Abstract
Yeast chorismate mutase (EC 5.4.99.5) shows homotropic activation by the substrate, allosteric activation by tryptophan, and allosteric inhibition by tyrosine. In this study mutants of chorismate mutase have been found that remain sensitive to one allosteric effector (tryptophan) but insensitive to the other (tyrosine). These mutations are located in the catalytic domain: loop 220s (212–226) and helix 12 (227–251). The first example starts with the Thr-266 → Ile mutant that had previously been shown to be locked in the activated R state. The additional mutation Ile-225 → Thr unlocks the R state and restores the activation by tryptophan but not the inhibition by tyrosine. The second example refers to a molecular trigger for the switch between the T and R state: a hydrogen-bonded system, which stabilizes only the T state, from Tyr-234 to Glu-23 to Arg-157. Various mutants of Tyr-234, especially Tyr-234 → Phe, are unresponsive to tyrosine but are activated by tryptophan. This separation of activation from inhibition may indicate a pathway for activation that is independent of the allosteric transition and may also be consistent with an intermediate structure between T and R states.
Keywords: Claisen rearrangement, site-directed mutagenesis, allosteric regulation
When the velocity of an enzyme-substrate reaction is plotted versus substrate concentration, a hyperbolic curve is usually present (if substrate inhibition is not large) when cooperativity among active sites is absent. On the other hand, when cooperativity is present, a sigmoidal curve (1–3) is usually found, thus implying two (or more) structural states that may have different affinities for substrate (K system) or different rates of catalysis (V system). This cooperative enzyme may also respond to inhibition or activation at a regulatory site that is distinct (2) from the active site. The regulatory ligand usually has a different structure (allosteric) from that of the substrate, and the enzyme itself is usually referred to as allosteric.
Models for allosteric behavior include the symmetry model (3), the progressive distortion model (4), and the sequential models (5). In the simplest symmetry model the equilibrium between the less-active T state and the more-active R state is changed by substrate at the active sites or (and) by inhibitor or activator at distant allosteric sites. In the present study we consider the simplest models of allosteric behavior in relation to the allosteric enzyme chorismate mutase (chorismate pyruvatemutase, EC 5.4.99.5) from Saccharomyces cerevisiae.
Chorismate mutase catalyzes the conversion of chorismate to prephenate. This isomerization is the first committed step toward phenylalanine and tyrosine (6). The enzyme from Saccharomyces cerevisiae exhibits sigmoidal dependence of activity as substrate concentration is varied and also shows allosteric feedback inhibition by tyrosine and activation by tryptophan (7, 8). An oversimplified description is that substrate causes the sigmoidal behavior of this dimeric enzyme, and that tyrosine and tryptophan modulate the T-to-R equilibrium in opposite directions (3, 7, 8). A mutation of Thr-226 → Ile yields an activated enzyme locked in the R state and insensitive to either allosteric effector, Tyr or Trp (8). Seven other replacements at position 226 yield mutant enzymes that have regulatory behavior between the wild type and the Thr-226 → Ile mutant (9).
Three-dimensional structures have been determined for the R state (Thr-226 → Ile plus tryptophan) (10), and for the T state (wild type plus tyrosine) (11). This T-to-R transition is characterized by a rotation of the major part of one subunit relative to the other by 15°. When a transition state inhibitor (12) is present in the wild-type enzyme plus tryptophan or tyrosine, this relative rotation becomes 22°, to yield a super-R state (13). Although position 226 is not at the regulatory site, it is nearby, but not at, the active site in a “220s” loop [212–226] between two helices, H11 [195–211] and H12 [227–251], both of which contribute binding residues to the active site.
In the present study two new mutants are examined. The Thr-226 → Ile mutant, which is “frozen” R state, has been subjected to a second mutation of Ile-225 → Thr. This double mutant restores activation by tryptophan, although it remains insensitive to tyrosine. This loss of sensitivity to the allosteric inhibitor tyrosine is also shown by mutations of Tyr-234, which remain sensitive to activation by tryptophan. Tyr-234 is hydrogen bonded only in the T state to Glu-23. In the activated R state the hydrogen-bonding system from Tyr-234 to Glu-23 to the active site residue Arg-157 is not present (10, 11, 22). It will be argued here that the separation of activation from inhibition is inconsistent with the simplest models of allosteric transitions.
MATERIALS AND METHODS
Yeast Strains, Media, Plasmids, and Transformation.
All yeast strains used are derivatives of the S. cerevisiae laboratory strain X2180-1A (MATa, gal2, SUC2, mal CUP1) and X2180–1B (MATα, gal2, SUC2, mal, CUP1). Derivatives of plasmid pME605 (9) were used for overexpression of chorismate mutase enzymes in strain RH1242 (Mata, aro7, leu2–2). Plasmid pME781 was constructed by inserting a blunt-ended XhoI/BamHI fragment from pGEM7Zf+ (Promega) carrying the ARO7 gene as EcoRI fragment into the XbaI/HindIII digested and flushed 2-μ derivative pJDB207 (14). The 5′ or 3′ terminal portions of the gene were replaced by a PCR-mutagenized (15) NdeI/XbaI or XbaI/BamHI fragment, respectively. The NdeI site has been introduced to the vector by site-directed mutagenesis at the ATG start codon of the ARO7 ORF. Transformation was carried out by the LiAc method (16). Minimal vitamins minimal medium for the cultivation of yeast was described earlier (17). Strains requiring amino acids were supplemented with 30 mg/liter l-tyrosine or 50 mg/liter l-phenylalanine.
Purification of Yeast Chorismate Mutase.
Yeast cells were grown at 30°C in 10-liter rotating fermentors under aeration. Cells were harvested in mid-log phase at an OD546 of 4–6, washed twice with 50 mM K-phosphate buffer, and stored in l ml buffer/g wet cells at −20°C containing protease inhibitors [0.1 mM phenylmethylsulfonyl fluoride (PMSF), 0.2 mM EDTA, and l mM DTT]. For purification, 80–110 g of cell paste was thawed and run three times through a French Pressure Cell (18,000 psi). Cell debris was sedimented by centrifugation at 30,000 × g for 20 min. The enzyme was purified by ammonium sulfate precipitation, hydrophobic interaction chromatography on ethylamino Sepharose, anion exchange chromatography on MonoQ, and gel filtration on Superdex 200 according to the procedure described by Schmidheini et al. (8). Additionally, PMSF was added to the equilibration buffer for the ethylamino Sepharose column, dialysis was used to desalt protein extracts, and chorismate mutase was applied to a second MonoQ column (HR 5/5). Chorismate mutase was detected by SDS/PAGE (18), by enzyme assays, and by immunoblotting with rabbit antibody raised against purified yeast chorismate mutase and a second antibody with alkaline phosphatase activity. Measurements of protein concentrations were performed by using the Bradford assay calibrated with BSA (19).
Enzyme Assays (7).
A stop assay as described previously was used for measuring enzymatic activity of yeast chorismate mutase. The assay was standardized by keeping enzymatic reactions at 30°C and equilibrating the spectrophotometer cell to the same temperature. Reaction volumes of 250 μl containing 100 mM Tris at pH 7.6, 2 mM EDTA, 20 mM DTT, optionally 0.1 mM tyrosine or 0.01 mM tryptophan, chorismate mutase enzyme, and chorismate in a range from 0.25 to 10 mM were used. The reaction was started by adding a mix of all ingredients to the prewarmed chorismate solution. Reaction was stopped by adding 250 μl of 1 M HCl. After an incubation time of 10 min, 4 ml of l M NaOH was added and OD320 was measured against H2O. Blanks of increasing chorismate concentrations without enzyme were prepared and their absorbances were subtracted from optical densities measured for enzyme activities. A calibration curve was measured with different, known phenylpyruvate concentrations that had been treated in the same way as the enzyme reactions. The molecular extinction coefficient at 30°C was determined as 13,095 M1 cm−1. The collected data were transformed to international units (μmol/min) per mg enzyme. The maximum velocity, Vmax, the Hill coefficient, nH, and the substrate concentration at half-maximal velocity, S0.5 or Km, were determined by using a computer program applying the Quasi-Newton method (Davidon–Fletcher–Powell algorithm) to fit optimal curves to the data (20). For substrate saturation curves data were fitted either to the Michaelis–Menten equation [v = Vmax [S] (Km + [S])−1] or to the Hill equation [v = Vmax [S]n ([S]n + S′)−1], where S′(l/n) = S0.5. Eadie–Hofstee plots (v[S]−1 vs. v) were drawn to decide to which equation a set of kinetic data had to be applied. Enzyme kinetics without cooperativity result in a linear curve, whereas even small degrees of cooperativity result in concave curvatures of the kinetic data (21). Hill plots (log (v (Vmax − v)−1)) vs. log [S] were used to calculate Hill coefficients. The resulting Vmax values were transformed to catalytic constants [kcat = Vmax Mr E0−1 (60s)−1; substrate turnover per enzyme dimer]. The numerical values turned out to be identical because of the use of 60,000 as molecular weight for chorismate mutase.
RESULTS
Additional Replacement of Ile-225 → Thr Unlocks the Ile-226 R State and Restores Tryptophan Activation of the Unregulated Ile-226 Chorismate Mutase.
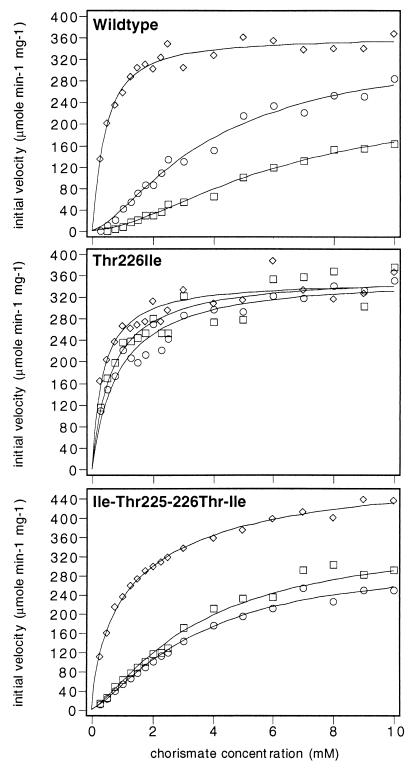
Loop 220s of yeast chorismate mutase and the adjacent helix 12 are located within the catalytic domain of yeast chorismate mutase. For example, amino acid residue Glu-246 of helix 12 is part of the catalytic center (10, 13, 22). Thr-226 is the last residue of loop 220s and connects loop 220s to helix 12. A replacement of Thr-226 by isoleucine results in an unregulated, constitutively activated enzyme (8, 9). Fig. 1 (Top) shows the substrate saturation curves for the purified wild-type chorismate mutase. The three substrate saturation curves were determined in the absence of effectors, in the presence of the inhibitor tyrosine (100 μM), or in the presence of the activator tryptophan (10 μM). Catalytic constants derived from the analysis of the steady-state kinetics are summarized in Table 1. Yeast chorismate mutase exhibits positive cooperativity for chorismate and thus shows a typical sigmoid substrate saturation curve. In the presence of tryptophan, substrate affinity is elevated (Fig. 1 and Table 1), and this activated enzyme follows noncooperative Michaelis–Menten kinetics. When the inhibitor tyrosine is added (in the absence of tryptophan), chorismate mutase reaches half-maximal velocity at higher chorismate concentrations (Fig. 1 and Table 1). Here, the increasing flatness of the substrate saturation curve makes it difficult to extrapolate the curves to find Vmax. The substrate saturation curves of the Thr-226 → Ile mutant (Fig. 1) under all effector conditions are comparable to the curve for activated wild-type chorismate mutase. This mutant enzyme shows no substrate cooperativity and no regulation by either of the effectors, tyrosine or tryptophan. In the wild-type chorismate mutase an isoleucine is located at position 225. Therefore, an additional substitution at position 225 by threonine yields an enzyme in which 225 and 226 of the wild-type enzyme are interchanged. The question under investigation was whether a second site mutation is able to suppress the phenotype of the Ile-226 mutant. This novel enzyme with the amino acid sequence Thr-Ile-225–226 instead of the wild-type Ile-Thr-225–226 regains tryptophan activation similar to that shown by wild-type chorismate mutase (Fig. 1). Remarkably, this enzyme is insensitive to the inhibitor amino acid tyrosine. The unliganded doubly mutated enzyme exhibits cooperativity as chorismate is varied and the tryptophan-activated enzyme has the characteristics of a Michaelian enzyme. Moreover, the catalytic constant is increased compared with wild-type enzyme (Table 1). A threonine at position 225 therefore is able to unlock the constitutive R state of Ile-226 chorismate mutase. Because the enzyme does not respond to tyrosine, loop 220s might play an essential role in the stabilization of the inhibited T state of yeast chorismate mutase. Thr-226 is the last residue of the 220s loop next to helix 12. Subsequently, we analyzed mutants in helix 12 to examine their participation in the allosteric inhibition of yeast chorismate mutase.
Figure 1.
Substrate saturation plots of wild-type, Thr-226 → Ile, and Ile-225 → Thr-Thr-226 → Ile mutant chorismate mutase. The enzymes were assayed with 10 μM tryptophan (diamonds), without effectors (circles), or in the presence of 100 μM tyrosine (squares). The data were fitted to functions describing cooperative or Michaelis–Menten-type saturation.
Table 1.
Kinetic parameters of wild-type and mutant chorismate mutases
| Protein | Value of protein with amino acid replacement
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Inhibited (100 μM tyrosine)
|
Unliganded
|
Activated (10 μM tryptophan)
|
|||||||
| kcat, s−1 | Km, S0.5, mM | nH | kcat, s−1 | Km, S0.5, mM | nH | kcat, s−1 | Km, S0.5, mM | nH | |
| Wild type | 240 | 6.2* | 1.7 | 342 | 4.0 | 1.5 | 361 | 0.4 | 1.1† |
| Thr-226 → Ile | 362 | 0.7 | 0.6† | 361 | 0.9 | 0.6† | 351 | 0.4 | 0.7† |
| Ile-225 → Thr- | 366 | 3.5 | 1.3 | 320 | 3.6 | 1.3 | 535 | 1.5 | 0.7† |
| Thr-226 → Ile | |||||||||
| Glu-23 → Asp | 655 | 9.5* | 1.2† | 630 | 4.5 | 1.1† | 625 | 1.6 | 1.0† |
| Glu-23 → Gln | 14 | 5.4* | 1.4 | 31 | 10.7* | 1.3 | 171 | 6.0 | 1.1† |
| Glu-23 → Ala | ‡ | ‡ | ‡ | ‡ | ‡ | ‡ | 14 | 5.2 | 1.2 |
| Tyr-234 → Phe | 457 | 1.2 | 1.4 | 420 | 0.9 | 1.2 | 565 | 0.7 | 1.0† |
| Tyr-234 → Ala | 243 | 1.2 | 1.1† | 228 | 1.1 | 1.2† | 252 | 0.8 | 1.3 |
| Tyr-234 → Ser | 66 | 2.6 | 1.3 | 72 | 3.2 | 1.3 | 74 | 1.3 | 1.7 |
| Tyr-234 → Glu | 44 | 7.0* | 1.1† | 47 | 7.3* | 1.0† | 44 | 6.3* | 1.1† |
Values for kcat, Km, and S0.5 were determined by fitting initial velocity data to equations describing hyperbolic or cooperative saturation, respectively. Hill coefficient (nH) values were calculated from Hill plots by linear regression.
Values had uncertainty intervals of more than 10% according to the fitting procedure.
Hyperbolic saturation was indicated by linearity of the Eadie–Hofstee plots.
No measurable chorismate mutase function was detected.
Helix 12, Helix 2, and the Active Site Residue Arg-157 Interact in the Crystal Structure of the T State.
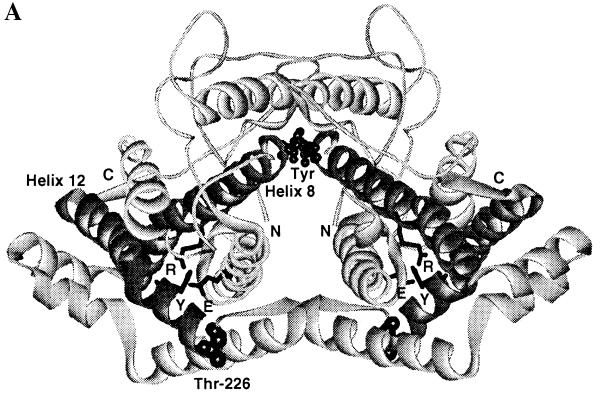
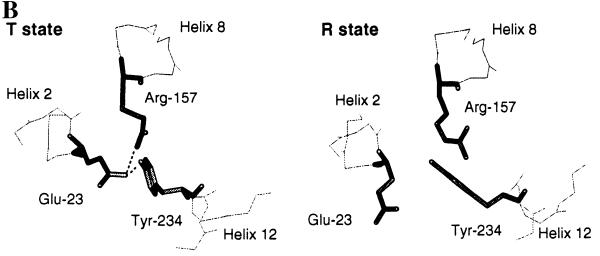
The x-ray structure of the R and T states of yeast chorismate mutase have been described (10, 11, 13). These crystal structures describe the global structural changes that occur when the enzyme switches between the inhibited and activated conformations (11, 13). Position 226 is clearly distant from the binding sites for tyrosine and tryptophan in the x-ray structures (Fig. 2A). Thr-226 is the last residue of the 220s loop next to helix 12, which has residues that show significant shifts in the T-to-R transition. In the T state, crystal Tyr-234, which is part of helix 12, is hydrogen bonded to Glu-23 (Fig. 2B). Glu-23 is part of helix 2 and interacts in the T state with the active site residue Arg-157 (11, 13, 22). Arg-157 is part of the long helix 8, which connects the active site to the binding site for tyrosine and tryptophan (refs. 11 and 13; Fig. 2A). Replacements of Arg-157 by other amino acids resulted in inactive enzymes (22). The x-ray structures show that in the T-to-R transition, Glu-23 of helix 2 moves 5.0 Å further away from the active site and no longer interacts with Arg-157 (Fig. 2B).
Figure 2.
(A) Structure of the T state of chorismate mutase drawn in ribbon style. The N termini and C termini are labeled as N and C, respectively. The two tyrosine molecules located at the interface of the dimer are indicated as Tyr. Helix 8, which connects the allosteric and active sites, and helix 12 are drawn in a dark color. The side chains of Glu-23 (E) and Tyr-234 (Y), which are involved in the hydrogen-bonding interaction with the active residue Arg-157 (R), are drawn in stick model. Thr-226, which connects the 220s loop and helix 12, is indicated. (B) Residues Glu-23, Tyr-234, and Arg-157 in the crystal structures of the R and T states of yeast chorismate mutase. Hydrogen-bonding interactions are indicated by dotted lines. Glu-23 and Tyr-234 move toward the active site upon the transition from the R to the T state. In the T state, Glu-23 is in hydrogen-bonding distance to the active site residue Arg-157, and the OH of Tyr-234 is 2.7 Å from Oɛ2 of Glu-23; in the R state this distance increases to 4.7 Å.
Analysis of the Interaction Tyr-234 → Glu-23 → Arg-157 in the Catalytic and Regulatory Properties of Yeast Chorismate Mutase.
To examine the catalytic and allosteric significance of the structural changes of Tyr-234 and Glu-23 in the T-to-R transition, and the interaction with the active site residue Arg-157, we have mutagenized the ARO7 gene to code for phenylalanine, alanine, serine, and glutamate at the wild-type position Tyr-234. All engineered chorismate mutases were expressed in yeast cells from high-copy plasmids. The ARO7 alleles Tyr234Phe, Tyr234Ala, Tyr234Ser, and Tyr234Glu encoded functional chorismate mutases and complemented the auxotrophy of an aro7 minus strain. However, we determined very low in vitro activity of the mutant Tyr-234 → Glu chorismate mutase in crude extracts. We further replaced Glu-23 by alanine, glutamine, aspartate, and arginine. The alleles were named Glu23Ala, Glu23Arg, Glu23Gln, and Glu23Asp. Glu23Arg is not able to complement the nutritional auxotrophy of an aro7 minus strain. Glu23Ala, Glu23Gln, and Glu23Asp encoded functional chorismate mutases. All functional enzymes were purified to homogeneity by fast protein liquid chromatography technique and characterized by initial rate kinetics. No changes in the stability or in the elution behavior were observed on any chromatography column of the various mutant enzymes, indicating that no serious damage had occurred to the overall structure of the protein.
Tyr-234 → Phe Chorismate Mutase Is Insensitive Toward Tyrosine and Exhibits an Increased Kcat Value.
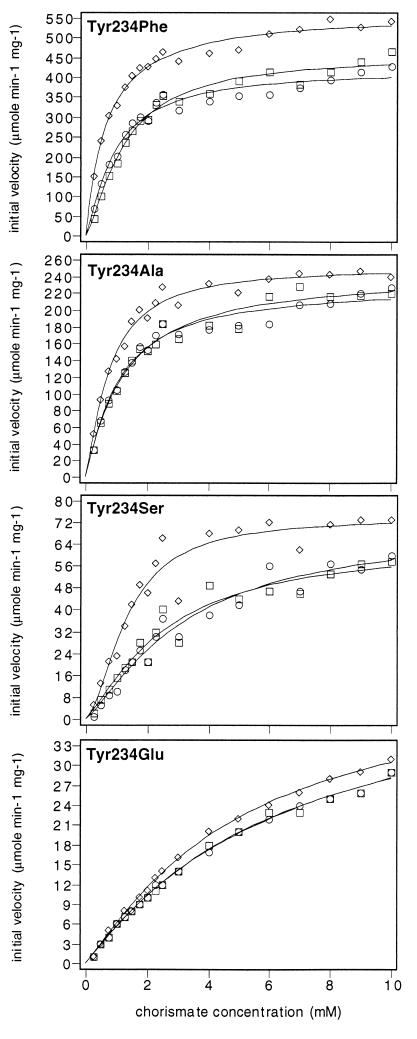
X-ray crystallographic data show that Tyr-234 interacts with Glu-23 in the inhibited T state of yeast chorismate mutase by its phenolic hydroxyl group. To test the significance of this interaction for the enzyme in solution, we replaced Tyr-234 by phenylalanine. Because phenylalanine differs from tyrosine in that it lacks the phenolic hydroxyl group, this replacement should prevent the T state interaction with Glu-23. Tyr-234 → Phe is insensitive toward the inhibitor tyrosine: the substrate saturation curve with tyrosine corresponds to the curve determined for the enzyme without effectors (Fig. 3). However, the enzyme still responds toward the activator tryptophan. The Km value of 0.9 mM determined for the unliganded enzyme is much lower than the value for wild-type chorismate mutase (Table 1). This result indicates that Tyr-234 → Phe is shifted toward an active structure by the amino acid substitution. Replacement of Tyr-234 by phenylalanine also results in increased maximal velocity compared with wild-type chorismate mutase (Table 1).
Figure 3.
Substrate saturation plots of mutant chorismate mutases carrying amino acid substitutions at position Tyr-234. The enzymes were assayed with 10 μM tryptophan (diamonds), without effectors (circles), or in the presence of 100 μM tyrosine (squares). The data were fitted to functions describing cooperative or Michaelis–Menten-type saturation.
Replacements of Tyr-234 by Alanine and Serine Show in Increased Km Values and a Loss of Catalytic Activity.
Enzymes carrying replacements of Tyr-234 by serine and alanine are insensitive toward the inhibitor tyrosine (Fig. 3). The replacements cause a reduction of the turnover number for the Tyr-234 → Ala enzyme and even more for Tyr-234 → Ser (Table 1). Substrate affinity is increased for the nonactivated enzymes and cannot be shifted to a low value by the addition of tryptophan as found for wild-type chorismate mutase. Substrate cooperativity for Tyr-234 → Ser is reduced compared with wild-type enzyme and is not observed for Tyr-234 → Ala.
Tyr-234 → Glu Chorismate Mutase Exhibits Low in Vitro Activity.
Because Tyr-234 interacts with Glu-23 we speculated that a negative charge at position 234 might cause major defects. Determination of the Tyr-234 → Glu chorismate mutase activity in yeast crude extracts indicated very low activity of this mutant enzyme (data not shown). However, the activity is sufficient for the growth of yeast even if the allele is expressed from the singular chromosomal copy. The substrate saturation curves show no significant difference between the enzyme without effectors and the enzyme incubated with tyrosine (Fig. 3). Tryptophan slightly activated this mutant enzyme. A low maximum activity of about 45 s−1 was determined for purified Tyr-234 → Glu (Table 1). Here, substrate affinity is reduced compared with wild-type chorismate mutase.
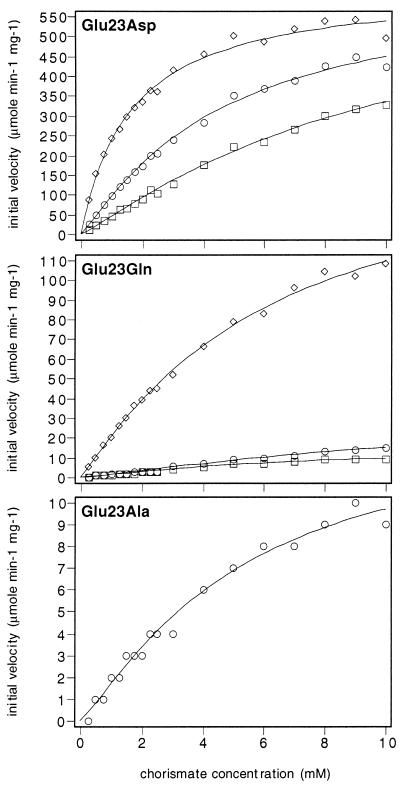
Substitution of Glu-23 by Aspartate Results in Increased Chorismate Mutase Activity and a Loss of Cooperativity.
Glu-23 was exchanged for aspartate, which also has a negative charge but a shorter side chain. The resulting enzyme exhibits high chorismate mutase activity (Fig. 4 and Table 1). The Km values, between 625 and 655 s−1, exceed by far the values determined for wild-type chorismate mutase (Table 1). Glu-23 → Asp is regulated by both effectors, tyrosine and tryptophan (Fig. 4). However, the range at which substrate affinity is modulated is smaller than that of wild-type chorismate mutase. No substrate cooperativity was determined for this mutant enzyme under any effector concentration.
Figure 4.
Substrate saturation plots of mutant chorismate mutases carrying amino acid substitutions at position Glu-23. The enzymes were assayed with 10 μM tryptophan (diamonds), without effectors (circles), or in the presence of 100 μM tyrosine (squares). The data were fitted to functions describing cooperative or Michaelis–Menten-type saturation.
Glu-23 → Gln and Glu-23 → Ala Show Reduced Catalytic Activity.
We further mutagenized the ARO7 gene to code for a glutamine or alanine at the wild-type position Glu-23. Glutamine has a similar shape as glutamate but has an uncharged side chain. Alanine is different in shape and charge and should result in a rupture of all hydrogen-bonding interactions. Purified Glu-23 → Gln and Glu-23 → Ala chorismate mutases exhibit a drop in in vitro chorismate mutase activity. The kcat value of the activated Glu-23 → Gln chorismate mutase is about half of the value of 360 s−1 (Table 1) for the activated wild-type chorismate mutase. The Km value of Glu-23 → Gln is increased. The loss of activity was even more pronounced for the nonactivated states, where only residual activity was determined for the Glu-23 → Gln enzyme. The curves are extremely flat and hardly allow a reliable calculation of the enzyme kinetic data. However, residual response toward the inhibitor tyrosine is detected (Fig. 4). For the Glu-23 → Ala chorismate mutase, activity can be measured only in the presence of the activator tryptophan (Fig. 4). For the activated Glu-23 → Ala chorismate mutase the algorithm provided a low kcat value of 14 s−1 and a Km value of 5.2 mM compared with 0.4 mM for the activated wild-type enzyme (Table 1).
Replacements of Glu-23 by arginine, glutamine, and alanine have severe effects on the catalytic function of yeast chorismate mutase. The replacement by arginine resulted in a nonfunctional enzyme not able to complement an aro7 mutation.
DISCUSSION
The substitution of threonine by isoleucine at position 226 of yeast chorismate mutase leads to a complete loss of both cooperativity and regulation by the effector amino acids, tyrosine and tryptophan (8, 9). The enzyme is frozen in the activated R state. Seven other residues with different side-chain characteristics have been placed earlier at this position (9). All mutant enzymes showed a regulatory behavior between the wild-type and constitutively activated enzyme. An additional amino acid replacement from isoleucine to threonine at position 225 in the constitutively activated Ile-226 mutant is able to restore tryptophan activation. This enzyme remains insensitive toward tyrosine. Amino acids 225/226 are the last residues of loop 220s at the connection to helix 12. The unregulated phenotype of Ile-226 presumably is not caused by steric problems of an isoleucine at position 226 but rather by the lack of a threonine function at this position. Thr-226 might be involved in different hydrogen-bonding interactions in the T and R states, which could change the conformational orientation of the 220s loop, where Thr-226 resides (11). This loop moves in the transition between the T and R state structures and might affect helix 12 (11).
Replacements of the helix 12 residue Tyr-234, which is separated from position 226 by only seven residues, also resulted in functional enzymes that have specifically lost the ability to respond toward tyrosine. This demonstrates an important role of Tyr-234 in the allosteric inhibition of yeast chorismate mutase. A comparison of our results with the structural studies revealed changes in the side-chain conformation for the active site residue Arg-157. In the x-ray structure of the inhibited T state, this residue is in hydrogen-bond distance to the side chain of Glu-23, which, in the T state, is 5 Å closer to the active site cavity than in the R state. Molecular modeling suggested that in the inhibited enzyme the hydroxyl group of Tyr-234 may interact with Glu-23 by hydrogen bonding. This result is in agreement with the T state structure, which shows a distance of 2.7 Å between the OH of Tyr-234 and a carboxylate oxygen of Glu-23. Therefore, the Tyr-234/Glu-23/Arg-157 interaction seems to be a prerequisite for the establishment of the low-affinity state of the active site. The interaction of Glu-23 with the active site residue Arg-157 is presumably directly involved in modulating the enzyme activity by reducing the electrostatic field or changing the position of the guanidinium group of Arg-157. The Glu-23 → Gln mutant enzyme shows only residual tyrosine inhibition, but strong activation by tryptophan. Therefore, a negative charge at position 23 seems to be necessary for strong inhibition of yeast chorismate mutase. Ramilo et al. (23) have previously suggested a possible role of Tyr-234 in regulation of yeast chorismate mutase. This suggestion was based on the occurrence of a local conformational change upon activation and has been demonstrated by two-dimensional NMR spectroscopy (23). It was suggested that Tyr-234 is either part of or adjacent to the tryptophan binding site. Our results confirm and specify the regulatory role of Tyr-234 and are consistent with the crystal structures that show that Tyr-234 is located in the general vicinity of the active site residue Arg-157 but far from the allosteric binding site (11, 13).
The concerted transition theory, whether the symmetry (3) or sequential (15) models, suggests that regulation of an enzyme is achieved by shifting the equilibrium of two different conformational states of an enzyme. However, the simplest two-state model of Monod et al. (3) does not satisfactorily account for our experimental data. We describe enzymes that have specifically lost the ability to respond to the inhibitory amino acid, tyrosine, whereas the activator, tryptophan, still increases the activity. Only the conformational change to the T state seems to be blocked. The two-state model would lead to the expectation that an enzyme in which the T state is destabilized or in which a shift to the T state cannot be achieved would favor the more active R state. However, the results of our studies show that activation and inhibition can be separated and do not correspond to opposite displacements of two different structures in equilibrium. Furthermore, some of the mutant enzymes can be activated by tryptophan even without any detectable cooperativity. This result indicates that a global transition of the enzyme structure is not necessary for all aspects of allosteric regulation. It is possible that more than two stable structures exist and that the unligated enzyme occupies a stable intermediate structure between T and R state. Another possibility is that the homotropic and heterotropic interactions proceed by different mechanisms or that at least parts of the allosteric transitions do not require a global change of the overall enzyme structure.
Structures of these and other mutant yeast chorismate mutase variants may identify displacements of features in the molecule that lead to an understanding of the nonconcerted aspects described in this study. Indeed, the known structures show that the long helix H8 (140–171) extends from the regulatory site near 139–145 to the active site (Arg-157, Lys-168). In addition, there is a cascade of changes in the T-to-R transition between helices H2 (14–33) and H11 (195–211) extending from the active site (near Glu-23) to the interface between subunits and, therefore, involved in the cooperativity of the T-to-R transition. It remains to be shown whether helix H8 is associated with the independence of the activation by tryptophan in these mutants.
Binding of tyrosine to the allosteric site causes structural changes, which include alternative conformations around Tyr-234. In the T state, interaction with the active site residue Arg-157 via Glu-23 is the trigger for decreased substrate affinity. Changes in the adjacent loop 220s can affect this molecular trigger and ultimately result in enzymes that are unresponsive to tyrosine. We suggest that the molecular trigger of the T state induced by tyrosine and activation of the enzyme by tryptophan are independent processes and use alternative pathways within the molecule. It remains to be elucidated whether besides R and T state, a third structure of the enzyme will be identified corresponding to a unoccupied allosteric site.
Finally, the results found here may be compared with a somewhat similar situation in aspartate transcarbamylase from Escherichia coli, in which some modified forms separate homotropic (substrate) effects from the heterotropic (activation by ATP, inhibition by CTP) effects (24). Some mutants are known that respond to ATP, but not to CTP, and others show the reversed response (24). In the wild-type enzyme, the CTP inhibition influences the R-to-T transition, but the ATP stimulation is independent of the R-to-T transition (25).
Acknowledgments
Roney Graf and Thomas Gerstberger were involved in the initial phase of the work. We thank them for discussion, and Eric Kübler for critical reading of the manuscript. This work was supported by a grant from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Volkswagen-Stiftung. W.N.L. acknowledges the contribution of N. Sträter and support by National Institutes of Health Grant GM06920.
References
- 1.Bohr C, Hasselbalch K, Krough A. Scand Arch Physiol. 1904;16:402–412. [Google Scholar]
- 2.Monod J, Changeux J P, Jacob F. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 3.Monod J, Wyman J, Changeux J-P. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 4.Hathaway J A, Atkinson D E. J Biol Chem. 1963;238:2875–2881. [PubMed] [Google Scholar]
- 5.Koshland D E, Nemethy G, Filmer D. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 6.Braus G H. Microbiol Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidheini T, Sperisen P, Paravicini G, Hütter R, Braus G. J Bacteriol. 1989;171:1245–1253. doi: 10.1128/jb.171.3.1245-1253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidheini T, Mösch H-U, Evans J N S, Braus G. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 9.Graf R, Dubaquié Y, Braus G H. J Bacteriol. 1995;177:1645–1648. doi: 10.1128/jb.177.6.1645-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue Y, Lipscomb W N, Graf R, Schnappauf G, Braus G. Proc Natl Acad Sci USA. 1994;91:10814–110818. doi: 10.1073/pnas.91.23.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sträter N, Kakänsson K, Schnappauf G, Braus G, Lipscomb W N. Proc Natl Acad Sci USA. 1996;93:3330–3334. doi: 10.1073/pnas.93.8.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartlett P A, Johnson C R. J Am Chem Sci. 1985;107:7792–7793. [Google Scholar]
- 13.Sträter N, Schnappauf G, Braus G, Lipscomb W N. Structure. 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]
- 14.Beggs J D. Nature (London) 1978;275:104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- 15.Giebel L B, Spritz R A. Nucleic Acids Res. 1990;18:4947. doi: 10.1093/nar/18.16.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito H, Jukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miozzari G, Niederberger P, Hütter R. J Bacteriol. 1978;134:48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lämmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Fletcher R, Powell M J D. Comput J. 1963;6:163–168. [Google Scholar]
- 21.Newell J O, Schachmann H K. Biophys Chem. 1990;37:183–196. doi: 10.1016/0301-4622(90)88018-n. [DOI] [PubMed] [Google Scholar]
- 22.Schnappauf G, Sträter N, Lipscomb W N, Braus G H. Proc Natl Acad Sci USA. 1997;94:8491–8496. doi: 10.1073/pnas.94.16.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramilo C, Braus G, Evan J N S. Biochem Biophys Acta. 1993;1203:71–76. doi: 10.1016/0167-4838(93)90037-r. [DOI] [PubMed] [Google Scholar]
- 24.Lipscomb W N. Chemtracts Biochem Mol Biol. 1991;2:1–15. [Google Scholar]
- 25.Fetler L, Tauc P, Hervé G, Moody M F, Vachette P. J Mol Biol. 1995;251:243–255. doi: 10.1006/jmbi.1995.0432. [DOI] [PubMed] [Google Scholar]