0 | 0 |
| (AG)15 | |||||
| NGA168 | 0 | 0 | 0 | 0 | 0 |
| (CT)20 | |||||
| NGA8 | 3.1ef (7.8) | 3.1f (9.4) | 3.1e (9.4) | 0 | 0 |
| (AG)25 | |||||
| NGA172 | 1.6f (1.6) | 6.2ef (9.4) | 3.1f (3.1) | 0 | 0 |
| (TC)26 | |||||
| NGA1107 | heth | 6.2ef (12.5) | 3.1e (9.4) | 0 | 0 |
| (CT)27 | |||||
| NGA151 | 3.1ef (14.1) | Heth | 6.2ei (9.4) | 0 | 0 |
| (TC)29 | |||||
| NGA6 | heth | 6.2ef (6.2) | 3.1e (9.4) | 0 | 0 |
| (AG)31 | |||||
| NGA139 | 3.1ef (7.8) | 3.1f (6.2) | 3.1e (9.4) | 3.6e (7.1) | 0 |
| (TC)33 | |||||
a Thirty-two progeny (64 alleles per locus) examined.
b Sixteen progeny (32 alleles per locus) examined.
c Fourteen progeny (28 alleles per locus) examined.
d Eighteen progeny (36 alleles per locus) examined.
e (+2) shift.
f (−2) shift.
g (+4) shift.
h Parent already heterozygous at this locus.
i (−4) shift.
Figure 3.
Frequency (%) of repeat length-shifted alleles in endogenous microsatellite loci. Frequencies were calculated by dividing numbers of unique length shifts (A) or total numbers of all alleles showing non-parental lengths (B) in the progeny of two AtMSH2+/+ plants or of three AtMSH2-/- plants or of four individual AtMSH2(RNAi) plants by the total number of alleles tested in each group. Data represent sums for all nine loci analyzed or for the six loci with ≥25 repeat units. No length shifts were detected in progeny of two plants transformed with the empty binary vector pFGC5941.
Inactivation of AtMSH2 by RNAi
We also inactivated MMR via double-stranded RNAi. A fragment of AtMSH2 DNA was ligated in sense and antisense orientations on opposite sides of an 1.4-kb stuffer (intron) fragment, creating binary vector pMSH2(RNAi). This was transformed into Arabidopsis in parallel with transformation with vector-only (pFGC5941) controls. T2 progeny carrying at least one copy of the transgene were analyzed for insertions/deletions at the same endogenous microsatellite loci as described above.
Line-to-line phenotypic variability is well documented in RNAi-mediated suppression in plants (Chuang and Meyerowitz, 2000), so we expected to detect microsatellite alterations more frequently in some RNAi-transformed lines than in others. Progeny of four lines (independent transformations) were analyzed for endogenous microsatellite instability by the PCR-electrophoresis technique (Table III). One line, MSH2(RNAi-3), showed no repeat length shifts in the 288 alleles assayed. However, length shifts were detected in the remaining three lines at varying frequencies. In 16 progeny of MSH2(RNAi-1), MSH2(RNAi-2), and MSH2(RNAi-4), there were eight, four, and three unique length shifts, respectively, equivalent to frequencies of 2.8%, 1.4%, and 1.0%. The frequencies of unique shifts in progeny of lines MSH2(RNAi-1) and MSH2(RNAi-2) approached those observed in AtMSH2-/- insertion-mutant plants (Table III; Fig. 3). As in earlier measurements, mutations were more frequent at loci with ≥25 repeat units. For repeat lengths ≥25 units, the frequencies of unique and total length-shifted alleles for the two high RNAi lines were 3.1% and 6.8%, respectively, similar to those for AtMSH2-/- plants—no length shifts occurred in loci with repeats less than 20 in length.
Table III.
Effects of AtMSH2 RNA-interference on stability of nucleotide-repeat sequence (microsatellites) alleles
Frequencies of unique and (total) repeat-length shifts at indicated loci in progeny of indicated plants.
| Locus
|
AtMSH2 (RNAi)
|
Empty Vectora
|
||||
|---|---|---|---|---|---|---|
| 1b | 2b | 3b | 4b | 1b | 2b | |
| AtEAT | 0 | 0 | 0 | 0 | 0 | 0 |
| (AT)11 | ||||||
| T3F17 | 0 | 0 | 0 | 0 | 0 | 0 |
| (AG)15 | ||||||
| NGA168 | 0 | 0 | 0 | 0 | 0 | 0 |
| (CT)20 | ||||||
| NGA8 | 0 | 0 | 0 | 0 | 0 | 0 |
| (AG)25 | ||||||
| NGA172 | 0 | 6.2cd (25.0) | 0 | 0 | 0 | 0 |
| (TC)26 | ||||||
| NGA1107 | 9.4cde (25.0) | 3.1d (3.1) | 0 | 6.2cf (6.2) | 0 | 0 |
| (CT)27 | ||||||
| NGA151 | 0 | 0 | 0 | 3.1c (3.1) | hetg | 0 |
| (TC)29 | ||||||
| NGA6 | 9.4cde (9.4) | 0 | 0 | 0 | 0 | 0 |
| (AG)31 | ||||||
| NGA139 | 6.2cd (15.6) | 3.1d (3.1) | 0 | 0 | 0 | 0 |
| (TC)33 | ||||||
a Transformed with empty vector pFGC5941.
b Sixteen progeny (32 alleles per locus) examined.
c (+2) shift.
d (−2) shift.
e (+4) shift.
f (+4) shift.
g Parent already heterozygous at this locus.
To ensure that the observed length shifts were the result of RNAi interference and were not an artifact of T-DNA transformation, we analyzed shifts in two lines transformed with empty vectors. The progeny of one plant segregated for two alleles at locus NGA 151, indicating that the parent was heterozygous; as in the case of AtMSH2::TDNA plants heterozygous for two microsatellite alleles (see above), these data were not included in our calculations. No other shifts were detected in any of the 544 other alleles assayed, indicating that the observed shifts in the RNAi lines were due to interference with AtMSH2 mRNA stability by the product of the dsRNA-producing vector pMSH2(RNAi).
DISCUSSION
Arabidopsis and other higher plants whose genomes have been analyzed thus far encode suites of MSH and MLH proteins highly similar to other eukaryotic MMR proteins, plus an extra mismatch recognition component apparently unique to plants, MSH7 (Hays, 2002). Initial biochemical studies of mismatch recognition by MSH2·MSH3, MSH2·MSH6, and MSH2·MSH7 heterodimers (Culligan and Hays, 2000) suggest but do not prove that MSH/MLH systems correct DNA replication errors and antagonize homeologous recombination in plants, as in other organisms. Here, we have analyzed the effects of inactivation in Arabidopsis of AtMSH2—the constant component of all mismatch recognition complexes—by gene disruption or RNAi. The dramatic increases in insertion-deletion mutation of endogenous nucleotide repeat sequences (microsatellite instability) measured in the progeny of AtMSH2-deficient plants are similar to those seen in MMR-defective mammalian and yeast cells, for the first time directly implicating plant MSH proteins—and by extension MLH proteins—in maintenance of plant genomic stability. AtMSH2 disruption also increases insertion-deletion mutation of synthetic repeat sequences in GUS reporter transgenes, mostly in leaves, but to a considerably lesser extent than increases seen for similar repeat sequences in yeast and mammalian cells.
AtMSH2::TDNA homozygotes were recovered in Mendelian ratios from segregating populations and exhibited wild-type seed sets and germination rates. This apparent absence of reduced fitness is consistent with the normal viability of MSH2-deficient mice (Reitmair et al., 1995) and yeast (Reenan and Kolodner, 1992). Nonetheless, it would seem that if MMR functions in plants as it does in other organisms, these and other MMR-defective plants would accumulate errors detrimental to overall fitness over the course of generations. Such a cumulative decrease in fitness has recently been described in MMR-defective Caenorhabditis elegans (Degtyareva et al., 2002), and we are carrying out similar long-term studies.
Instability of microsatellite sequences—that is, expansion/contraction of long repeats of one to four nucleotides—has long been a hallmark of MMR deficiency, particularly in mammalian cells. In the absence of a reserved plant germ line, the nearest thing to a measure of germ line mutation rates would seem to be frequencies of microsatellite length shifts among the progeny of a single parental plant, reflecting events not only during floral development, but also during division of meristematic cells in the lineage of floral precursors. We cannot distinguish between specific single parental microsatellite repeat length shifts that give rise to multiple shifted progeny and multiple independent parental shifts of the same type, so the minimum estimate of instability would be the number of unique shifts—(+2), (-2), etc.—among the total alleles (progeny × loci×2) tested: 2.3% for all loci and 3.6% for all repeats of 25 units or more for AtMSH2::TDNA plants; 0.2% and 0.3%, respectively, for AtMSH2+ plants. For the six loci with repeat lengths of 25 or greater (NGA6, NGA8, NGA139, NGA151, NGA172, and NGA1107), the progeny of all three AtMSH2::TDNA plants analyzed showed at least one shift at every locus, except for the two cases—progeny of plant 2 at locus NGA151 and progeny of plant 1 at loci NGA6 and NGA1107—where parental plants were already heterozygous, indicative of shifts earlier in their ancestry; analysis was impossible in these cases. In comparison, 11.5% of 771 dinucleotide-repeat alleles analyzed in sperm from MMR-defective male mice showed repeat length shifts (Baker et al., 1995), and 74 of 816 (9.5%) of maternal alleles analyzed in (MMR-proficient) progeny of a MMR-deficient female crossed with a wild-type male mouse showed length shifts (Gurtu et al., 2002). These frequencies for MMR-defective mice again reflect both unique events and single length shift events yielding multiple progeny, and so appear similar to the 5.1% (all loci) and 8.2% (≥25 repeat loci) frequencies that we observed for MMR-defective plants. In addition, Gurtu et al. (2002) observed two shifts in 816 paternal alleles analyzed from progeny of a wild-type male and a MMR-deficient female, similar to the one unique (two total) repeat length shift(s) at locus NGA139 that we observed in 578 alleles analyzed in progeny from wild-type parents. Thus MMR deficiency appears to have similar effects on germ line genomic stability in plants and mammals.
Previous measurements of AtMSH2, AtMSH3, and AtMSH7 mRNA levels (Ade et al., 1999) have suggested that expression of MMR genes is quite low in leaves (and perhaps other differentiated tissues), perhaps because DNA replication fidelity is not critical during the last few cell divisions in these tissues or during the DNA endoreduplication that follows (Galbraith et al., 1991). Because GUS mutation-reversion events were mostly scored as blue-staining spots in leaves, apparent spontaneous mutation rates might therefore appear higher than those observed in other systems—mammalian cells dividing in culture for example. Kovalchuk et al. (2000) reported rates of base-substitution reversion of 10-7 to 10-8 per generation in wild-type plants, 2 orders of magnitude higher than rates estimated for other organisms, although the large differences in rates for the same alleles in independent sub-lines made it difficult to determine true basal rates. If in fact MMR control of genomic fidelity is relaxed in leaves, then nucleotide-repeat sequences (microsatellites), whose insertion-deletion rates typically show dramatic increases in MMR-defective cells, would provide a sensitive test. For example, Tran et al. (1997) reported increases of more than 6,000- and 400-fold in mutation of (A)10 and (A)7 runs respectively in MMR-deficient yeast. An (A)14 run mutates at a rate of 1.6 × 10-7 in wild-type yeast cultures, as estimated by standard fluctuation test techniques (Tran et al., 1997). In 2-week-old (G)13GUS-Arabidopsis plants containing approximately 6.7 × 106 cells (see “Materials and Methods”), we measured an average of 11.2 GUS+ spots per wild-type plant, a mutation rate of 2 × 10-6 per division. Although our G runs might mutate slightly more rapidly than the yeast A runs (Boyer et al., 2002), the apparent plant (leaf) insertion-deletion mutation rate was thus an order of magnitude higher than in yeast. Similarly, the basal rate of reversion of (G)7GUS transgenes, about one per plant (2 × 10-7), was 2 orders of magnitude greater than the rate of reversion of an (A)7 run in yeast. Thus the relatively small increase in reversion of (G)7GUS in AtMSH2-/- plants—only 5-fold versus 100-fold for (A)7 runs in yeast—may reflect the lack of efficient MMR in differentiated tissues, so that reversion rates are already high even in the leaves of MMR-proficient plants.
Our studies thus indicate that by the criterion of parent-to-progeny microsatellite stability, plant germ line cells use MMR to correct primer-template slip-mispairs—and by extension other premutational substrates—as efficiently as do yeast and mammals. However, in leaf and perhaps other plant differentiated cells, differences between MMR-deficient and-proficient plants are not as dramatic as in yeast or mammalian cells. In mammalian somatic cells, mutations that give rise to tumorigenesis pose a threat to survival of the organism, so the considerable energy cost of MMR—synthesis of multiple large proteins and replacement of hundreds of nucleotides per correction event—would seem justified, but this may not be the case in plant somatic cells.
MATERIALS AND METHODS
Growth of Plants
All plants were ecotype Columbia-0 Arabidopsis. For measurement of mutation rates, plants were grown in 100-× 2.5-mm petri dishes containing one-half-strength Murashige and Skoog salts, B5 vitamins, and 3% (w/v) Suc, solidified with 0.75% (w/v) agar. For selection of transformed plants, kanamycin was included at 25 mg L-1. We compared mutation rates of plants grown on the same medium with 1% (w/v) Suc and found no significant difference; therefore 3% (w/v) Suc was used for all experiments. Seeds were surface-sterilized by treatment with 95% (v/v) ethanol for 1 h, and 50% (v/v) household bleach for 10 min, followed by four to five washes with sterile water. Sterilized seeds were evenly distributed in a grid pattern of 49 seeds per 100-mm petri dish, stratified at 4°C for 24 h, and placed in a growth chamber at 24°C under constant illumination with cool-white lamps at approximately 60 μmol m-2 s-1. DNA was extracted with an efficiency of recovery of approximately 75% from six 2-week-old plants grown on 3% (w/v) Suc medium, using the method of Draper and Hays (2000). Plants grown under these conditions yielded an average of 1.4 μg DNA plant-1, corresponding to an estimated 6.7 × 106 cells, similar to the estimates of Kovalchuk et al. (2000). Plants were grown for 14 d before scoring reversion of the GUS microsatellite transgene. For endogenous microsatellite assays, seeds from individual soil-grown plants were sterilized and plated on one-half-strength Murashige and Skoog media and grown for 10 to 12 d; whole plants were used for DNA preparations.
Construction of Microsatellite Reporter Genes and Transformation into Plants
The GUS microsatellite reversion reporters were derived from binary vector pBI221 (BD Biosciences Clontech, Palo Alto, CA). Restriction endonucleases BamHI, EcoRI, SalI, SwaI, AscI, XbaI, and MfeI were purchased from Invitrogen (Carlsbad CA). A 2,165-bp BamHI-EcoRI fragment of pBI221 encoding GUS and a nopaline synthesis terminator was ligated into (BamHI+EcoRI)-digested plasmid pBluescript SK+ (Stratagene) forming pGUS3. After removing the SalI recognition site in the polylinker region of pGUS3, an in-frame SalI restriction site was introduced immediately downstream of the initiating ATG codon of the GUS gene by PCR mutagenesis, as follows. A PCR product was templated by pBI221 DNA, using primer OGUS3 (5′-CCGATCCACCATGTCGACTTTACGTCCTGTAGAAACC-3′) encoding a BamHI restriction site, a Kozak consensus translation signal upstream of the initiating ATG codon of GUS, and a SalI restriction site immediately downstream, and primer OGUS1 (5′-CTTCCTGATTATTGACCCACAC-3) 288 bp downstream of the GUS ATG. This product was the source of a BamHI-MfeI fragment used to replace the 5′ end of GUS in (BamHI+MfeI)-digested pGUS3, yielding pGUS5. Oligonucleotides with complementary regions were annealed to produce double-stranded DNA with ends complementary to SalI-restricted pGUS5. To introduce an out-of-frame (G)7 run, a double-stranded DNA fragment with staggered ends complementary to SalI-restriction ends, made by annealing oligonucleotides G7SAL (5′-TCGAAGGGGGGGC-3′) with C7SAL (5′-TCGAGCCCCCCCT-3′), was ligated into SalI-restricted pGUS5. (G)10 and (G)13 runs were introduced in an identical manner by annealing together oligonucleotides with internal G and C repeats. PCR analysis and DNA sequencing of the products confirmed orientation of the introduced nucleotides. Restriction of the sequence-confirmed intermediate plasmids with BamHI and EcoRI released approximately 2.2-kb fragments—encoding the mutated GUS and terminator sequence—that were used to replace the wild-type GUS sequence in (BamHI+EcoRI)-digested binary vector pBI221. Binary vectors were transformed into Agrobacterium tumefaciens strain EHA105 and maintained by kanamycin selection. Plants were transformed using the dipping method (Clough and Bent, 1998), and primary transformants were selected by plating 2,000 seeds plate-1 onto media containing 50 mg L-1 kanamycin.
UV-C Irradiation of Plants
Nine-day-old plants grown in petri dishes were irradiated with 1,200 J m-2 of UV-C light produced by a bank of six 15-W Sylvania germicidal lamps at a distance of 125 cm. Dosage was calculated from measurements made with a Spectroline DRC-100X digital radiometer (Spectronics, West-bury, NY). Plants were immediately returned to growth chambers for 5 d and then histochemically stained, in parallel with unirradiated control plants to detect spots of GUS+ (reverted) cells.
Histochemical Staining
Staining of 2-week-old plants was as described by Kovalchuk et al. (2000). In brief, plants were vacuum-infiltrated for 10 min with a solution of 333 mg L-1 5-bromo-4-chloro-3-indolyl-β-d-glucuronide in 100 mm phosphate buffer, pH 7.0, and 0.1% (v/v) Triton X-100. After 48 h of incubation at 37°C, plants were decolorized in ethanol. Blue spots were counted manually under a stereo dissecting microscope. The exact tissue layers contributing to blue staining were not resolvable at this level of magnification.
Genotyping of Transgenic Plants
A modified cetyl-trimethyl-ammonium bromide method, as described by McGarvey and Kaper (1991), was used to prepare DNA from single leaves (for F2 genotyping) or single cotyledons (for mutation rate analyses); DNA was resuspended in 10 or 30 μL of DNA buffer (10 mM Tris, pH 7.0 1mM EDTA), respectively. In PCR reactions templated with 1 μL of DNA, AtMSH2-specific primer OMSH30 (5′-CCAGTTGCCCTACTCCATACTG-3′) was paired with T-DNA-specific primer OLBA1 (5′-TGGTTCACGTAGTGGGCCATC-3′), yielding a 544-bp fragment, or with AtMSH2-specific primer OMSH5 (5′-TTTCAGTGTCAATGTGAGCG-3′), yielding a 1,107-bp product. Typical PCR conditions were 94°C for 3 min followed by 30 cycles of 15 s at 94°C, 15 s at 54°C, and 1 min at 72°C. PCR products were separated by electrophoresis on 0.8% (w/v) agarose.
DNA Analyses
For DNA-blot (Southern) analysis of SALK_002708, 1 μg of EcoRV-digested wild-type or AtMSH2::TDNA genomic DNA was electrophoresed through 0.8% (w/v) agarose gels and transferred onto Magnacharge (Osmonics, Minnetonka, MN) nylon membrane. A 1.1-kb probe was PCR-amplified from Col-0 DNA using AtMSH2-specific primers OMSH21 (5′-GAGTGCTCTGTAGATCTTGACC-3′) and OMSH29 (5′-CAACTTGAGCCATCAGCACAATC-3′) and then 32P-radiolabeled using a DECA-prime II kit (Ambion, Austin, TX). After overnight hybridization at 65°C, the blot was washed in 0.1% (w/v) SDS and 2× SSC for 30 min at room temperature followed by a final 65°C 1-h wash in 0.1% (w/v) SDS, 1× SSC. Southern-blot analysis of lines transformed with GUS-microsatellite alleles was identical except that 1 μg of DNA from each line that segregated 3:1 for kanamycin resistance was digested with EcoRI or BamHI before electrophoresis and blotted as described above. Blots of DNA from GUS-microsatellite-transformed lines were probed with a 32P-radiolabeled 534-bp HincII fragment of GUS excised from pBI221 beginning 605 bp past the initiating GUS ATG codon.
Analysis of Microsatellite Instability
All microsatellite loci in this study (except T3F17, developed by us) were chosen from the list of nucleotide repeat sequences and respective primer pairs for PCR amplification of Bell and Ecker (1994). In some cases, new primers were designed, using Primer 3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi), to alter the length of the resultant PCR products so that a mixture of PCR products from all nine loci could be resolved by differential mobility and/or fluorescence in a single electrophoretic capillary tube. Oligonucleotides, one for each locus-pair labeled with hexachloro-6-carboxyfluoroscein or 6-carboxyfluoroscein fluorescent dyes, were obtained from MWG Biotech (Highpoint, NC). The following forward and reverse primers were used: AtEAT1, 5′-TGCGTGAATGATATGTAACTGG-3′ and 5′-TTTGATTGCCTTATTTGTCTGC-3′; T3F17, 5′-CTGAGTTTATACCGTTGCATCC-3′ and 5′-AAAAAGTGGAAAAACCCTACCC-3′; NGA6, 5′-GACTAAAGTGGGTCCCTTGG-3′ and 5′-CACACCCAAAACTCGTAAAGC-3′; NGA8, 5′-TGGCTTTCGTTTATAAACATCC-3′ and 5′-GAGGGCAAATCTTTATTTCGG-3′; NGA139, 5′-GGTTTCGTTTCACTATCCAGG-3′ and 5′-AGAGCTACCAGATCCGATGG-3′; NGA151, 5′-CCAGAGCTTGTTTTGGGAAG-3′ and 5′-TTTGATGAAACGGAATATAGAAAGC-3′; NGA168, 5′-GAGCTTGTGCCAGTGTCG-3′ and 5′-GGAGCCTCGAATGATGAAGC-3′; NGA172, 5′-CATCCGAATGCCATTGTTC-3′ and 5′-AGCTGCTTCCTTATAGCGTCC-3′; and NGA1107, 5′-CGACGAATCGACAGAATTAGG-3′ and 5′-GGCTACAATAGTGGGAAAACG-3′. The reverse primers for loci ATEAT1, NGA151, NGA172, and NGA1107 were labeled with FAM fluorescent dye; the reverse primers for loci T3F17, NGA6, NGA8, NGA139, and NGA168 were labeled with HEX fluorescent dye. Initial denaturation at 94°C for 3 min was followed by 30 cycles of PCR amplification: 94°C for 30 s, 54°C for 30 s, and 72°C for 30 s, then a final 1-h incubation at 72°C. Up to nine locus-specific PCR reaction products were diluted as empirically found appropriate for subsequent analysis, mixed, and analyzed by capillary electrophoresis and measurement of DNA-band fluorescent intensities, using the ABI Prism 3100 Genetic Analyzer and associated software (Applied Biosystems, Palo Alto, CA). Product sizes were precisely and reproducibly assigned by comparison with an internal lane standard using Genes-can software (GS400HD Rox, Applied Biosystems), although absolute size measurements differed from genome-predicted sizes. Electropherograms were generated using Genotyper software. Initially, DNA samples from 30 progeny of an ATMSH2-/- plant were amplified with radiolabeled primers for loci NGA6, NGA168, or T3F17, electrophoresed on a polyacrylamide gel and visualized by autoradiography, or were amplified with fluorescently labeled primers and analyzed with the ABI 3100 as described above. The two analytical methods identically detected shifts (or not) in the product band distributions generated by PCR slippage (see Fig. 2); all subsequent analyses employed the capillary-electrophoresis Genetic Analyzer technology. In all cases where product distributions appeared shifted, assays were repeated with an independent PCR amplification of DNA from the same plant.
Construction of dsRNA Vector
A dsRNA vector designed to silence AtMSH2 was created as described on the Web site (http://www.chromdb.org/plasmids/pFGC5941.html) by inserting two identical fragments of AtMSH2 in opposite orientations, flanking a 1.4-kb chalcone synthase intron under the control of a cauliflower mosaic virus 35S promoter into binary vector pFGC5941 (Arabidopsis Biological Resource Center, accession no. CD3-447). In brief, a 735-bp fragment of AtMSH2 beginning at bp 829 of the coding sequence, was amplified from an AtMSH2 cDNA clone (Culligan and Hays, 1997) using one primer designed to add SwaI and BamHI recognition sites to the 5′ end of the PCR product paired with a downstream primer designed to add XbaI and AscI sites to the 3′ end of the PCR product. A (SwaI+AscI)-digested PCR product was ligated into the SwaI and AscI sites of pFGC5941. An XbaI-BamHI fragment of the original PCR product was subsequently ligated into the BamHI and XbaI sites of the resultant intermediate plasmid. The final construct, pMSH2(RNAi), or empty vector pFGC5941 was transformed into A. tumefaciens strain EHA105 and used for parallel plant transformations.
Acknowledgments
We thank Kevin Culligan for bringing to our attention the T-DNA mutant in the database. We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and the Arabidopsis Biological Resource Center Stock Center (Ohio State University, Columbus) for providing seeds.
This research was supported by the National Science Foundation (grant no. MCB 0078262 to J.B.H.) and by a National Institute of Environmental Health Science Training Grant (grant no. 1P42 ES10338 to S.R.B.).
References
- Ade J, Belzile F, Philippe H, Doutriaux M-P (1999) Four mismatch repair paralogues coexist in Arabidopsis thaliana: AtMSH2, AtMSH3, AtMSH6-1 and AtMSH6-2. Mol Gen Genet 262: 239-249 [DOI] [PubMed] [Google Scholar]
- Alani E, Sokolsky T, Studamire B, Miret JJ, Lahue RS (1997) Genetic and biochemical analysis of MSH2p-MSH6p: role of ATP hydrolysis and Msh2p subunit interactions in mismatch base pair recognition. Mol Cell Biol 17: 2436-2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Bronner CE, Zhang L, Plug AW, Robatzek M, Warren G, Elliott EA, Yu J, Ashley T, Arnheim N et al. (1995) Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell 82: 309-319 [DOI] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137-144 [DOI] [PubMed] [Google Scholar]
- Boyer JC, Yamada NA, Roques CN, Hatch SB, Riess K, Farber RA (2002) Sequence dependent instability of mononucleotide microsatellites in cultured mismatch repair proficient and deficient mammalian cells. Human Mol Genet 11: 707-713 [DOI] [PubMed] [Google Scholar]
- Chambers SR, Hunter N, Louis EJ, Borts RH (1996) The mismatch repair system reduces homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol 16: 6110-6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97: 4985-4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735-743 [DOI] [PubMed] [Google Scholar]
- Culligan KM, Hays JB (1997) DNA mismatch repair in plants: an Arabidopsis thaliana gene that predicts a protein belonging to the MSH2 subfamily of eukaryotic MutS homologs. Plant Physiol 115: 833-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Hays JB (2000) Arabidopsis MutS homologs-AtMSH2, At-MSH3, AtMSH6, and a novel AtMSH7-form three distinct protein heterodimers with different specificities for mismatched DNA. Plant Cell 12: 991-1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupples CG, Cabrera M, Cruz C, Miller JH (1990) A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics 125: 275-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyareva NP, Greenwell P, Hofmann ER, Hengartner MO, Zhang L, Culotti JG, Petes TD (2002) Caenorhabditis elegans DNA mismatch repair gene msh-2 is required for microsatellite stability and maintenance of genome integrity. Proc Natl Acad Sci USA 99: 2158-2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Whitford R, Langridge P (2002) A DNA mismatch repair gene links to the Ph2 locus in wheat. Genome 45: 116-124 [DOI] [PubMed] [Google Scholar]
- Draper CK, Hays JB (2000) Replication of chloroplast, mitochondrial and-nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J 23: 255-265 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Knapp SJ (1991) Systemic endopolyploidy in Arabidopsis thaliana. Plant Physiol 96: 985-989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtu VE, Verma S, Grossmann AH, Liskay RM, Skarnes WC, Baker SM (2002) Maternal effect for DNA mismatch repair in the mouse. Genetics 160: 271-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays JB (2002) Arabidopsis thaliana, a versatile model system for study of eukaryotic genome-maintenance functions. DNA Repair 1: 579-600 [DOI] [PubMed] [Google Scholar]
- Horwath M, Kramer W, Kunze R (2002) Structure and expression of the Zea mays mutS-homologs Mus1 and. Mus2 Theor Appl Genet 105: 423-430 [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Marsischky GT (1999) Eukaryotic DNA mismatch repair. Curr Opin Genet Dev 9: 89-96 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I, Kovalchuk O, Hohn B (2000) Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J 19: 4431-4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Bebenek K (2000) DNA replication fidelity. Annu Rev Biochem 69: 497-529 [DOI] [PubMed] [Google Scholar]
- Loeb LA (1994) Microsatellite instability: marker of a mutator phenotype in cancer. Cancer Res 54: 5059-5063 [PubMed] [Google Scholar]
- Matic I, Rayssuguier C, Radmann M (1995) Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80: 507-515 [DOI] [PubMed] [Google Scholar]
- McGarvey P, Kaper JM (1991) A simple and rapid method for screening transgenic plants using the PCR. Biotechniques 11: 428-432 [PubMed] [Google Scholar]
- Parsons R, Li G-M, Longley MJ, Fang W-H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P (1993) Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell 75: 1227-1236 [DOI] [PubMed] [Google Scholar]
- Reenan RA, Kolodner RD (1992) Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics 132: 975-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmair AH, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker HW, Wakeham A, Liu B et al (1995) MSH2 deficient mice are viable and susceptible to lymphoid tumours. Nat Genet 11: 64-70 [DOI] [PubMed] [Google Scholar]
- Strand M, Prolla TA, Liskay RM, Petes TD (1993) Destabilization of tracts of repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365: 274-276 [DOI] [PubMed] [Google Scholar]
- Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA (1997) Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading mutants. Mol Cell Biol 17: 2859-2865 [DOI] [PMC free article] [PubMed] [Google Scholar]