Abstract
Plants are increasingly used as production platforms of various heterologous proteins, but rapid protein turnover can seriously limit the steady-state expression level. Little is known about specific plant proteases involved in this process. In an attempt to obtain potato (Solanum tuberosum cv Desirée) plants resistant to Colorado potato beetle (Leptinotarsa decemlineata Say) larvae, the protease inhibitor equistatin was expressed under the control of strong, light-inducible and constitutive promoters and was targeted to the secretory pathway with and without endoplasmic reticulum retention signal. All constructs yielded similar stepwise protein degradation patterns, which considerably reduced the amount of active inhibitor in planta and resulted in insufficient levels for resistance against Colorado potato beetle larvae. Affinity purification of the degradation products and N-terminal sequencing allowed the identification of the amino acid P1-positions (asparagine [Asn]-13, lysine-56, Asn-82, and arginine-151) that were cleaved in planta. The proteases involved in the equistatin degradation were characterized with synthetic substrates and inhibitors. Kininogen domain 3 completely inhibited equistatin degradation in vitro. The results indicate that arginine/lysine-specific and legumain-type Asn-specific cysteine proteases seriously impede the functional accumulation of recombinant equistatin in planta. General strategies to improve the resistance to proteases of heterologous proteins in plants are proposed.
The expression of many traits in transgenic plants can be severely hampered by the individual characteristics of foreign genes and proteins not adapted to the specific subcellular environment of the new host. Significant progress has been made in optimizing the rate of transcription and translation in plant cells (Koziel et al., 1996; Dai et al., 2000; Outchkourov et al., 2003). However, proteolytic degradation of heterologously expressed proteins is still a limiting factor in the accumulation of many foreign proteins in plants (Dolja et al., 1998; Stevens et al., 2000; Sharp and Doran, 2001a, 2001b). A generally adopted approach to increase heterologous protein accumulation levels in plants is to change the compartmentalization of the expressed proteins by targeting to and retention in the endoplasmic reticulum (ER; Wandelt et al., 1992; Schouten et al., 1996) or chloroplasts (Wong et al., 1992). The targeting to the endomembrane system of plants, however, leads to an unpredictable final subcellular location of the expressed protein even when the ER retention signal is added (Vitale and Galili, 2001).
In different plant cellular compartments, a wide variety of proteases are involved in the processing and degradation of proteins (Vierstra, 1993, 1996). The levels of these plant proteases are affected by many developmental factors like seed maturation and germination (Bottari et al., 1996) or leaf senescence (Ueda et al., 2000), combined with environmental conditions such as carbon starvation (Brouquisse et al., 1998) or infection and wounding (Guevara et al., 2002). In general, subtilisin/kexin-like serine and metallo-proteases in the secretory pathway are known to process pro-proteins (Viale et al., 1999; Raharjo et al., 2001), and cysteine proteases are known to process mis-folded proteins (Otsu et al., 1995) or to mobilize seed storage proteins (Schlereth et al., 2001). The legumain-type cysteine proteases enter the secretory pathway as inactive precursors and are activated in protein bodies (protein storage vacuoles) and the cell wall (Hiraiwa et al., 1999; Kinoshita et al., 1999; Fischer et al., 2000; Muntz and Shutov, 2002). Together with papain-like cysteine proteases, they were shown to be involved in the degradation of plant storage proteins (Fischer et al., 2000; Schlereth et al., 2001). They function in the processing of storage protein precursors during seed development and storage protein mobilization during seed germination (Hara Nishimura et al., 1991, 1995, 1998; Hara Nishimura, 1993; Rotari et al., 2001). Legumain-type proteases are also known to be upregulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions (Kinoshita et al., 1999). Several cysteine proteinases, pathogen-related proteins, and chitinase and wound-inducible proteinase inhibitors are proposed as putative targets for activation by the action of legumain, Asn-specific proteases (Kinoshita et al., 1999).
The expression of heterologous protease inhibitors in plants has been used as an approach to induce plant resistance to insects (Ryan, 1990). Many host plant-derived inhibitors are unsuitable, however, due to the fact that many insects have evolved resistance (Jongsma and Bolter, 1997). Extensive screening of non-host plant-derived inhibitors has resulted in novel candidate proteins that in vitro and in vivo demonstrated promising levels of resistance against various insect pests (Gruden et al., 1998; Harsulkar et al., 1999). The expression of these proteins in planta, however, brings them into a subcellular environment that may be far from optimal for the stable accumulation of the protein, whereas the effectiveness of protease inhibitors against insects depends on an expression level of around 1% of total soluble protein (Ryan, 1990; Jongsma and Bolter, 1997).
Equistatin is a protease inhibitor from the sea anemone Actinia equina and consists of three thyroglobulin type I domains. The first N-terminal domain acts as a Cys protease inhibitor (Ki for papain of 0.57 ± 0.04 nm), and the second as an aspartic protease inhibitor (Ki for cathepsin D of 0.3 ± 0.15 nm); the function of the third domain is not yet known (Lenarcic and Turk, 1999; Strukelj et al., 2000). The protein was previously found to be highly active against the gut proteases of Colorado potato beetle (CPB; Leptinotarsa decemlineata Say), and on artificial diets, the protein caused complete growth inhibition and mortality among the larvae (Gruden et al., 1998). The coding part of the equistatin gene was modified for maximal expression in potato (Solanum tuberosum cv Desirée) by site-directed mutagenesis (accession no. AY166597), and functional protein was expressed at levels of grams per liter in the yeast Pichia pastoris (Outchkourov et al., 2002). In potato leaves, accumulation of only partly functional equistatin up to levels of 7% of total soluble protein was achieved (Outchkourov et al., 2003). For functional expression, it is necessary, however, that the expressed protein is properly folded and essentially undegraded.
In this study, equistatin expressed in the ER of potato was found to be cleaved and further degraded by endogenous plant proteases. Due to the light-regulated Rubisco promoter this resulted in a cyclic presence of higher and lower levels of equistatin that were differentially degraded. As a result, insufficient levels of functional protein were maintained for resistance against CPB. The cleavage sites were determined by sequencing of the peptide fragments. The plant proteases responsible for equistatin degradation were partly characterized with specific substrates and inhibitors. The results may have significant implications in engineering the stability of heterologously expressed proteins in plants.
RESULTS
Equistatin Expression Analysis
Potato was transformed with three different constructs that contained the equistatin gene optimized for expression in plants under the control of the cauliflower mosaic virus (CaMV) 35S promoter, the Lhca.3.St1 promoter from potato, and the RbcS1 Rubisco small subunit promoter from chrysanthemum (Fig. 1). To enhance the protein accumulation, an additional construct was prepared in which the equistatin gene was also given an ER retention signal (KDEL) and placed under the control of the RbcS1 promoter. The expression of at least 30 transgenic plants per construct was analyzed by immunological detection, and on some of the pLhca.3.St1/EIM transformants, northern blots were carried out. The different constructs resulted in strongly different levels of expression. The highest population mean of expression for the equistatin gene without KDEL was obtained with the RbcS1 promoter (0.36% of total soluble protein), followed by the Lhca.3.St1 promoter (0.10%) and the 35S promoter with very low levels of less than 0.01% (Table I). The addition of the four amino acid ER retention signal (KDEL) at the C terminus of equistatin under the control of the RbcS1 promoter improved the expression 5-fold, raising the mean expression level to 1.9% (Table I). A large proportion of the pRbcS1/EIM/KDEL lines expressed equistatin at levels between 1% and 7% of total soluble protein.
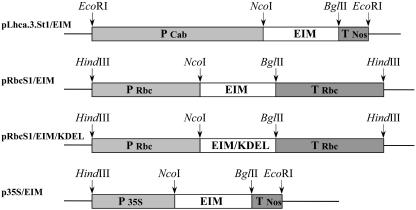
Figure 1.
Schematic representation of the constructs used in this study. P Lhca.3.St1 is the light-inducible light-harvesting complex photo-system I promoter from potato; P RbcS1 is the Rubisco small subunit promoter from chrysanthemum; P 35S is the double CaMV35S promoter; T RbcS1 is the terminator of small subunit of Rubisco; and T Nos is the terminator of the nopaline synthase gene. EIM is the modified equistatin coding part. EIM/KDEL is the modified equistatin coding part plus four amino acids at the C terminus, KDEL, respectively.
Table I.
Expression levels of the different constructs measured by dot-blot immunological detection on samples collected between 9:30 and 10 AM
| Constructs | Plants | Mean Expression |
|---|---|---|
| % of total protein | ||
| PLhca.3.St1/EIM | 30 | ∼0.10 |
| PRbcS1/EIM | 30 | 0.36(±0.12)a |
| PRbcS1/EIM KDEL | 30 | 1.9 (±0.39)a |
| p35/EIM | 30 | <0.01 |
a ses. The low signal of the plants generated with pLhca.3.St1/EIM and p35/EIM constructs did not allow precise estimation of the mean value.
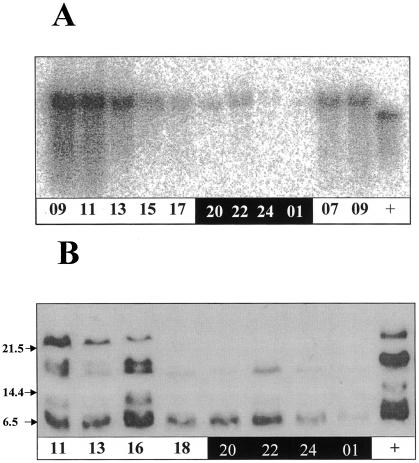
Considering that the light-regulated promoters Lhca.3.St1 and RbcS1 performed 10- to 40-fold better than the constitutive 35S promoter, it seemed that the known circadian expression of these promoters might also possibly affect our results. Twenty-four-hour time-course samples of both RNA and protein were taken to investigate the relationship between equistatin synthesis and degradation in the plants transformed with the pLhca.3.St1/EIM construct (Fig. 2). The northern blot confirmed that the Lhca.3.St1 promoter is transcriptionally active during the morning and nearly silent during the night, in agreement with Nap et al. (1993). At the protein level (Fig. 2B) intact equistatin (top band) only represented about one-third of the signal even at 11 am at the peak of transcription (9-11 am). Most of the protein was degraded into increasingly smaller fragments. After 6 pm, only the lowest 5-kD bands were visible, and with very low intensity after 7 h of darkness (1 am). The defined sets of bands indicated that degradation is initiated by proteases that first cleave the peptide chain of equistatin at specific sites. This degradation did not occur during the extraction procedure because, first, a cocktail of synthetic inhibitors including E-64 was added to the extraction buffer to prevent degradation. We knew from earlier experiments that E-64 could prevent equistatin degradation in extracts. Second, the patterns of degradation were clearly different depending on the time of the day, the promoter, and the targeting used (Figs. 2 and 3). Third, all samples were prepared and extracted in the same way and at the same time. It would be unusual for a protease to be present at high concentrations late at night and to be gone at dawn. Instead, it is shown that the high concentrations of intact protein in the morning derive from the strong transcription of the gene at that time, which disappears during the rest of the day.
Figure 2.
A, Northern-blot analysis of the equistatin transcript expressed in potato cv Desirée transformed with pLhca.3.St1/EIM construct. Numbers represent the hours of the day, and black and white shading represents the dark and the light part of the photoperiod. +, Positive control mRNA used in the experiment. B, Western-blot analysis of the equistatin protein accumulated in potato cv Desirée transformed with pLhca.3.St1/EIM construct. Numbers represent the hours of the day, and black and white shading represents the dark and the light part of the photoperiod. +, Positive control protein, produced in Escherichia coli and used in the experiment.
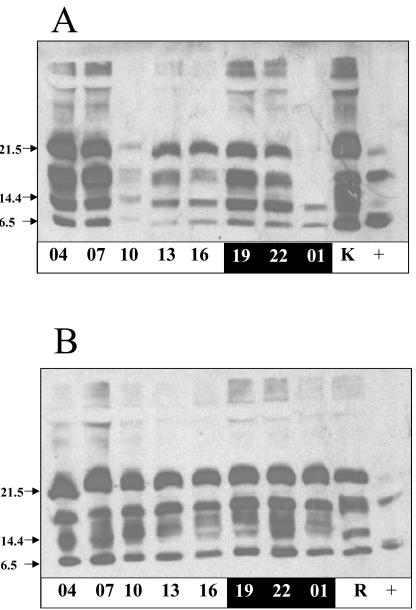
Figure 3.
Western-blot analysis of the equistatin protein accumulated in potato cv Desirée transformed with pRbcS1/EIM (A) and pRbcS1/EIMKDEL (B) construct. Numbers represent the hours of the day, and black and white shading represents the dark and the light part of the photoperiod. +, Positive control protein, produced in E. coli and used in the experiment. K is protein extract from KDEL plant, and R is protein extract from non-KDEL plant, both taken at 7 am.
Western-blot analysis of the plants that expressed the pRbcS1/EIM construct (Fig. 3A) revealed a diurnal rhythm of protein expression and a pattern of equistatin degradation similar to the plants expressing pLhca.3.St1/EIM. A notable difference is that a higher level of transcription is apparently maintained during daylight and early night resulting in the presence of undegraded protein during the whole day and the first part of the night. Only after 7 h of darkness have all intact bands disappeared, and only degraded protein remains.
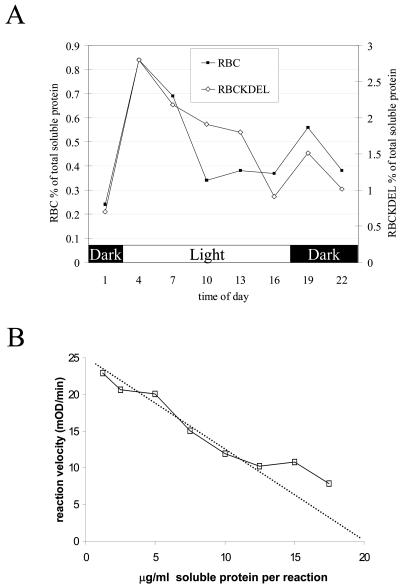
The effect of adding a KDEL retention signal was a 5-fold improvement of the amount of accumulated protein (degraded and undegraded), but it was not yet clear whether qualitative changes had occurred in the type of degradation of the protein. The differences in degradation of equistatin with and without the KDEL signal can be observed by comparing Figure 3, A and B. Four bands were observed for the -KDEL plants: 21.5 kD (intact protein), approximately 18 kD, approximately 12 kD, and approximately 6 kD. The +KDEL construct resulted in a slightly different degradation pattern: Instead of the defined approximately 12-kD band, a number of bands with slower mobility appeared. The western blots on Figure 3 were, however, overexposed to show all degradation bands. To precisely quantify the qualitative diurnal variation of accumulated equistatin protein, two plant lines of the pRbcS1/EIM construct, lines 24 and 36, and two lines of the pRbcS1/EIMKDEL construct, lines 21 and 28, were selected for a precise quantification. Time-course samples were taken every 3 h during a 24-h period, and the precise amount of equistatin protein was quantified by dot-blot immunological analysis. In this way, all immunoreactive equistatin, degraded and undegraded, was measured and compared. The results for the two different constructs were compared by aligning the two data sets on the basis of the highest protein accumulation point at 4 am (Fig. 4A). The two data sets are following a very similar diurnal fluctuation despite the fact that the average protein expression level was 5-fold higher in the presence of KDEL. The amount of accumulated protein in both cases peaks to a 4-fold higher level 1 h after light is switched on and then gradually decreases during the day to a 3-fold lower level at 4 pm. One hour after the light is switched off, there is a smaller secondary peak, finally leading to the lowest accumulation level at 1 am after a 7-h dark period. The increased equistatin accumulation in KDEL plants, therefore, seems to be due to an overall slow down of degradation of the protein because it is unlikely that transcription or translation itself would be affected by the downstream KDEL element. The graphs suggest that the sampling moment at 10 am to determine the population average may have affected the ratio found of a 5-fold improvement due to KDEL and would have been less pronounced at other moments during the day.
Figure 4.
A, Diurnal equistatin protein quantifications based on dot-blot analysis. Mean values found on plant lines pRbcS1/EIM 24 and 36, indicated as RBC, are shown against mean values of pRbcS1/EIM/KDEL lines 21 and 28, indicated as RBCKDEL. B, Titration of 15 nm papain activity against potato leaf extract from transgenic line pRbcS1/EIM/KDEL22 using Z-Phe-Arg-pNA as a synthetic substrate as described in “Materials and Methods.” The x axis indicates the concentration of total soluble plant protein from the transgenic plant required to inhibit 15 nm papain; at the x axis intercept (dotted line), all papain is inhibited by 15 nm equistatin (0.32 μg mL-1), which converts to 1.6% (w/w) of total soluble protein.
The cleavage and degradation of equistatin was expected to directly affect the amount of active protein. This was investigated by taking plant material harvested at 10 am, and titrating it against papain (Fig. 4B). The selected line KDEL 22 was found to express 1.6% of active protein, whereas according to the immunological dot-blot detection, it expressed equistatin at 5.2%. This indicated that a large fraction of the expressed equistatin was probably inactivated by the specific cleavages observed on the western blots.
Isolation and Characterization of in Planta Equistatin Degradation Products
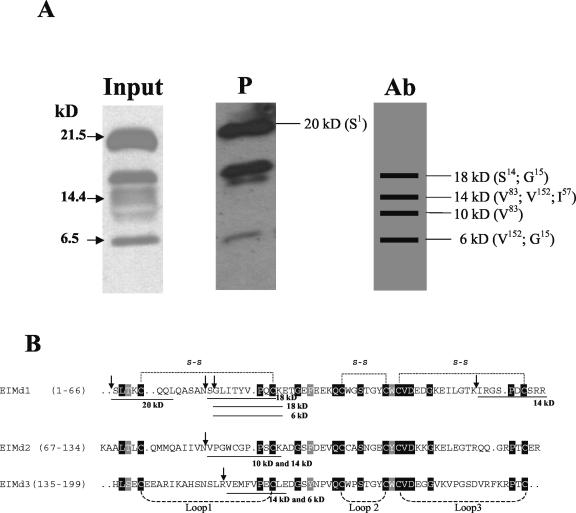
The defined set of equistatin degradation bands in planta indicated that specific proteases initiate the degradation of equistatin at specific positions in the polypeptide chain. To investigate the sites of cleavage, we isolated the protein and its degradation products by means of immuno-affinity and papain-affinity chromatography. The isolated products were separated on gel, blotted to polyvinylidene difluoride membrane, and N-terminally sequenced (Fig. 5).
Figure 5.
Isolation from plants and N-terminal sequencing of the equistatin degradation products. A, Purification on either papain Sepharose (P) or antibody affinity column (Ab) as described in “Materials and Methods.” For the P column, the fractions containing equistatin were analyzed by western blot and are shown. For the Ab column, horizontal bars indicate the sequenced bands found on the Coomassie-stained membrane. Molecular mass and the N-terminal amino acids of the different peptides are indicated. B, An alignment of the three domains of equistatin is shown. The positions of the disulfide bridges and protein loops as deduced from the solved structure of the homologous thyropin p41 (Gun[caron]car et al., 1999) are marked above or below the alignment. The N-terminal sequences of the in planta degradation bands are shown underlined.
By immuno-affinity, only the immunologically weaker bands could be recovered. The intact equistatin band remained on the column probably due to the multiple clonal IgG proteins present in the serum that cooperatively bind many different epitopes on the intact protein. The papain-affinity column was expected to result in the purification of those equistatin fragments, which still contained a functional first domain, because only that domain inhibits papain. Interestingly, the papain column led to the purification of the intact band (22 kD), the largest degraded fragment (approximately 16 kD), and a small fragment (approximately 5 kD), but not of any of the weaker other bands.
All bands from the immuno-affinity column and the intact band of the papain column were sequenced by Edman degradation. We were unsuccessful in sequencing the degraded papain-purified bands, because the papain column yielded a smear of proteins in which only the undegraded band could be identified for sequencing. Some of the bands yielded different peptide fragments, probably due to complex trimming of the polypeptide both at the N and C termini, resulting in similar sized peptides. In addition, we obtained mixed N-terminal sequences for the cleavage in loop 1 of domain 1 (1.1): two from the fragment with a size of 18 kD (N terminus: S14 and G15) and one from the fragment of 6 kD (N terminus: G15; Fig. 5B). The fragments differ by one amino acid, which suggests N-terminal trimming by aminopeptidase activity. Also, the first loops of the other equistatin domains (2.1; 3.1) were cleaved at nearly identical topological positions (N terminus: V83 and V152; Fig. 5). A fourth cleavage site was identified in the third loop of the first domain (1.3). The specificity of proteases is usually mainly determined by the P1 amino acid, N-terminal to the hydrolyzed peptide bond. These were characterized by Asn-13 (1.1), Asn-82 (2.1), Lys-56 (1.3), and Arg-151 (3.1).
The fast shift in size from intact protein to the first major degradation band with an N terminus at Asn-13 (1.1) indicates that probably the Asn-13 cleavage occurs at a similar rate to a cleavage at possibly Arg-151, which can explain the approximately 18-kD peptide product. The cleavage Lys-56 (1.3) we considered as less efficient in planta than the other three, because the corresponding peptide was found only in minor quantities among two other peptides, whereas the corresponding position of the band was weaker when analyzed by western blot.
Proteolytic Activities in Potato Plant Extract
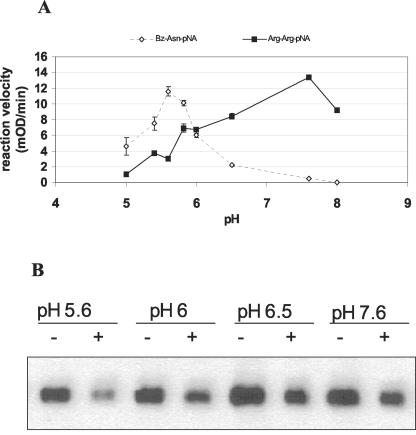
Equistatin was incubated in potato leaf extract in buffers of different pH. In addition, we used an Asn-specific substrate Bz-Asn-pNA, which is specific for legumain-type proteases, and an Arg-specific substrate Z-Arg-Arg-pNA, which is known to be hydrolyzed by both cysteine (papain-like) and serine proteases (kexin/subtilisin-like). The two substrates were chosen on the basis of the in planta cleavage points behind Asn and Arg/Lys P1 residues. The results are presented in Figure 6. In our in vitro experiment, we could only observe the disappearance of the intact equistatin band, and we could not see the equistatin degradation products as in the in planta degradation. Apparently due to tissue grinding, many plant peptidases were released from their subcellular compartment and degraded the equistatin degradation products faster than they could be observed. It was found that the optimal pH for degradation of equistatin in potato leaf extract was pH 5.6 or below (not determined). This agrees with the optimum of proteolytic activity known for legumain proteases, which was confirmed with the Bz-AsnpNA substrate. In contrast, the Z-Arg-Arg-pNA substrate showed an optimum at pH 7.8, which did not coincide with the optimum of degradation of equistatin. This suggested that legumain-like proteases may be crucial in the initiation of degradation of equistatin in planta.
Figure 6.
Determination of the pH optima of specific potato proteolytic activities. A, Proteolytic activities of potato cv Desirée leaf extract (60 μg protein) toward two different substrates, Bz-Asn-pNA and Z-Arg-Arg-pNA, at various different pH values. B, Leaf extract of potato cv Desirée (60 μg) incubated with equistatin (125 ng) for 1 h at various different pH values and subsequently analyzed for remaining equistatin by western blot (+). -, Negative control of pre-heated potato extract in the corresponding buffer.
Proteinaceous Inhibitors of Equistatin Degradation
The pH optimum of equistatin degradation below pH 6 and the fast Arg- and Asn-specific cleavages in the protein suggested the involvement of cysteine proteases. Papain-like cysteine proteases can be inhibited by all types of cystatins, but plant legumains are inhibited only by type II (egg-white cystatin) and type III cystatins (kininogen; Abe et al., 1993; Rotari et al., 2001). The inhibition of porcine legumain by human cystatin C (similar to egg-white cystatin) was found to be due to a second reactive site situated at the “back” of the inhibitor. Thus, cystatin C and some other cystatins possess two separate inhibitor active sites that enable them to inhibit both legumain and papain proteases simultaneously (Chen et al., 1997; Alvarez Fernandez et al., 1999).
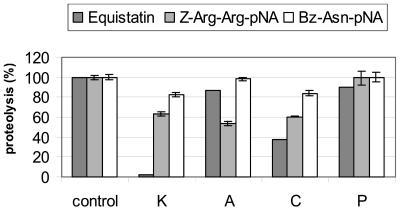
To test whether the equistatin degradation could be selectively inhibited by different cystatins in a plant extract, we tested representative members of all known cystatin types: stefin A, type I; cystatin C, type II; kininogen D3, type III; and potato cystatin, phytocystatin (Fig. 7). The two inhibitors without Cys bridges and with no known legumain inhibitory activity, stefin A and potato cystatin, inhibited the equistatin degradation by only 10% to 12%, relative to the control without inhibitor. In contrast, cystatin C inhibited equistatin degradation up to 60%, and full inhibition was obtained using kininogen domain III as inhibitor. In line with our previous experiments, when the synthetic substrate Bz-Asn-pNA was added to the plant extract, the highest inhibition of the proteolysis was achieved by using kininogen D3 and cystatin C. In contrast, when the Z-Arg-ArgpNA substrate was used, stefin A was the strongest inhibitor, but this had no significant effect on preventing equistatin degradation. This suggests that at least in a crude plant extract, legumain-like plant proteases initiate the degradation of equistatin, whereas the papain-like cysteine proteases contribute only about 10% of equistatin degradation.
Figure 7.
Hydrolysis of a leaf extract at pH 5.6 was measured using Z-Arg-Arg-pNA, Bz-Asn-pNA, and equistatin as substrates in the presence/absence of the indicated inhibitors. control, No inhibitor; K, kininogen domain 3; A, stefin A; C, cystatin C; P, potato cystatin. Error bars represent the se of the mean from four simultaneous measurements. The proteolysis of equistatin was measured by western-blot analysis. The amount of disappearance of intact protein in the absence of inhibitors (equistatin in preheated extract minus equistatin in plant extract) was taken as 100% proteolysis.
Insect Feeding Trials
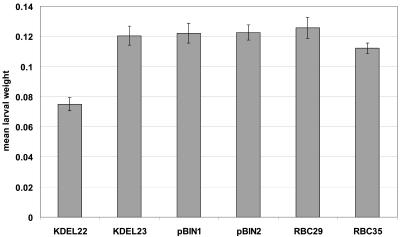
The efficacy of equistatin to give rise to resistance against CPB larvae when expressed in potato was checked in leaf feeding trials. In total, eight different bioassays were conducted. Figure 8 shows a representative bioassay. Newly hatched larvae were fed with leaves from several lines originating from pRbcS1/EIM and pRbcS1/EIM/KDEL and using pBINPLUS transformed lines as a control. Line KDEL22, which at 10 am showed an expression level of 5.2% total protein (dot blot) and 1.6% functional protein (papain titration), was found to suppress larval growth of CPB by 38%. Larval development was not altered when the larvae were fed KDEL23 plants that expressed equistatin at 10 am at 3.3% total protein (dot blot). In the given experiment, no significant differences were observed in larval mortality. However, between the eight different experiments the effects on larval growth and mortality tended to vary strongly (from high mortality to no effect on the larvae fed on transgenic plants). This may have been due to the observed fluctuation in the levels of expressed functional protein, which between assays are probably also affected by developmental (plant age), stress, and seasonal (light intensity) factors.
Figure 8.
Representative CPB bioassay. Mean larval weight (n = 6) after 3 d of feeding. Expression levels: KDEL22, 5.2%; KDEL23, 3.3%; pBINPLUS1 and 2, 0%; RBC29, 0.17%; and RBC35, 0.58%.
DISCUSSION
Equistatin Degradation in Potato cv Desirée
In this study, we aimed to obtain potato plants resistant to CPB larvae via stable and high-level expression of recombinant equistatin in the leaves of potato cv Desirée plants. Three different promoters (CaMV35S, Lhca.3.St1, and RbcS1) were analyzed for their ability to drive maximal expression of equistatin. The rbcS1 promoter from chrysanthemum (Outchkourov et al., 2003) yielded the highest average expression level (0.36%), and this was further improved 5-fold by the addition of the KDEL retention signal (1.9%). Some plant lines were found to express equistatin at levels of up to 7% of total soluble protein as measured by dot-blot detection. Protein degradation and the circadian expression of the RbcS1 promoter, however, affected the functionality and accumulation level of the accumulated protein. The distinct bands of degradation indicated that equistatin was cleaved at specific positions in planta. Two-thirds of the proteins were found to be degraded into an inactive form at 10 am, and during the night, only 20% of the initial protein accumulated in the morning remained. Thus, during the night, the level of functional protein in the best plant was probably at least 15-fold lower: around 0.5%. Probably as a consequence of the degradation of equistatin in plants, we did not consistently observe resistance to CPB larvae in these plants. Apparently, the level of expression of functionally active equistatin was too strongly affected by the endogenous proteases of potato.
Identification of the Plant Proteases Involved in Equistatin Proteolysis
The cleavage sites of the equistatin degrading proteases in plants were investigated by affinity purification and N-terminal sequencing of the degraded equistatin fragments. Lenarcic et al. (1997) isolated intact equistatin protein from its natural host A. equina. Lenarcic and Turk (1999) showed that in vitro equistatin is susceptible to cleavage by the serine protease trypsin at Lys-67-Ala-68 and Arg-151-Val-152, producing fragments of 7 and 14 kD. In Escherichia coli, equistatin was found to be cleaved at Val-152-Glu-153 (Strukelj et al., 2000). In planta, four putative cleavage positions were identified: Asn-13, Ser-14, Lys-56, Asn-82, and Arg-151. The different cleavage positions (Asn-13, Ser-14, Lys-56, and Asn-82) clearly indicate the involvement of plant-specific proteases because they do not occur in any of the other hosts. In planta, we suspect that in the case of the linked Asn-13 and Ser-14 cleavages, the primary cleavage site is at Asn-13 rather than Ser-14 and that the cleavage at Ser-14 is the result of aminopeptidase activity that removed the Ser-14 residue. An Asn residue (Asn-148) is also found in domain 3 in a corresponding position to the cleaved Asn residues in the other two domains. From the sequence alignment, it seems likely that Asn-148 in domain 3 is the actual endopeptidase cleavage site, but due to aminopeptidase activity, we have possibly misidentified Arg-151 as the primary P1 residue (Fig. 5B).
Furthermore, equistatin was most efficiently degraded at pH 5.6, suggesting that Asn-specific proteases may initiate the degradation of the protein. The only known family of Asn-specific proteases in plants is known as the legumain family (Muntz and Shutov, 2002), and the pH 5.6 optimum is in agreement with the characteristics of this enzyme family (Rotari et al., 2001).
Until now, there were no reported legumain inhibitors, to our knowledge, from plants. In contrast, cystatins derived from the animal kingdom have been shown to inhibit some legumains from both plants and animals. It was demonstrated that the addition of domain 3 of human kininogen (type III) at 10 μm concentration could completely inhibit the degradation of equistatin in vitro in potato cv Desirée leaf extract. The known inhibitor of the mammalian legumain human cystatin C (type II) inhibited equistatin degradation up to 60% at the same concentration. In contrast, there was very little effect (approximately 10%) of stefin A (type I) and potato cystatin (phytocystatin). Legumains from jack bean (Canavalia ensiformis; Abe et al., 1993) seeds were also found to be sensitive to inhibition by kininogen and not by stefin A. Using synthetic substrates, potato Asn-specific activity was also partially inhibited by kininogen and cystatin C, and not by stefin A and potato cystatin. Arg-specific activity was inhibited most strongly by stefin A, less by kininogen and cystatin C, and not by potato cystatin. The combination of results on the pH optimum of degradation, the cleavage sites, and the specific inhibitors of degradation of equistatin and synthetic substrates, therefore, indicate that the third domain of kininogen is preventing equistatin degradation in vitro by inhibiting both potato legumains and Arg-specific proteases. The activity of kininogen against potato legu-main is confirmed by the presence on domain III of a potential legumain-binding back-side loop similar to the one on type II cystatins, which is not present on domain I/II of kininogen (Alvarez Fernandez et al., 1999). In the study of Alvarez Fernandez et al. (1999), kininogen was not active toward pig legumain, but structural differences between pig and potato/kidney bean legumain may be involved in the different sensitivity to kininogen (Rotari et al., 2001).
Plant Endosomal Proteases as an Impediment for Protein Production in Plants
The function of the ER as the initial compartment of the secretory pathway is to ensure proper folding, posttranslational modifications (peptide processing, glycosylation, and disulfide formation), and oligomerization of the secretory proteins. For functional expression, many heterologous proteins depend on the secretory pathway, yet it is evident that the protein accumulation levels may be seriously affected by the proteases resident in the endosomal system of potato.
The accumulation of other proteins like antibodies, targeted to the ER, is known to be severely affected by plant proteases (Stevens et al., 2000). The study of Sharp and Doran (2001a) indicates that the degradation of a murine IgG1 is most likely initiated along the secretory pathway outside the ER, and it is further degraded in the extracellular space. In our particular case, it is likely that equistatin ends up in the vacuole or the cell wall, where it is further degraded. Legumain activity resides both in the vacuole and the cell wall (Muntz and Shutov, 2002). The KDEL signal seems to only partly retain equistatin in the ER because nearly identical patterns of degradation were found in both plants expressing equistatin with and without KDEL signal. This inability of the KDEL signal to retain proteins completely in the ER has been described before with proteins ending up in either the vacuole (Toyooka et al., 2000) or the cell wall (Inohara et al., 1989).
Furthermore, the significance of proteolytic degradation in plants is highlighted by a number of studies that describe different proteolytic fragments in different plant species and plant cell cultures (Hiatt et al., 1989; De Neve et al., 1993; Dolja et al., 1998; Khoudi et al., 1999). Thus, plant proteases can be a serious hurdle for the production of functional intact protein in plants, and strategies to effectively avoid this degradation are poorly developed.
Strategies to Reduce the Degradation of Heterologous Proteins in Plants
Reduction of the degradation of ER-targeted equistatin in potato and of proteins in the secretory pathway in general may be achieved in two different ways. Replacement of the amino acids presently cleaved by functionally equivalent amino acids, which preserve the biological activity of the protein but prevent cleavage, may allow the design of stable molecules for expression in plants. Alternatively, the co-expression of protease inhibitors that prevent protein degradation in vitro may also stabilize the protein in planta. A rational mutagenesis approach will only be possible with proteins for which precise information on susceptible cleavage sites is available. In most cases, protein degradation occurs too rapidly to identify the initial cleavage points. As a general approach, the co-expression of cysteine protease inhibitors like kininogen domain III may therefore more effectively stabilize foreign proteins in plants. Future studies are needed to investigate the feasibility of such approaches.
MATERIALS AND METHODS
Plant Material and Insects
Potato (Solanum tuberosum cv Desirée) plants were used in all experiments. Transgenic potato plants were grown in the greenhouse with supplementary high-pressure sodium light under 16-h/8-h light/dark rhythm and temperature regime of 21°C/18°C. CPB larvae were obtained from an established colony at the Department of Entomology, Wageningen University and Research Center (The Netherlands).
Preparation of Constructs for Plant Transformation
Four plant transformation cassettes were prepared to express the equistatin gene (B. Rogelj, B. Strukelj, and M.A. Jongsma, unpublished data). The coding part of equistatin was excised out of the plasmid pEIM8 with NcoI-BglII restriction enzymes and then inserted into pUCLhca.3.St1 (Nap et al., 1993), pUCRbcS1 (Outchkourov et al., 2003), and pUCAP35S (van Engelen et al., 1995) to create pUCLhca.3.St1EIM, pUCRbcS1EIM, and pUCAP35S. pUCRbcS1/EIM/KDEL was constructed by PCR amplification of the equistatin gene using pUCLhca.3.St1/EIM as the template and the primers Cabd (5′-cttttgtatttaatttattcttg-3′) and EIMKDEL (5′-ttttttagatctttaaagttcatccttagcgcatgtgggacgtttgaatc-3′) to add the KDEL amino acid sequence to the C-terminal end of equistatin. The obtained fragment was cut with NcoI-BglII restriction enzymes and inserted into the linearized pUCRbcS1 vector. These four expression cassettes containing a promoter, equistatin, and a terminator were then excised and inserted into the multiple cloning site of the pBINPLUS vector (van Engelen et al., 1995) to create pLhca.3.St1/EIM, pRbcS1/EIM, pRbcS1/EIMKDEL, and p35S/EIM (Fig. 1).
Potato Transformation
All pBINPLUS constructs and empty pBINPLUS vector as control were introduced into Agrobacterium tumefaciens AGL0 by electroporation. Potato plants were transformed using the stem segment transformation method (Visser et al., 1989).
Northern Blot
Total RNA was isolated from plant tissue according to Verwoerd et al. (1989). Samples of 10 μg of total RNA were denatured and separated on a 1.2% (w/v) agarose gel as previously described (Kevil et al., 1997). As a positive control, RNA from the equistatin gene synthesized with Riboprobe in vitro Transcription Systems (Promega, Madison, WI) was used. Band intensities were visualized by autoradiography using a BAS-2000 phosphor imaging scanner (Fuji Photo Film, Tokyo).
Dot-Blot Immunological Detection and Quantification of Equistatin in Transgenic Potato Plants
Unless otherwise indicated, leaf samples were collected between 9:30 and 10 am. Leaf discs were punched into separate wells of a 96-well microtiter plate and to each well, 200 μL of protein extraction buffer (80 mm Tris-Cl, pH 7.6, containing 25 mm diethyldithiocarbamate, and 50 mm Na2EDTA) was added. The samples were then three times frozen to -20°C and thawed to room temperature. The extract was pipetted to a new microtiter plate, separating it sufficiently from the insoluble leaf material. For dot-blot analysis 2.5 μg of protein extract obtained with the freeze-thaw extraction procedure was transferred to Trans-Blot nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA) using the SRC 96 D Dot-blot apparatus (Schleicher & Schuell, Dassel, Germany). Subsequently, the membranes were blocked in 10 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% [v/v] Tween 20 containing 2% (w/v) non-fat milk powder for 1 h and then incubated with rabbit anti-EI antibodies (Eurogentec, Seraing, Belgium). The blots were subsequently washed and incubated with goat anti-rabbit IgG conjugated with horseradish peroxidase (Jackson Immunoresearch Laboratories, West Grove, PA). The membranes were visualized with Lumi-Light western blotting substrate and scanned in the Lumi-Imager F1 under the control of Lumi-Analist software (Roche Diagnostics, Mannheim, Germany). Purified equistatin as a quantitative protein reference was obtained from the study of Strukelj et al. (2000). Based on dot-blot analysis and polyclonal sera, the total amount of equistatin present in extracts was determined. This method measured both intact and degraded nonfunctional protein.
Western-Blot Analysis
A piece of a fully expanded young leaf (200-300 mg) was placed in a 1.5-mL Eppendorf tube and ground in liquid nitrogen to a fine powder. The powder was resuspended in 300 μL of extraction buffer (100 mm Tris-HCl, pH 7.6, 25 mm sodium-diethyldithiocarbamate, 50 mm EDTA, 10% [w/v] polyvinylpolypyrrolidone [PVPP], and 1 tablet 25 mL-1 protease inhibitors cocktail Complete [Roche Diagnostics]), and this crude extract was twice centrifuged for 5 min at 14,000 rpm and 4°C. Each time, the supernatant was replaced into a new tube. SDS-PAGE was performed using a 15% (w/v) precast resolving gel (Bio-Rad Laboratories) on a mini-Protean II slab cell apparatus (Bio-Rad Laboratories). The gels were run according to the manufacturer's instructions. The separated protein samples from the SDS-PAGE gels were transferred to Trans-Blot (Bio-Rad Laboratories) nitrocellulose membranes using the mini-Protean II electrotransfer apparatus (Bio-Rad Laboratories). Blocking of the membranes and the detection procedure was the same as described for the dot blots.
Assay of Inhibitory Activity
Plant protein extracts were tested for the presence of equistatin by the inhibition of papain (2× crystallized, Sigma-Aldrich, St. Louis). Buffer solution of 50 μL (50 mm MES, pH 6.5, with 5 mm Cys and 0.1 mg mL-1 bovine serum albumin fraction V) was mixed with 20 μL of 150 nm papain (final concentration 15 nm) in the wells of a microtiter plate. Increasing quantities (0-80 μL) of total soluble leaf protein (50 μg mL-1) from transgenic line pRbcS1/EIM/KDEL22 were mixed with decreasing quantities (80-0 μL) of total soluble leaf proteins (50 μg mL-1) from control plants transformed with the empty pBINPLUS vector to maintain a constant final concentration of plant protein in each measurement (20 μg mL-1 in 200 μL reaction volume) and to control for endogenous potato inhibitors. After 30 min of pre-incubation at room temperature, the reaction was started by adding 50 μL of substrate consisting of 0.9 mg mL-1 Z-Phe-Arg-pNA (Bachem, Bubendorf, Switzerland) in 50 mm MES buffer, pH 6.5 (15 mg mL-1 stock in MeOH). The reaction was spectrophotometrically recorded at 405 nm on a Benchtop microtiter plate reader (Bio-Rad Laboratories). On the basis of the x axis intercept (20 μg mL-1 total soluble protein), the equimolar concentration of inhibitor contained therein (15 nm or 0.32 μg mL-1 equistatin based on molecular weight of 21,500 g mol-1) was experimentally determined (1.6%).
Purification of the Equistatin Degradation Products from Transgenic Potato Plants
Papain and anti-equistatin antiserum were coupled to cyanogen bromide-activated Sepharose 4B (Amersham Biosciences, Uppsala) according to the manufacturer's requirements. An additional carboxymethylation step was conducted for papain after the coupling to the cyanogen bromide-activated Sepharose 4B. The papain Sepharose was first activated by the addition of 5 mm l-Cys in 0.1 m sodium-phosphate, pH 7.0, for 15 min at room temperature. Then the excess buffer was removed on a sintered glass filter (porosity G3), and the gel was placed in carboxymethylation buffer (0.1 m iodoacetate and 0.1 m sodium-phosphate, pH 7.0) at 37°C for 1 h. The gel was then washed with a 0.1 m sodium-phosphate buffer, pH 7.0. The papain and antisera-Sepharose affinity gels were packed in 1- × 6-cm columns.
Fully expanded young leaves were collected and ground in liquid nitrogen to a fine powder. The powder was resuspended in 1:1 ratio of extraction buffer (100 mm Tris-HCl, pH 7.6, 25 mm sodium-diethyldithiocarbamate, 50 mm EDTA, and 10% [w/v] PVPP). This crude extract was twice centrifuged for 5 min at 14,000 rpm and 4°C. The supernatant was then heated for 3 min at 80°C and centrifuged again. This material was filtered through 22-μm filters and was used as input for the immunoaffinity and papain-affinity columns. The proteins in above buffers were loaded on either column at a speed of 1 mL min-1. The immuno-affinity column was washed with 1 m NaCl and 50 mm Tris, pH 7.5, and the papain column with 1 m NaCl and 50 mm MES, pH 6.5. Bound proteins were eluted using 20 mm Tris, pH 10.7, which contained 20% (v/v) dimethyl sulfoxide (DMSO). Fractions (1 mL) were collected and the immediately neutralized with 0.2 mL of 1 m sodium-phosphate, pH 7.0. The fractions were analyzed by western-blot analysis. The ones that contained the equistatin bands were separated on 15% (w/v) SDS-PAGE and were blotted on polyvinylidene difluoride membrane (Millipore, Bedford, MA). The membranes were washed once with double distilled water, then saturated with 100% methanol for a few seconds, and finally stained with 0.1% (w/v) Coomassie Blue R-250 in 40% (v/v) methanol and 1% (v/v) acetic acid. The bands were visualized after destaining with 50% (v/v) methanol. The bands were cut using surgical blades and placed in separate 1.5-mL Eppendorf tubes and sent for N-terminal sequencing to ProSeq, Protein Microsequencing (Boxford, MA).
pH Optimum of the Proteolytic Activities in Potato Leaf Extracts toward Bz-Asn-pNA, Z-Arg-Arg-pNA,and Equistatin
Fully expanded leaves were ground using a prechilled mortar in the presence (2:1, w/v) of double distilled water that contains 10% (w/v) PVPP. This crude extract was twice centrifuged for 5 min at 14,000 rpm and 4°C, and each time, the supernatant was replaced into a new tube. Buffers (50 mm MES-NaOH, pH 5.6, 6.0, and 6.5, respectively, and 50 mm Tris-HCL, pH of 7.6) were prepared and mixed in a ratio of 1:1 (v/v) with the above prepared supernatant of the plant extract. These mixtures were centrifuged again to remove the insoluble materials, and the supernatant was used to carry out the enzymatic analysis. In a 96-well microtiter plate, plant extracts that contain 60 μg of protein were mixed with buffers with a corresponding pH to final volume of 150 μL. The reaction was started by adding 50 μL of each of the two substrates consisting of 0.9 mg mL-1 Bz-Asn-pNA and Z-Arg-Arg-pNA (Bachem) in water (15 mg mL-1 stock in DMSO for Bz-Asn-pNA and in MeOH for Z-Arg-Arg-pNA). The reaction was spectrophotometrically recorded at 405 nm on a Benchtop microtiter plate reader (Bio-Rad Laboratories). Equistatin was also incubated in the above prepared plant extracts in the different pH buffers, and the degradation of the intact protein was monitored by western-blot analysis.
Inhibitors of Equistatin Degradation
Leaf extract of potato cv Desirée (60 μg) was incubated with equistatin (125 ng) for 1 h at pH 5.6, plus or minus inhibitors (10 μm kininogen domain 3, stefin A, cystatin C, and potato cystatin), and subsequently, the remaining equistatin was analyzed by western blot. In parallel, plant extract (60 μg, pH 5.6) was mixed with a MES-NaOH buffer, pH 5.6, to a final volume of 130 μL in wells of a microtiter plate, then 20 μL of each inhibitor (100 μM stock), and 50 μL of each of the two substrates consisting of 0.9 mg mL-1 Bz-AsnpNA and Z-Arg-Arg-pNA (Bachem) in water (15 mg mL-1 stock in DMSO for Bz-Asn-pNA and in MeOH for Z-Arg-Arg-pNA). The reaction was spectrophotometrically recorded at 405 nm on a Benchtop microtiterplate reader (Bio-Rad Laboratories). Purified inhibitors kininogen domain 3, stefin A, and cystatin C were provided by Dr. M. Abrahamson (Department of Clinical Chemistry, University of Lund, Sweden). Potato cystatin as a quantitative protein reference was obtained from the study of Annadana et al. (2002).
Insect Feeding Trials
Young leaves about 5 cm long and having 0.5 to 1 cm of stem were cut off from chosen transformed and control lines and inserted into an Eppendorf tube that contained 0.3% (w/v) agar in water. Each leaf was placed on filter paper in a closed petri dish and inoculated with six newly hatched CPB larvae. The bioassays were conducted under the following conditions: 16-h/8-h light/dark, 25°C, and 70% relative humidity. Leaves were changed every day at 10 am. Surviving larvae were counted every day, and from the 3rd d onward, they were also weighed to measure growth rates. Most of the high-expressing transgenic plant lines were screened in eight different bioassays.
Acknowledgments
We thank Dr. Lucas H. Stevens for careful reading of the manuscript, and Dr. M. Abrahamson for the generous gift of purified cystatins.
This work was supported by the European Union Research and Technical Development program (grant no. FAIR6-CT98-4239) and by the Dutch Ministry of Agriculture, Nature, and Fisheries (program nos. 282 and 338).
References
- Abe Y, Shirane K, Yokosawa H, Matsushita H, Mitta M, Kato I, Ishii S (1993) Asparaginyl endopeptidase of jack bean seeds: purification, characterization, and high utility in protein sequence analysis. J Biol Chem 15: 3525-3529 [PubMed] [Google Scholar]
- Alvarez Fernandez M, Barrett Alan J, Gerhartz B, Dando Pam M, Ni J, Abrahamson M (1999) Inhibition of mammalian legumain by some cystatins is due to a novel second reactive site. J Biol Chem 274: 19195-19203 [DOI] [PubMed] [Google Scholar]
- Annadana S, Schipper B, Beekwilder MJ, Outchkourov NS, Udayakumar M, Jongsma MA (2003) Cloning, expression in Pichia pastoris, and purification of potato cystatin and multicystatin. J Biosci Bioeng 95: 118-123 [DOI] [PubMed] [Google Scholar]
- Bottari A, Capocchi A, Galleschi L, Jopova A, Saviozzi F (1996) Asparaginyl endopeptidase during maturation and germination of durum wheat. Physiol Plant 97: 475-480 [Google Scholar]
- Brouquisse R, Gaudillere JP, Raymond P (1998) Induction of a carbon-starvation-related proteolysis in whole maize plants submitted to light/dark cycles and to extended darkness. Plant Physiol 117: 1281-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, Hewitt E, Watts C, Barrett AJ (1997) Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J Biol Chem 272: 8090-8098 [DOI] [PubMed] [Google Scholar]
- Dai Z, Hooker BS, Anderson DB, Thomas SR (2000) Improved plant-based production of E1 endoglucanase using potato: expression optimization and tissue targeting. Mol Breed 6: 277-285 [Google Scholar]
- De Neve M, De Loose M, Jacobs A, Van Houdt H, Kaluza B, Weidle U, Van Montagu M, Depicker A (1993) Assembly of an antibody and its derived antibody fragment in Nicotiana and Arabidopsis. Transgenic Res 2: 227-237 [DOI] [PubMed] [Google Scholar]
- Dolja VV, Peremyslov VV, Keller KE, Martin RR, Hong J (1998) Isolation and stability of histidine-tagged proteins produced in plants via potyvirus gene vectors. Virology 252: 269-274 [DOI] [PubMed] [Google Scholar]
- Fischer J, Becker C, Hillmer S, Horstmann C, Neubohn B, Schlereth A, Senyuk V, Shutov A, Muntz K (2000) The families of papain- and legumain-like cysteine proteinases from embryonic axes and cotyledons of Vicia seeds: developmental patterns, intracellular localization and functions in globulin proteolysis. Plant Mol Biol 43: 83-101 [DOI] [PubMed] [Google Scholar]
- Gruden K, Strukelj B, Popovic T, Lenarcic B, Bevec T, Brzin J, Kregar I, HerzogVelikonja J, Stiekema WJ, Bosch D et al. (1998) The cysteine protease activity of Colorado potato beetle (Leptinotarsa decemlineata Say) guts, which is insensitive to potato protease inhibitors, is inhibited by thyroglobulin type-1 domain inhibitors. Insect Biochem Mol Biol 28: 549-560 [DOI] [PubMed] [Google Scholar]
- Guevara MG, Oliva CR, Huarte M, Daleo GR (2002) An aspartic protease with antimicrobial activity is induced after infection and wounding in intercellular fluids in potato tubers. Eur J Plant Pathol 108: 131-137 [Google Scholar]
- Gun[caron]car G, Punger[caron]ci[caron]c G, Klemen[caron]ci[caron]c I, Turk V, Turk D (1999) Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J 18: 793-803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Nishimura I (1993) Vacuolar processing enzyme [in Ricinus communis, rice, maize, soyabeans, pumpkins and spinach] responsible for proprotein processing of various seed proteins. Rep Soy Protein Res Committee Jpn 14: 122-126 [Google Scholar]
- Hara Nishimura I, Inoue K, Nishimura M (1991) A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett 294: 89-93 [DOI] [PubMed] [Google Scholar]
- Hara Nishimura I, Shimada T, Hiraiwa N, Nishimura M (1995) Vacuolar processing enzyme responsible for maturation of seed proteins. J Plant Physiol 145: 632-640 [Google Scholar]
- Hara Nishimura I, Shimada T, Kuroyanagi M, Yamada K (1998) Vacuolar sorting machinery and processing mechanism for storage proteins. Soy Protein Res Jpn 1: 6-12 [Google Scholar]
- Harsulkar AM, Giri AP, Patankar AG, Gupta VS, Sainani MN, Ranjekar PK, Deshpande VV (1999) Successive use of non-host plant proteinase inhibitors required for effective inhibition of Helicoverpa armigera gut proteinases and larval growth. Plant Physiol 121: 497-506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiatt A, Cafferkey R, Bowdish K (1989) Production of antibodies in transgenic plants. Nature 342: 76-78 [DOI] [PubMed] [Google Scholar]
- Hiraiwa N, Nishimura M, Hara Nishimura I (1999) Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett 447: 213-216 [DOI] [PubMed] [Google Scholar]
- Inohara N, Shimomura S, Fukui T, Futai M (1989) Auxin-binding protein located in the endoplasmatic reticulum of maize shoots: molecular cloning and complete primary structure. Proc Natl Acad Sci USA 86: 3564-3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma MA, Bolter C (1997) The adaptation of insects to plant protease inhibitors. J Insect Physiol 43: 885-895 [DOI] [PubMed] [Google Scholar]
- Kevil CG, Walsh L, Laroux FS, Kalogeris T, Grisham MB, Alexander JS (1997) An improved, rapid Northern protocol. Biochem Biophys Res Commun 238: 277-279 [DOI] [PubMed] [Google Scholar]
- Khoudi H, Laberge S, Ferullo JM, Bazin R, Darveau A, Castonguay Y, Allard G, Lemieux R, Vezina LP (1999) Production of a diagnostic monoclonal antibody in perennial alfalfa plants. Biotechnol Bioeng 64: 135-143 [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Yamada K, Hiraiwa N, Kondo M, Nishimura M, Hara Nishimura I (1999) Vacuolar processing enzyme is up-regulated in the lytic vacuoles of vegetative tissues during senescence and under various stressed conditions. Plant J 19: 43-53 [DOI] [PubMed] [Google Scholar]
- Koziel MG, Carozzi NB, Desai N (1996) Optimizing expression of transgenes with an emphasis on post-transcriptional events. Plant Mol Biol 32: 393-405 [DOI] [PubMed] [Google Scholar]
- Lenarcic B, Ritonja A, Strukelj B, Turk B, Turk V (1997) Equistatin, a new inhibitor of cysteine proteinases from Actinia equina, is structurally related to thyroglobulin type-1 domain. J Biol Chem 23: 13899-13903 [DOI] [PubMed] [Google Scholar]
- Lenarcic B, Turk V (1999) Thyroglobulin type-1 domains in equistatin inhibit both papain-like cysteine proteinases and cathepsin D. J Biol Chem 274: 563-566 [DOI] [PubMed] [Google Scholar]
- Muntz K, Shutov A (2002) Legumains and their functions in plants. Trends Plant Sci 7: 340-344 [DOI] [PubMed] [Google Scholar]
- Nap JP, van Spanje M, Dirkse WG, Baarda G, Mlynarova L, Loonen A, Grondhuis P, Stiekema WJ (1993) Activity of the promoter of the Lhca3. St. 1 gene, encoding the potato apoprotein 2 of the light-harvesting complex of photosystem I, in transgenic potato and tobacco plants. Plant Mol Biol 23: 605-612 [DOI] [PubMed] [Google Scholar]
- Otsu M, Urade R, Kito M, Omura F, Kikuchi M (1995) A possible role of ER-60 protease in the degradation of misfolded proteins in the endoplasmic reticulum. J Biol Chem 270: 14528-14961 [DOI] [PubMed] [Google Scholar]
- Outchkourov NS, Peters J, de Jong J, Rademakers W, Jongsma MA (2003) The promoter-terminator of chrysanthemum rbcS1directs very high expression levels in plants. Planta 216: 1003-1012 [DOI] [PubMed] [Google Scholar]
- Outchkourov NS, Stiekema WJ, Jongsma MA (2002) Optimization of the expression of equistatin in Pichia pastoris. Protein Exp Purif 24: 18-24 [DOI] [PubMed] [Google Scholar]
- Raharjo SB, Emoto N, Ikeda K, Sato R, Yokoyama M, Matsuo M (2001) Alternative splicing regulates the endoplasmic reticulum localization or secretion of soluble secreted endopeptidase. J Biol Chem 276: 25612-25620 [DOI] [PubMed] [Google Scholar]
- Rotari VI, Dando PM, Barrett AJ (2001) Legumain forms from plants and animals differ in their specificity. Biol Chem 382: 953-959 [DOI] [PubMed] [Google Scholar]
- Ryan CA (1990) Protease inhibitors in plants: genes for improving defences against insects and pathogens. Annu Rev Phytopathol 28: 425-449 [Google Scholar]
- Schlereth A, Standhardt D, Mock HP, Muntz K (2001) Stored cysteine proteinases start globulin mobilization in protein bodies of embryonic axes and cotyledons during vetch (Vicia sativa L.) seed germination. Planta 212: 718-727 [DOI] [PubMed] [Google Scholar]
- Schouten A, Roosien J, Engelen FA, Jong GAM, Borst Vrenssen AWM, Zilverentant JF, Bosch D, Stiekema WJ, Gommers FJ, Schots A (1996) The C-terminal KDEL sequence increases the expression level of a single-chain antibody design to be targeted to both the cytosol and the secretory pathway in transgenic tobacco. Plant Mol Biol 30: 781-793 [DOI] [PubMed] [Google Scholar]
- Sharp JM, Doran PM (2001a) Characterization of monoclonal antibody fragments produced by plant cells. Biotechnol Bioeng 73: 338-346 [DOI] [PubMed] [Google Scholar]
- Sharp JM, Doran PM (2001b) Strategies for enhancing monoclonal antibody accumulation in plant cell and organ cultures. Biotechnol Prog 17: 979-992 [DOI] [PubMed] [Google Scholar]
- Stevens LH, Stoopern GM, Elbers IJW, Molthoff JW, Bakker HAC, Lommen A, Bosch D, Jordi W (2000) Effect of climate conditions and plant developmental stage on the stability of antibodies expressed in transgenic tobacco. Plant Physiol 124: 173-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strukelj B, Lenarcic B, Gruden K, Pungercar J, Rogelj B, Turk V, Bosch D, Jongsma MA (2000) Equistatin, a protease inhibitor from the sea anemone Actinia equina, is composed of three structural and functional domains. Biochem Biophys Res Commun 269: 732-736 [DOI] [PubMed] [Google Scholar]
- Toyooka I, Okamoto T, Minamikawa T (2000) Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum derived vesicle is involved in protein mobilization in germinating seeds. J Cell Biol 148: 453-463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Seo S, Ohashi Y, Hashimoto J (2000) Circadian and senescence-enhanced expression of a tobacco cysteine protease gene. Plant Mol Biol 44: 649-657 [DOI] [PubMed] [Google Scholar]
- van Engelen FA, Molthoff JW, Conner AJ, Nap JP, Pereira A, Stiekema WJ (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res 4: 288-290 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Ortola C, Hervieu G, Furuta M, Barbero P, Steiner Donald F, Seidah Nabil G, Nahon Jean L (1999) Cellular localization and role of prohormone convertases in the processing of pro-melanin concentrating hormone in mammals. J Biol Chem 274: 6536-6545 [DOI] [PubMed] [Google Scholar]
- Vierstra RD (1993) Protein degradation in plants. Annu Rev Plant Physiol Plant Mol Biol 44: 385-410 [Google Scholar]
- Vierstra RD (1996) Proteolysis in plants: mechanisms and functions. Plant Mol Biol 32: 275-302 [DOI] [PubMed] [Google Scholar]
- Visser RGF, Jacobsen E, Hesseling Meinders A, Schans MJ, Witholt B, Feenstra WJ (1989) Transformation of homozygous diploid potato with an Agrobacterium tumefaciens binary vector system by adventitious shoot regeneration on leaf and stem segments. Plant Mol Biol 12: 329-337 [DOI] [PubMed] [Google Scholar]
- Vitale A, Galili G (2001) The endomembrane system and the problem of protein sorting. Plant Physiol 125: 115-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandelt CI, Khan MRI, Craig S, Schroeder HE, Spencer D, Higgins TJV (1992) Vicilin with carboxy-terminal KDEL is retained in the endoplasmic reticulum and accumulates to high levels in the leaves of transgenic plants. Plant J 2: 181-192 [DOI] [PubMed] [Google Scholar]
- Wong EY, Hironaka CM, Fischhoff DA (1992) Arabidopsis thaliana small subunit leader and transit peptide enhance the expression of Bacillus thurigiensis proteins in transgenic plants. Plant Mol Biol 20: 81-93 [DOI] [PubMed] [Google Scholar]