Abstract
The kinetic properties of an adenine DNA methyltransferase involved in cell cycle regulation of Caulobacter crescentus have been elucidated by using defined unmethylated or hemimethylated DNA (DNAHM) substrates. Catalytic efficiency is significantly enhanced with a DNAHM substrate. Biphasic kinetic behavior during methyl incorporation is observed when unmethylated or DNAHM substrates are used, indicating that a step after chemistry limits enzyme turnover and is most likely the release of enzyme from methylated DNA product. The enzyme is thermally inactivated at 30°C within 20 min; this process is substantially decreased in the presence of saturating concentrations of DNAHM, suggesting that the enzyme preferentially binds DNA before S-adenosylmethionine. The activity of the enzyme shows an unusual sensitivity to salt levels, apparently dissociating more rapidly from methylated DNA product as the salt level is decreased. The enzyme acts processively during methylation of specific DNA sequences, indicating a preferred order of product release in which S-adenosylhomocysteine is released from enzyme before fully methylated DNA. The kinetic behavior and activity of the enzyme are consistent with the temporal constraints during the cell cycle-regulated methylation of newly replicated chromosomal DNA.
DNA methylation is a ubiquitous biological process that occurs in diverse organisms ranging from bacteria to humans. During this process, DNA methyltransferases (MTases) catalyze the addition of a methyl group to the N6 position of adenine or the C5 or N4 position of cytosine for which S-adenosylmethionine (AdoMet) is the universal donor of the methyl group (reviewed in ref. 1). Methylation of adenine or cytosine residues in bacteria and eukaryotes is recognized as an important cellular function. For example, bacteria modify certain sequences in their genome to distinguish their own DNA from that of invading viruses; the methylated host sites resist cleavage by endogenous restriction endonucleases that inactivate the unmodified viral genome (reviewed in ref. 1). In addition, the methylation state of some promoter sequences regulates gene expression (2). In higher eukaryotes, DNA methylation plays a central role in X chromosome inactivation (3), genomic imprinting (4), and embryonic development (5) whereas aberrations in DNA methylation have recently been implicated in aging (6) and various diseases including cancer (7).
There are examples of at least two bacterial N6 adenine DNA MTases that lack a cognate restriction enzyme and function to coordinate cell cycle events. These are Escherichia coli DAM (8) that recognizes GATC and Caulobacter crescentus CcrM (M.CcrMI) (9) that recognizes GANTC sequences. Cell cycle-regulated methyltransferase (CcrM) differs from the E. coli DAM enzyme in that CcrM is essential for viability. Furthermore, CcrM activity is tightly regulated during the cell cycle as the enzyme is only present in the predivisional cell (10). If CcrM is expressed throughout the cell cycle, the normal patterns of DNA replication and cell division are disrupted (9, 11). The regulatory functions of DNA methylation by CcrM appear not be unique to C. crescentus. Homologs of CcrM are widespread in the entire α subdivision of Gram-negative bacteria, a diverse ensemble that includes among its members free-living stalked bacteria (such as C. crescentus), plant pathogens (Agrobacterium tumefaciens), nitrogen-fixing symbionts (Rhizobium meliloti), and animal pathogens (Brucella abortus) (10, 12). Recently, the R. meliloti CcrM homolog was shown to be essential for viability and overexpression of CcrM results in aberrant patterns of cell division in R. meliloti (12) and B. abortus (R.W., G. Robertson, M. Roop, and L.S., unpublished results).
In C. crescentus, DNA replication is initiated on a fully methylated chromosome in the stalked cell (9). Replication yields two hemimethylated chromosomes that are brought to their fully methylated state during a narrow window of the cell cycle, just before division. This is accomplished by turning on the expression of ccrM in the predivisional cell and then clearing the cell of CcrM by Lon-mediated proteolysis (11). Thus, CcrM has a limited amount of time to catalyze in the predivisional cell the methylation of two newly replicated, hemimethylated chromosomes, each with approximately 15,000 GANTC sites. To define the kinetic mechanism used by CcrM to perform this temporally restricted catalysis, the ccrM gene was overexpressed in E. coli, and the enzyme was purified to homogeneity. By elucidating the minimal kinetic mechanism of CcrM, we hope to further understand this enzyme’s role in cell cycle-mediated events, thus providing a useful paradigm for studies of MTases in higher organisms.
MATERIALS AND METHODS
Materials.
[3H-CH3]AdoMet (81 or 15 Ci/mmol), [γ-32P]ATP, and [α-32P]dATP were from New England Nuclear, and unlabeled AdoMet, Adoltcy, and ATP were from Sigma. Phosphoramidites for DNA synthesis were obtained from Glen Research (Sterling, VA) with the exception of the N6-methyldeoxyadenosine phosphoramidite, which was obtained from Pharmacia. All restriction and DNA-modifying enzymes used during molecular cloning and DNA manipulation experiments were from New England Biolabs, Promega, United States Biochemical, or Boehringer Mannheim. DE81 ion exchange paper disks and glass fiber filters were obtained from Whatman. All other materials were obtained from commercial sources and were of the highest available quality.
DNA Substrates.
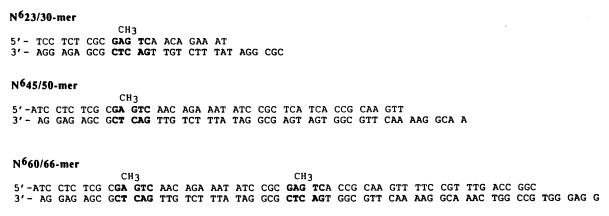
All synthetic oligonucleotides were synthesized with an Expedite BioSystems DNA synthesizer and were purified as previously described (13). Small duplex DNA substrates (<45/50-mers) were prepared by the protocol of Kuchta et al. (14), and larger DNA substrates (60/66-mer and N660/66-mer) were prepared by using a modification of the protocol established by Kaboord and Benkovic (15). Following purification of each respective large oligonucleotide, the two strands were annealed and purified by nondenaturing gel electrophoresis (14). All duplex DNA were quantitated as described by Kuchta et al. (14) and are depicted in Fig. 1.
Figure 1.
Defined DNA substrates used to measure CcrM activity in vitro. For convenience, only DNAHM substrates are depicted with the target DNA methylation in bold. Unmethylated DNA substrates contain identical sequences except that the target adenine residue is not methylated at the N6 position.
Enzyme Purification.
To overexpress CcrM in E. coli, the ccrM gene was subcloned into pET21b (Novagen) to create pCS255b, which allows transcription by the T7 RNA polymerase in E. coli. After induction with 1 mM isopropyl β-d-thiogalactoside, the cells were harvested by centrifugation and the pellet was retained. The cells were suspended in 50 mM Tris-HCl, pH 7.5/1 mM EDTA/5 mM 2-mercaptoethanol/50 mM NaCl (buffer A) with 0.1% phenylmethylsulfonyl fluoride and lysed by sonication. Cell debris was removed by centrifugation at 12,000 rpm for 20 min at 4°C followed by ultracentrifugation (30,000 rpm) for 90 min at 4°C. The supernatant was diluted 1:1 with buffer A and applied to a 50-ml DEAE-Sephacel column connected in series to a 30-ml phosphocellulose column, both preequilibrated with 1 liter of buffer A. The protein was washed with 500 ml of buffer A and then eluted from the phosphocellulose column by using a linear gradient of buffer A with 50–750 mM NaCl. CcrM elutes at approximately 250 mM NaCl, and these fractions were collected and analyzed for protein composition by SDS/PAGE, which revealed resolution of protein with an apparent molecular weight of 42 kDa. After elution of the protein from the phosphocellulose column, the enzyme was concentrated by using an Amicon apparatus employing a YM-30 molecular weight cut-off membrane. After concentration, the protein was determined to be >95% pure based on SDS/polyacrylamide gel electrophoresis (data not shown). The extinction coefficient of the protein was calculated to be 61,920 M−1 cm−1 as deduced from the predicted amino acid composition (9) by using the method of Gill and von Hippel (16). The concentration of CcrM based on measuring the A280 of the protein agrees with the concentration based on Bradford determinations (17).
Kinetic Assays.
MTase activity of CcrM was measured by monitoring the incorporation of [3H]CH3 from [3H]AdoMet into DNA. A typical assay consists of 250 nM CcrM/5 μM DNA (hemi- or unmethylated)/6 μM [3H]AdoMet in a 50-μl total volume at 30°C. A reaction buffer consisting of 50 mM Hepes, pH 7.5/5 mM 2-mercaptoethanol/150 mM potassium acetate was typically used in each assay. Following the enzymatic incorporation of [3H]CH3 from [3H]AdoMet into DNA, a 5-μl aliquot of the reaction was spotted onto DE81 anion-exchange filter paper at various times. The filters were then washed with 200 ml of 0.3 M ammonium formate (pH 8) for 10 min for three times to remove all unreacted [3H]AdoMet. The filters were then briefly washed twice with 95% ethanol followed by once with anhydrous ether. The filters were air dried and counted by standard liquid scintillation techniques. The specific activity of the reaction is determined by measuring the radioactivity present in 5 μl of the reaction spotted on glass filter fibers without washing. The amount of methyl incorporation into the DNA substrate is determined by dividing the counts per min of the washed reaction of AdoMet samples by the specific activity of the total reaction mixture. All data are corrected for nonspecific binding of [3H]AdoMet to the washed filter.
Processive Methylation Assays.
The second in vitro assay is based on resistance of DNA to restriction endonuclease digestion because of the enzymatic production of the dimethylated DNA (DNADM) product. The amount of DNA that is resistant to cleavage by HindII digest resulting from full methylation of the small, synthetic DNA substrate containing two GAGTC sites can be accurately monitored. If the GAGTC site is fully methylated by CcrM, HindII will be unable to cleave the fully methylated DNA molecule, and full-length starting material will be obtained. If the DNA is cleaved by the restriction enzyme, smaller DNA fragments will be obtained, directly indicating a lack of methyl incorporation into the DNA substrate.
The N660/66-mer containing two GAGTC sites was 5′-labeled by using T4 polynucleotide kinase and [γ32P]ATP according to the manufacturer’s protocol (United States Biochemical). Methylation assays were performed by using 250 nM CcrM/2 μM 5′-labeled N660/66-mer/6 μM [3H]AdoMet in the appropriate reaction buffer at 30°C. Five microliters of reaction was quenched and subjected to the filter binding assay monitoring [3H]CH3 incorporation from [3H]AdoMet into duplex DNA as described above.
Concomitantly, 20-μl reaction aliquots were quenched by either heat denaturation (65°C) of CcrM or by the addition of 50 μl of phenol/chloroform at times varying from 15 s to 20 min. The quenched reactions were then subjected to HindII digestion consisting of 10 μl of the quenched DNA in a 20-μl reaction with the appropriate reaction buffer and 1 μl of HindII (Promega). After 3 h of HindII digestion at 37°C, 10 μl of this reaction was quenched with 10 μl of gel loading dye. DNA fragments were then resolved by 16% denaturing gel electrophoresis, and gel images were obtained with a Molecular Dynamics PhosphorImager.
RESULTS AND DISCUSSION
CcrM Activity with Unmethylated or Hemimethylated Duplex DNA.
Our goals are to ultimately define the kinetic, chemical, and regulatory mechanisms of the DNA methyltransfer reaction catalyzed by CcrM. To facilitate such studies, small, synthetic defined DNA sequences were constructed that mimic a specific target DNA sequence within the C. crescentus genome, GAGTC, in either unmethylated or hemimethylated form (Fig. 1).
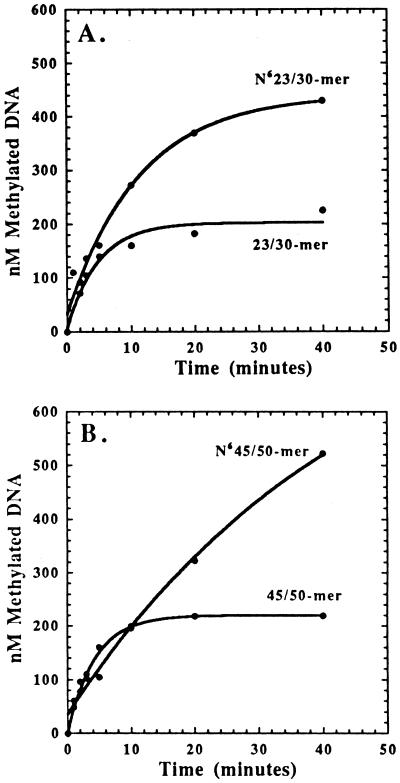
The integrity and performance of CcrM on these defined DNA sequences were addressed by monitoring [3H]methyl incorporation into duplex DNA. For all DNA substrates tested, either unmethylated or hemimethylated, the amount of methylated DNA produced as a function of time was not linear as one would typically expect for an enzyme displaying Michaelis-Menten kinetic behavior. Only one turnover of the enzyme is detected by using unmethylated DNA as the substrate, and the time required for this initial turnover is approximately 5–10 min (Fig. 2). The amount of methylated product formed is directly proportional to enzyme concentration, consistent with an initial burst in product formation followed by a rate-limiting step after methyl transfer (described in more detail below).
Figure 2.
Preference for DNAHM vs. unmethylated DNA. Reaction mixtures of 50 μl contained 250 nM CcrM preincubated with 5 μM unmethylated or DNAHM and initiated by the addition of 50 μM AdoMet. Data with DNAHM were fit to the equation, y = Ae−kt + Bx + C, where A is the burst amplitude, k is the observed rate constant for the burst amplitude, B is the steady state rate, and C is a defined constant. Data with unmethylated DNA were fit to the equation, y = Ae−kt + C, where A is the burst amplitude, k is the observed rate constant for the burst amplitude, and C is a defined constant.
By using hemimethylated DNA (DNAHM) as the substrate, more than one turnover of the enzyme was detected characterized by an initial burst in product formation followed by a second, slower phase after chemistry, specifically methyl transfer (Fig. 2). The higher turnover of CcrM with DNAHM indicates that the enzyme has a distinct preference for hemimethylated vs. unmethylated DNA as a substrate, consistent with its in vivo activity. Interestingly, the steady-state portion of the time course continues briefly before a third even slower phase in product formation becomes dominant at reaction times greater than 20 min (data not shown). The mechanistic ramifications of the triphasic time course are discussed later.
Active site titrations of CcrM purified on three separate occasions were performed by preincubating a variable concentration of CcrM (125, 250, and 500 nM) with 10 μM N645/50-mer followed by the addition of 50 μM AdoMet (data not shown). These assays included 150 mM potassium acetate, which, based on studies of CcrM activity in E. coli (I.L. and S.J.B., unpublished results), is most likely physiologically relevant. All time courses displayed a rapid burst in product formation followed by a slower rate of product formation. The burst amplitudes obtained were dependent on the concentration of CcrM, consistent with at least a two-step kinetic mechanism in which binding and methylation occurs in a first rapid step followed by regeneration of enzyme in a second rate-limiting step, which is most likely the release of CcrM from DNADM product (see below). Extrapolation of the steady-state rate back to time 0 yielded burst amplitudes of 1 mol of methylated DNA product per mol of CcrM. The enzyme is 100% active, and its activity does not vary from preparation to preparation because the burst amplitudes are equal to that of enzyme concentration. The rate constant, kobs, for the initial burst in product formation is 0.2 min−1, whereas a kcat value of 0.02 min−1 is obtained from the linear portion of the time courses, which reflects methylation under steady-state conditions. Thus, under optimal conditions, kobs is at most 10 times faster than kcat.
Time-Dependent Thermal Inactivation of CcrM Activity.
The first two phases of the triphasic time course for methyl incorporation result from a burst of one enzyme turnover followed by a second, slower rate that reflects subsequent enzyme turnover. The appearance of a third phase during the time course is indicative of several possibilities including, but not limited to, substrate depletion, product inhibition, and/or enzyme instability. It is unlikely that substrate depletion results in triphasic behavior because high levels of both substrates are utilized. Accumulation of either DNADM or Adoltcy product may also inhibit MTase activity. Whereas both products are inhibitors of CcrM, inhibition is only detected at far higher concentrations than would be present under these initial velocity conditions (A.J.B. and S.J.B., unpublished results).
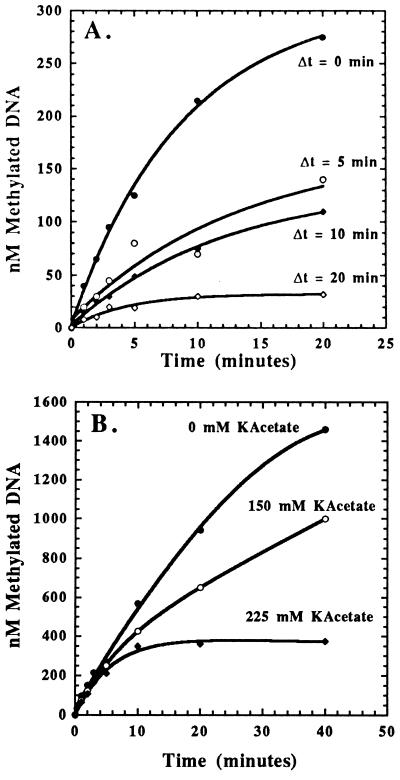
The possibility of the enzyme being unstable during the course of the reaction was first addressed by incubating the enzyme at either 4 or 30°C for 20 min before the addition of DNA and AdoMet. The enzyme preincubated at 4°C was fully active, whereas the enzyme preincubated at 30°C was completely inactive (data not shown). The rate of thermal inactivation of CcrM was then measured as a function of time by preincubating the enzyme at 30°C for times varying from 0 to 20 min before the addition of DNA and AdoMet (Fig. 3A). The amount of product formation decreases as a function of preincubation time, and the resulting data were plotted as log of product formation (at t = 20 min) vs. preincubation time (data not shown) to yield an apparent inactivation rate constant of 0.14 min−1.
Figure 3.
(A) Time-dependent, thermal inactivation of CcrM. Reaction mixtures of 50 μl contained 250 nM CcrM incubated for variable time and initiated with the simultaneous addition of 5 μM N623/30-mer and 50 μM AdoMet. Data were fit as previously described in Fig. 2. (B) Effect of increasing potassium acetate concentrations on CcrM activity. Reaction mixtures of 50 μl contained 250 nM CcrM preincubated with 5 μM N645/50-mer and variable concentrations of potassium acetate before initiation by the addition of 50 μM AdoMet. The burst amplitudes and the observed rate constant for initial product formation are independent of varying salt concentration, whereas the steady-state rate decreases as the salt concentration is increased.
The effect of DNA “stabilizing” CcrM was examined in an identical manner, except that CcrM was first preincubated with N623/30-mer for variable times (0–20 min) before the addition of AdoMet (data not shown). Although a decrease in product formation was detected at long incubation times, the lifetime of the functional enzyme was increased by 5-fold on preincubation with saturating concentrations of DNA. The increased stability of the enzyme in the presence of DNA suggests a preferred order of substrate binding in which the enzyme binds DNA before AdoMet.
Effect of Salt Concentration on CcrM Activity.
The effect of salt concentration on CcrM activity was addressed by preincubating the enzyme with DNA in the appropriate reaction buffer with variable potassium acetate concentrations (0–225 mM) for 1 min before initiation of the reaction by the addition of AdoMet (Fig. 3B). The burst amplitudes and the observed rate constant for initial product formation are independent of varying salt concentration, indicating that the affinity of enzyme for substrate DNA is not altered nor are any steps before the rate-limiting step along the reaction pathway. However, the steady-state rate of MTase activity is highly sensitive to salt concentration because the steady-state rate decreases as the salt concentration is increased. Because the steady-state rate of MTase activity reflects enzyme dissociation from the DNADM product, the rate of dissociation decreases as the salt concentration is increased. This behavior is highly unusual because most DNA-metabolizing enzymes such as DNA repair enzymes (18) and replicative polymerases (19) display the opposite behavior as the salt concentration is increased, i.e., these enzymes tend to dissociate more rapidly from DNA as the salt concentration is increased.
Processive Methylation by CcrM.
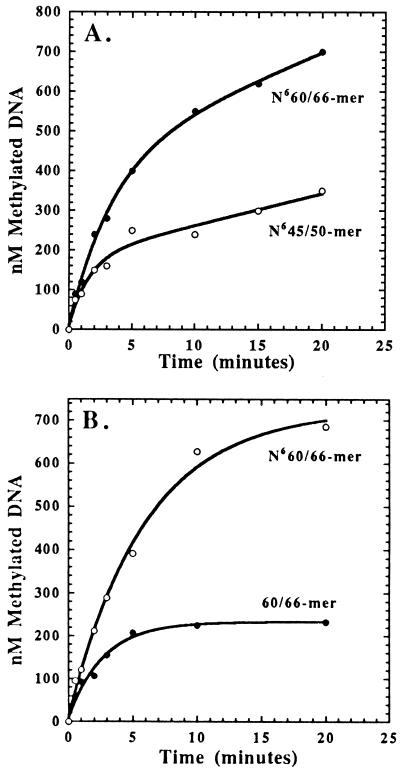
The above data suggest that CcrM, under physiological salt conditions, possesses a high intrinsic affinity for DNA, and this increased affinity for DNA may indicate that CcrM is a processive MTase as opposed to the more frequently reported distributive MTases (20). A DNAHM substrate containing two GANTC methylation sites designated N660/66-mer (Fig. 1) was used to address the processivity of CcrM. Assays were performed by preincubating a limiting concentration of CcrM (250 nM) with saturating N660/66-mer (5 μM) and initiating the reaction by the addition of 6 μM AdoMet (Fig. 4A). As controls, assays were also performed by using N623/30-mer and N645/50-mer as the DNA substrates under identical conditions (Fig. 4A).
Figure 4.
(A) Time course for product formation with N660/66-mer and N645/50-mer DNA as substrates. One mole equivalent of [3H]CH3 is incorporated into N645/50-mer, whereas 2 mol are incorporated into N660/66-mer, suggesting that CcrM processively methylates DNA. (B) Time course for product formation with N660/66-mer and 60/66-mer DNA as substrates. One mole equivalent of [3H]CH3 is incorporated into 60/66-mer, whereas 2 mol are incorporated into N660/66-mer, indicating that CcrM is capable of processively methylating DNAHM but not unmethylated DNA.
If CcrM is distributive, the enzyme will dissociate from the DNA molecule before methylating another site. CcrM must be distributive using substrates with only one possible methylation site per DNA molecule (N623/30-mer and N645/50-mer); in these cases, the enzyme dissociates from product DNA followed by subsequent turnovers. If the enzyme is processive, it is expected that two methyl groups will be incorporated into a single N660/66-mer molecule, which is due to methylation of both sites on a single molecule of DNA. A defined steady-state rate of enzyme activity will not be observed because the enzyme need not dissociate from the DNA before encountering another methylation site. The time courses for product formation by using N623/30-mer, N645/50-mer, and N660/66-mer reveal that this is indeed the case. As expected, a burst in product formation followed by a slow steady-state rate is obtained by using either N623/30-mer or N645/50-mer. However, the time course for methyl incorporation with N660/66-mer indicates that 2 mol eq of [3H]CH3 were incorporated with the same observed rate constant of 0.2 min−1. Furthermore, the lack of a defined steady-state rate with N660/66-mer is consistent with the proposed mechanism. Collectively, the data suggest that CcrM acts processively to methylate both sites of N660/66-mer by using low AdoMet concentrations.
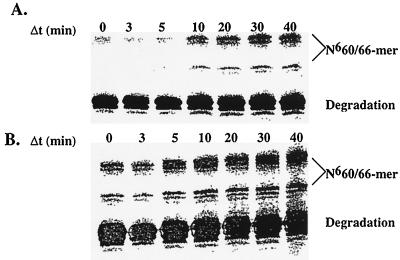
Additional evidence for the processive nature of CcrM catalysis was addressed by restriction digestion analysis of the reaction products. If CcrM is processive, both sites on a single DNA molecule will be methylated and thus resistant to HindII digestion, yielding full-length DNA after digestion. Conversely, if CcrM is distributive, only one site per DNA molecule will be methylated. After digestion with HindII, a mixture of products will be obtained, but full-length DNA will not be observed. Assays were performed preincubating 2 μM 32P-labeled N660/66-mer with 250 nM CcrM and initiating the reaction with 6 μM AdoMet. After quenching the reaction at variable times, aliquots were subjected to HindII digestion, and the reaction products were separated by 16% denaturing sequencing gel electrophoresis. Fig. 5A clearly displays an increase in full-length N660/66-mer, indicating that both sites are resistant to HindII digestion as a result of processive methylation. To conclusively eliminate distributive DNA methylation, an identical experiment was performed in which 10 μM DNA trap was added after initiation of the reaction. The DNA trap employed substitutes 2-aminopurine 2-deoxyribonucleoside for adenine in the target DNA sequence and is designated N623/30AP-mer. CcrM is incapable of methylating N623/30AP-mer because the amino group to be methylated is in the 2-position rather than the 6-position in adenine. N623/30AP-mer is an effective inhibitor of CcrM (A.J.B. and S.J.B., unpublished results) but does not interfere with the endonuclease activity of HindII (data not shown). If CcrM is distributive, inclusion of N623/30AP-mer will trap any free enzyme into a dead-end complex and prevent methylation of the second site of N660/66-mer after dissociation from the first site. Addition of a molar excess (10 μM) of N623/30AP-mer does not alter the production of full-length N660/66-mer (Fig. 5B), conclusively demonstrating that CcrM alone is capable of processive DNA methylation under physiological conditions.
Figure 5.
Visualization of processive DNA methylation by CcrM. (A) The increase in full-length N660/66-mer indicates that both sites are resistant to HindII digestion as a result of processive methylation. The majority of N660/66-mer is not utilized by CcrM and is therefore susceptible to restriction digestion by HindII. Unreacted N660/66-mer is cleaved into several distinct smaller DNA fragments, although only the largest of these fragments (≈52-mer) is shown for convenience. (B) Addition of a molar excess (10 μM) of N623/30AP-mer does not alter the production of full-length N660/66-mer, indicating that CcrM processively methylates DNA under physiological conditions.
The processive nature of CcrM activity was also addressed by using either unmethylated or DNAHM substrate containing two GANTC methylation sites designated 60/66-mer or N660/66-mer. Assays were performed and analyzed by filter binding techniques by using either substrate as previously described. By using the unmethylated 60/66-mer, one anticipates that at least 2 (or possibly 4) mol of [3H-CH3] will be incorporated into the 60/66-mer substrate because of methylation of both sites on a single molecule of DNA. Surprisingly, CcrM incorporates only 1 mol of [3H]CH3 into the 60/66-mer as compared with 2 mol of [3H]CH3 into the N660/66-mer substrate, indicating that CcrM does not act processively on unmethylated DNA substrates (Fig. 4B). This result is consistent with data obtained with the smaller, unmethylated DNA substrates containing only one possible methylation site and suggests that CcrM does not readily dissociate from a DNAHM site produced by its own activity.
Conclusions.
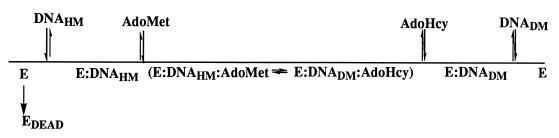
A minimal kinetic mechanism for CcrM is proposed based on the kinetic data obtained and is presented in Fig. 6. Because of the thermal, time-dependent inactivation of CcrM in the absence of DNA substrate, the enzyme most likely preferentially binds to DNA whether or not AdoMet is present. The biphasic nature of the enzyme-catalyzed reaction is consistent with a two-step kinetic mechanism in which binding of both substrates and methylation occur first in a rapid step followed by regeneration of enzyme in a second rate-limiting step. This kinetic behavior has also been observed in several other MTases, including EcoRI (21) and the murine DNA cytosine-C5 MTase (22). Both the observed rate constant for CcrM during initial product formation and kcat are substantially slower than the reported value for EcoRI but very similar to the value obtained for the murine C5 MTase. The slow kinetic rate constants displayed by CcrM may reflect a regulatory role during in vivo DNA methylation as has been proposed for the murine C5 MTase (22).
Figure 6.
Proposed minimal kinetic mechanism for CcrM. CcrM preferentially binds to duplex DNA followed by the binding of AdoMet. CcrM in the absence of DNA is thermally inactivated at a rate of 0.14 min−1.
Biphasic kinetic behavior during methyl incorporation is observed regardless of the methylation state of DNA substrate indicating that a step after methyl transfer, most likely release of CcrM from product DNA, limits enzyme turnover. Whereas CcrM can utilize unmethylated DNA as a substrate, the off-rate of the enzyme from the product DNA (DNAHM in this case) is much slower than that from DNADM and essentially limits the enzyme to only one turnover. Consistent with this model, only one turnover by CcrM is observed with the unmethylated 60/66-mer DNA substrate whereas both sites are processively methylated with the N660/66-mer substrate. The preference for DNAHM vs. unmethylated DNA as reflected by differences in catalytic efficiencies and processive methylation is consistent with the in vivo role of CcrM during the cell cycle methylation of genomic DNA (9, 10).
The processive nature of the enzyme with DNAHM yields important insight into the overall kinetic and regulatory mechanism of CcrM. Because CcrM is processive, AdoHyc must be released before fully methylated DNA, indicative of a preferred order in product release. The overall kinetic mechanism of CcrM thus appears to be symmetrical in which the enzyme binds DNA before AdoMet and then releases AdoHyc before release of methylated DNA. The processive nature of CcrM with DNAHM most likely reflects the role of CcrM in remethylation of the newly replicated chromosomes in the predivisional cell. After replication, C. crescentus has two chromosomes, each with a total of approximately 15,000 GANTC sites. Because the methyl transfer step is rate-limiting during processive DNA methylation (kobs = 0.2 min−1), approximately 12,000 sites will be methylated by the 3,000 CcrM molecules/C. crescentus cell (I.L. and S.J.B., unpublished results) within the 20-min time period of CcrM activity. Thus, on average, one molecule of CcrM must methylate at least three distinct GANTC sites. Thus, CcrM studied in vitro is sufficiently active and displays kinetic behavior consistent with maintaining the genomic methylation pattern of C. crescentus in vivo.
Acknowledgments
This work was supported by Research Grant MDA972-97-1-0008 from the Defense Advanced Research Planning Agency (to S.J.B. and L.S.) and National Institutes of Health Grant GM-52426 (to L.S.). J.K.C. was supported in part by National Institutes of Health Grant CA28097.
ABBREVIATIONS
- MTase
methyltransferase
- AdoMet
S-adenosylmethionine
- CcrM
cell cycle-regulated methyltransferase
- Adoltcy
S-adenosylhomocysteine
- DNAHM
hemimethylated DNA
- DNADM
dimethylated DNA
References
- 1.Cheng X. Annu Rev Biophys Biomol Struct. 1995;24:293–318. doi: 10.1146/annurev.bb.24.060195.001453. [DOI] [PubMed] [Google Scholar]
- 2.Braaten B, Nou X, Koltenbach L, Low D. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 3.Gartler S M, Riggs A D. Annu Rev Genet. 1983;17:155–190. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- 4.Swain J L, Stewart T A, Leder P. Cell. 1987;50:719–727. doi: 10.1016/0092-8674(87)90330-8. [DOI] [PubMed] [Google Scholar]
- 5.Li E, Bestor T, Jaenisch R. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 6.Nicholls R D, Rinchik E M, Driscoll D J. Semin Dev Biol. 1995;3:139–152. [Google Scholar]
- 7.Jones P A, Buckley J D. Adv Cancer Res. 1990;54:1–23. doi: 10.1016/s0065-230x(08)60806-4. [DOI] [PubMed] [Google Scholar]
- 8.Boye E, Lobner-Olesen A. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 9.Zweiger G, Marczynski G, Shapiro L. J Mol Biol. 1994;245:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]
- 10.Stephens C, Reisenauer A, Wright R, Shapiro L. Proc Natl Acad Sci USA. 1996;93:1210–1214. doi: 10.1073/pnas.93.3.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright R J, Stephens C, Zweiger G, Shapiro L, Alley M R K. Genes Dev. 1996;10:1532–1542. doi: 10.1101/gad.10.12.1532. [DOI] [PubMed] [Google Scholar]
- 12.Wright R J, Stephens C, Shapiro L. J Bacteriol. 1997;179:5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capson T L, Peliska J A, Kaboord B F, Frey M W, Lively C, Dahlberg M, Benkovic S J. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 14.Kuchta R D, Mizrahi V, Benkovic P A, Johnson K A, Benkovic S J. Biochemistry. 1987;26:8410–8417. doi: 10.1021/bi00399a057. [DOI] [PubMed] [Google Scholar]
- 15.Kaboord B F, Benkovic S J. Proc Natl Acad Sci USA. 1993;90:10881–10885. doi: 10.1073/pnas.90.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 17.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Bennett S S, Sanderson R J, Mosbaugh D W. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- 19.Williams A J, Wernette C M, Kaguni L S. J Biol Chem. 1993;268:24855–24862. [PubMed] [Google Scholar]
- 20.Surby M, Reich N O. Biochemistry. 1996;35:2201–2208. doi: 10.1021/bi951883n. [DOI] [PubMed] [Google Scholar]
- 21.Reich N O, Mashhoon N. J Biol Chem. 1993;268:9191–9193. [PubMed] [Google Scholar]
- 22.Flynn J, Glickman J F, Reich N O. Biochemistry. 1996;35:7308–7315. doi: 10.1021/bi9600512. [DOI] [PubMed] [Google Scholar]