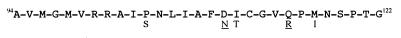Abstract
Phage exclusion is a form of programmed cell death in prokaryotes in which death is triggered by infection with phage, a seemingly altruistic response that limits multiplication of the phage and its spread through the population. One of the best-characterized examples of phage exclusion is the exclusion of T-even phages such as T4 by the e14-encoded Lit protein in many Escherichia coli K-12 strains. In this exclusion system, transcription and translation of a short region of the major head coat protein gene late in phage infection activates proteolysis of translation elongation factor Tu (EF-Tu), blocking translation and multiplication of the phage. The cleavage occurs between Gly-59 and Ile-60 in the nucleotide-binding domain. In the present work, we show that a 29-residue synthetic peptide spanning the activating region of the major head coat protein can activate the cleavage of GDP-bound EF-Tu in a purified system containing only purified EF-Tu and purified Lit protein. Lit behaves as a bona fide enzyme in this system, cleaving EF-Tu to completion when present at substoichiometric amounts. Two mutant peptides with amino acid changes that reduce the activation of cleavage of EF-Tu in vivo were also greatly reduced in their ability to activate EF-Tu cleavage in vitro but were still able to activate cleavage at a high concentration. Elongation factor G, which has the same sequence at the cleavage site and a nucleotide-binding domain similar to EF-Tu, was not cleaved by this system, and neither was heat-inactivated EF-Tu, suggesting that the structural context of the cleavage site may be important for specificity. This system apparently represents an activation mechanism for proteolysis that targets one of nature’s most evolutionarily conserved proteins for site-specific cleavage.
Phage-exclusion systems were discovered more than 40 years ago, and many of them are well characterized genetically (1, 2). The proteins involved are usually encoded by prophages, transposons, and plasmids and kill the cell upon infection by a different type of phage, preventing the multiplication of the infecting phage and its spread to other cells in the population. Well known examples include the exclusion of many phages by the rex gene products of λ, the exclusion of T7 by the pif gene product of the F plasmid, and the exclusion of T4 by the prrC gene product of a cyptic DNA element related to P1 phage in some clinical strains of Escherichia coli.
One of the best-understood phage exclusion mechanisms is caused by the defective prophage e14, a DNA element integrated in the isocitrate dehydrogenase gene of many E. coli K-12 strains (3, 4). The e14 element encodes a protein Lit (for late inhibitor of T4), which cleaves translation elongation factor Tu (EF-Tu) after infection by T4 and other T-even phages, thereby blocking translation and the multiplication of the infecting phage (3, 5–7). The proteolysis of EF-Tu is activated by the transcription and translation of a short region of only 75 bp within gene 23, the major head protein gene, of the infecting phage (5). This short region, named the gol region because it was first defined by mutations that allow the phage to grow on Lit protein-containing cells, is located about 300 bp downstream of the initiation codon of gene 23 (5, 6). The cleavage of EF-Tu occurs site-specifically between Gly-59 and Ile-60 in the effector (or L2) region of EF-Tu, one of the most conserved and functionally important parts of the molecule (cf. ref. 8). It is here that nucleotides bind and where the ribosome binds and stimulates hydrolysis of GTP. This is also one of the regions that shows similarity with other regulatory GTPases, including the oncogene, Ras (cf. ref. 9).
In our original in vivo experiments using plasmid clones of the gol region of gene 23, it was clear that transcription and translation of the gol region was required to activate cleavage of EF-Tu, but it was unclear whether the RNA or peptide, or both, from the gol region activates the cleavage and whether other cellular components are involved (7). Here we demonstrate the cleavage of EF-Tu in a purified in vitro system containing only Lit protein, chemically synthesized Gol peptide, and EF-Tu. We also show that the cleavage is highly specific for EF-Tu and may require structural features of the EF-Tu cleavage site.
MATERIALS AND METHODS
Expression and Purification of Lit.
To facilitate purification, the Lit protein was first overexpressed in a T7-based expression system. One to 2 liters of E. coli BL21 (DE3) containing the lit gene cloned in pET-11 was grown in Luria–Bertani (LB) broth containing ampicillin (100 μg/ml) at 37°C with shaking and overproduction of the protein induced by adding isopropyl β-d-thiogalactoside (IPTG) (1 mM final) for 2 hr at mid-log phase during growth. Cells were harvested by centrifugation, resuspended in 50 mM Tris⋅Cl, pH 8/10 mM EDTA/1 mM PMSF/10 mM 2-mercaptoethanol/200 μg/ml lysozyme, and disrupted by sonication. The suspension was then centrifuged at 12,000 × g for 10 min, the supernatant was discarded, and the pellet fraction containing Lit in the form of inclusion bodies was retained. The pellet was washed twice in a deoxycholate buffer (50 mM Tris⋅Cl, pH 8/10 mM EDTA/10 mM 2-mercaptoethanol/5% vol/vol glycerol/0.5% wt/vol deoxycholic acid) and solubilized using the anionic detergent sarkosyl (0.5% wt/vol) dissolved in 50 mM Tris⋅Cl, pH 8/10 mM EDTA/10 mM 2-mercaptoethanol/5% vol/vol glycerol/50 mM NaCl. This solubilized extract was then dialyzed against the same buffer but in the absence of sarkosyl, concentrated by ultrafiltration, and loaded onto a 360-ml Superdex-75 gel-filtration column (Pharmacia) equilibrated in 50 mM Tris⋅Cl, pH 8/1 mM EDTA/10 mM DTT/5% vol/vol glycerol/50 mM NaCl/0.05% wt/vol sarkosyl. Active Lit elutes as a monomer whereas inactive aggregate elutes in the void volume of the column. The Lit protein, >90% pure at this stage as judged by SDS/PAGE (data not shown), was concentrated by ultrafiltration, frozen, and stored at −20°C. Protein concentrations were determined by the Bradford method (10), using BSA as the calibrant. The yield of pure protein was typically 10 mg from 1 liter of culture.
Lit Activity Assays.
Activation of EF-Tu cleavage in crude cell extracts. Crude extracts of cells containing either Lit protein, expressed both because of an up-promoter lit mutation in e14 in the chromosome and a clone of the lit gene in the plasmid pTTQ18 (Amersham) (3) or the expressed gol region from the plasmid pUC84PZ1 (5), were prepared from 250 ml of mid-log cells in tryptone broth (10 g tryptone/5 g NaCl/liter) containing 50 μg/ml ampicillin and to which IPTG had been added to 1 mM, 30 min previously. The cells were centrifuged, washed once with saline, and resuspended in 3 ml of resuspension buffer (10 mM Tris-acetate, pH 8.2/60 mM KCl/14 mM MgOAc/1 mM DTT). They were then lysed by sonication and centrifuged at 12,000 × g for 10 min. The Gol peptide was dissolved in H20 at 3 mM and diluted to 0.3 mM in the extract. Cleavage of EF-Tu was monitored by SDS/PAGE and staining with Coomassie blue.
EF-Tu cleavage in a purified three-component system.
His-tagged EF-Tu (2 μM), purified as described previously (11), was incubated with either 2 μM or 0.2 μM Lit and differing concentrations of the 29-residue Gol peptide (2–200 μM) in 50 mM Tris⋅HCl, pH 8.0/10 mM 2-mercaptoethanol/5 mM MgCl2/10 μM GDP/30 μM sarkosyl in a final volume of 250 μl at 37°C for up to 180 min. Reactions were initiated with the addition of peptide and stopped by the addition of EDTA (to a final concentration of 10 mM) or by adding SDS sample buffer followed by boiling. Reaction products were then analyzed by 12% SDS/PAGE and Coomassie blue staining. EF-Tu cleavage was quantified by laser densitometry by using an LKB Ultroscan, and data were analyzed as the percentage of starting EF-Tu by using the gelscan xl software. Where cleavage of EF-G was being investigated, the assay was run as described above except that 5 μM EF-G, purified as described by Mester et al. (12), was added instead of EF-Tu. EF-Tu was heat-inactivated by a published procedure (13).
RESULTS
Lit-Mediated Cleavage of EF-Tu Is Activated by a 29-Residue Synthetic Peptide.
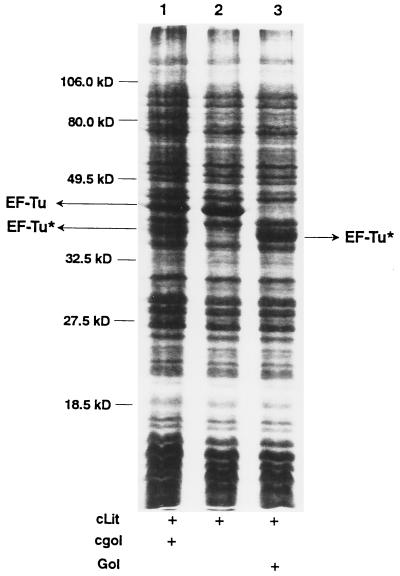
Previously we showed that extracts of E. coli expressing both Lit protein and the gol region of T4 were defective in translation and that the EF-Tu in these extracts was cleaved (7). To determine whether we could activate the cleavage in vitro, we prepared two extracts, one from cells expressing the gol region, and one from cells expressing Lit protein. When these extracts were mixed 1:1 and incubated for 30 min at 30°C, some EF-Tu was cleaved (Fig. 1, lane 1). No EF-Tu was cleaved when the Lit protein-containing extract was incubated alone (Fig. 1, lane 2). Apparently, the RNA or the peptide, or both, encoded by the gol region are required for the activation of cleavage of EF-Tu in extracts containing Lit protein. To determine whether the gol mRNA is the activating component, a 75-nt RNA corresponding to the gol region transcript was made in an in vitro transcription system using T7 RNA polymerase, and this RNA was added in lieu of the gol-expressing extract. No cleavage was observed, nor did pretreatment of the gol-expressing extract with RNase prevent the cleavage (data not shown), indicating that the RNA from the gol region is not required for the activation. To determine whether the peptide encoded by the gol region is solely responsible, we designed a 29-aa peptide (Fig. 2) based on the coding sequence from the PvuII site at 279 bp in T4 gene 23 to the endpoint of the Δ60 deletion, the minimum sequence required for gol region activity (5). When this chemically synthesized peptide was added in lieu of the gol-containing extract, all the EF-Tu in the extract was cleaved (Fig. 1, lane 3). Therefore, the Gol peptide, encoded by the gol region, is solely responsible for the activation of cleavage of EF-Tu.
Figure 1.
Activation of cleavage of EF-Tu in a crude cell lysate by Gol peptide. Two cell extracts, one containing Lit protein and one containing the expressed gol region of T4 phage, were prepared, incubated, and processed as described in Materials and Methods. A Coomassie blue-stained 12% SDS-polyacrylamide gel is shown. Lanes: 1, Gol and Lit-containing extracts mixed before incubation; 2, Lit-containing extract incubated alone; 3, synthetic Gol peptide added to Lit-containing extract before incubation. The positions of EF-Tu and one of the cleavage fragments of EF-Tu (EF-Tu*) are indicated by arrows.
Figure 2.
Sequence of the 29-residue synthetic Gol peptide from the major capsid protein of T4 phage. Amino acid changes from the five original mutations identified in the T4 gol region are shown below the sequence, and those chosen for use in this study are underlined. The numbers refer to the position of the amino acid in the gene 23 protein before cleavage during head formation.
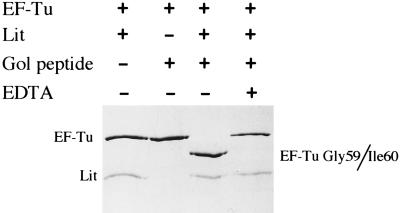
This experiment shows that the combination of the Lit protein and the Gol peptide in the crude extract is required to cleave EF-Tu, but does not eliminate the possibility that other components present in the crude extract are also required. To test this, the cleavage reaction was performed in a simple three-component system consisting of monomeric Lit protein and EF-Tu, purified as in Materials and Methods, as well as the chemically synthesized Gol peptide. As shown in Fig. 3, no cleavage of EF-Tu occurred if either Lit or the Gol peptide was omitted from the incubation but went to completion when all three components were present (Fig. 3, lane 3). Furthermore, the cleavage occurs between Gly-59 and Ile-60, the same site as in vivo, as revealed by N-terminal peptide sequencing of the blotted fragment (data not shown).
Figure 3.
EF-Tu can be site-specifically cleaved in a purified three-component system. Purified His-tagged EF-Tu (2 μM) was incubated in the presence or absence of Lit (1 μM) and the synthetic gol peptide (5 μM) in 50 mM Tris⋅HCl, pH 8.0 containing 10 mM 2-mercaptoethanol, 5 mM MgCl2, 10 μM GDP, and 30 μM sarkosyl at 37°C for 1 hr. Samples were then boiled in SDS sample buffer, resolved on a 12% SDS-polyacrylamide gel, and stained with Coomassie blue. Lanes: 1, no Gol peptide; 2, no Lit protein; 3, all three components; 4, all three components but with EDTA added to 10 mM before incubation.
Although these data clearly demonstrate that no other proteinaceous factors besides Lit, EF-Tu, and the Gol peptide are required for Lit-mediated cleavage of EF-Tu, other requirements of the cleavage reaction remain to be investigated. For example, there is an apparent requirement for metal ions because the cleavage reaction is very sensitive to the presence of the chelating agent EDTA (Fig. 3, lane 4). Lit has the consensus zinc metalloprotease motif His-Glu-Xaa-Xaa-His (14) (Fig. 4), so it may be a zinc metalloprotease, explaining the metal ion requirement. Alternatively, or in addition, the EDTA may be chelating the Mg2+ in the reaction buffer, which may be required to maintain the conformation of the EF-Tu substrate. Without Mg2+ in its nucleotide-binding site, EF-Tu rapidly loses activity and the conformation of the substrate may be important for the cleavage (see below). Also, the state of EF-Tu required for cleavage has not yet been investigated. The EF-Tu was purified in the presence of GDP to help stabilize the protein, so presumably most of the EF-Tu is in the GDP-bound state. Whether EF-Tu in a ternary complex with GTP and tRNA is also a substrate would have to be investigated. The EF-Tu of E. coli (and probably of all organisms) is also methylated at certain stages in the cell cycle. In E. coli, this methylation occurs on lysine 56 (15), only 4 aa removed from the cleavage site. However, it is unclear what influence the state of methylation of EF-Tu has on the cleavage, because no attempt was made to determine the extent of methylation of the EF-Tu in these preparations.
Figure 4.
A possible zinc metalloprotease motif in the Lit protein. The consensus histidine residues are putative metal ligation sites. Where there is some variation, the amino acids at those positions are shown. “X” means no apparent consensus at that position. The motif was found by using the GCG wisconsin motifs software program.
Lit Functions Enzymatically.
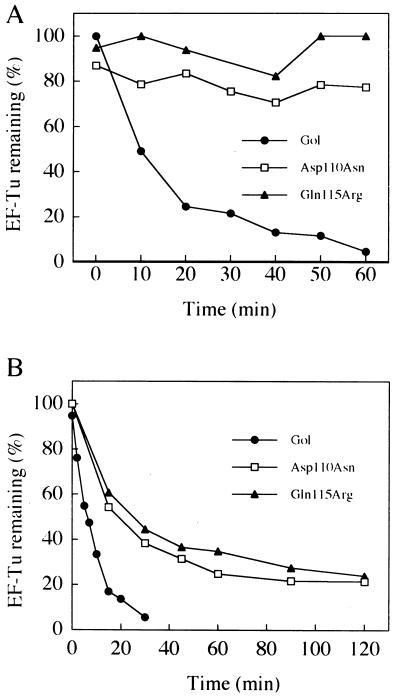
Stoichiometric amounts of Lit and EF-Tu were used in the cleavage reaction shown in Fig. 3, but complete cleavage of EF-Tu is also possible by using substantially lower concentrations of Lit compared with the substrate. For example, in the experiment shown in Fig. 5A, Lit and EF-Tu were incubated at a molar ratio of only 1:10. Nevertheless, the cleavage of EF-Tu went to completion, indicating that Lit turns over repeatedly, a characteristic of an enzyme. The rate of cleavage depends on the concentration of Gol peptide. When the rate of cleavage was compared at 2 μM (Fig. 5A) and 200 μM (Fig. 5B) of the peptide in two separate experiments, the cleavage displayed pseudo-first-order rates of 0.039 (+/−0.007) min−1 and 0.088 (+/−0.009) min−1, respectively.
Figure 5.
Enzymatic activity of Lit and the effect of gol mutations on EF-Tu cleavage. Time course of EF-Tu cleavage by Lit in the presence of wild-type and gol mutant peptides at 2 μM (A) and 200 μM (B) peptide. Incubations were as described in the legend to Fig. 3 and the Materials and Methods section except that the Lit concentration in both sets of data (0.2 μM) was 10% that of substrate EF-Tu. The data are shown as the percentage of starting EF-Tu as determined by laser densitometry of Coomassie blue-stained SDS gels.
Effect of Amino Acid Changes in the Gol Peptide on Lit-Mediated Cleavage of EF-Tu.
Some mutations in the gol region of T4 allow the multiplication of T4 in Lit protein-containing cells and/or allow transformation of Lit protein-containing cells by plasmids containing clones of the gol region (5, 6), presumably by preventing the activation of cleavage of EF-Tu. The expectation then was that mutant Gol peptides with the same amino acid changes caused by these mutations would be defective in the activation of the proteolysis in the purified three-component system. Five different amino acid changes in the Gol peptide due to such mutations have been identified (5, 6), two of which (Asp-110 → Asn and Gln-115 → Arg; Fig. 2) were selected for this in vitro study. The relative ability of the mutant Gol peptides to activate the cleavage was tested by comparing Lit-mediated EF-Tu cleavage in the presence of varying concentrations of wild-type and mutant Gol peptides (Fig. 5). At low concentrations (2 μM), both mutant peptides were essentially inactive (Fig. 5A); however, as the concentrations of the mutant peptides were raised (200 μM), significant activity was observed for both (Fig. 5B). Thus, even though these mutant peptides are severely defective in activating the proteolysis, they do retain some residual activating ability.
Structural Context Requirements for the Cleavage of EF-Tu.
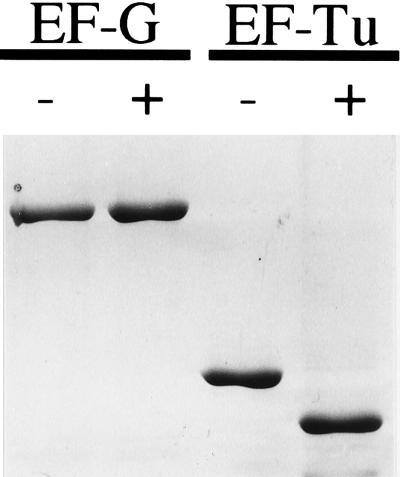
The Lit protease is obviously highly specific for EF-Tu, and the cleavage of other proteins in E. coli, besides EF-Tu, has not been detected. However, it is difficult to eliminate the possibility that other proteins are cleaved. The protein in E. coli that is most similar to EF-Tu in the cleavage region is EF-G, which has the same five-residue sequence RGITI that surrounds the cleavage site in EF-Tu. In addition, EF-G has been proposed to be a structural mimic of the ternary complex of EF-Tu, GTP, and amino acyl-tRNA (ref. 17 and references therein). Nevertheless, EF-G is not cleaved by the Lit system under conditions where EF-Tu is cleaved to completion (Fig. 6). Either the sequence requirements for the cleavage are more extensive than the 5 aa surrounding the cleavage site, or the primary sequence is not enough and the structural context of the cleavage site is also important for the cleavage. We have observed that heat-inactivated EF-Tu also is not a substrate for Lit-promoted cleavage (data not shown), supporting a role for the structural context in the cleavage.
Figure 6.
EF-G is not a substrate for Lit-mediated cleavage. Purified EF-G and EF-Tu (5 μM) were each incubated with Lit protein with or without Gol peptide (both 5 μM) at 37°C for 1 hr as described in the Materials and Methods. Samples were subjected to SDS/PAGE (10%) and stained with Coomassie blue. The Lit protein has run off the gel so it is not visible.
DISCUSSION
We have shown that site-specific proteolysis of EF-Tu in the presence of Lit protein is activated in a purified in vitro system by a 29-residue synthetic peptide normally internal to the major head coat protein gene of T4 phage. Lit functions enzymatically in the cleavage reaction and may be a metalloprotease. The cleavage is highly specific for EF-Tu and may depend on its native structure. High-resolution structural information is available for EF-Tu in many of its functional states (cf. refs. 8, 9, 18, and 19). The Lit-mediated cleavage occurs in the highly conserved RGITI motif common to all EF-Tus and EF-Gs of bacteria, and the corresponding EF-1αs and EF-2s of eukaryotes, respectively. A similar motif also occurs in other translation factors including initiation factor (IF)-2 (20). This motif occurs in a regulatory region called the L2 region, which in all regulatory GTPases is involved in binding the GTPase activating protein (in the case of EF-Tu the ribosome), thus triggering the conformational changes coincident with GTP cleavage. The threonine in this motif is common to most regulatory GTPases and binds to a Mg2+ ion as well as the γ-phosphate of GTP. From what is known of the structures of EF-Tu in its various functional states, the region of cleavage is relatively exposed and flexible, consistent with the fact that the adjacent Arg-Gly peptide bond is a hypersensitive site for trypsin cleavage of the native protein (cf. ref. 16). Cleaving EF-Tu in such a critical region could be predicted to have disastrous consequences for the activity of the protein. However, the detailed functional consequences on translation of the cleavage are not clear. Based on the current view of the properties of trypsinized Thermus thermophilus EF-Tu, in which the adjacent Arg-59 → Gly-60 bond (homologous to Arg-58 → Gly-59 in E. coli) is cleaved, it seems likely that the cleavage of EF-Tu by Lit may abolish the ability of ribosomes to stimulate the GTPase (16).
It is likely that cleavage of EF-Tu is solely responsible for the inhibition of translation in Lit-containing cells after infection with T4 phage. Although we have not rigorously excluded the possibility that other proteins in E. coli besides EF-Tu are cleaved, it is noteworthy that EF-G, the translation factor in E. coli that apparently most closely resembles EF-Tu in the cleavage region, is not cleaved. Furthermore, earlier experiments, in which the cellular content of EF-Tu was increased beyond what could be cleaved by Lit, had indicated that, if other proteins are cleaved, they could not be proteins whose activities are required for phage multiplication (7). In these experiments, the residual uncut EF-Tu supported some translation and phage were produced.
It is interesting to note that the shutting-down of translation in response to viral infection is not peculiar to the phage-exclusion systems of bacteria and that analogous mechanisms operate in both plant and animal cells. Host cell protein synthesis is inhibited late in adenovirus infection of human cells through the phosphorylation of the translation initiation factor IF-2 (21). The strategy of inducing cell death by targeting major cellular processes for site-specific proteolysis is also not without precedent. For example, poly (ADP ribose) polymerase, an enzyme involved in DNA repair in eukaryotic cells, is site-specifically cleaved in response to environmental stress by the protease CED-3, a necessary event in apoptosis (ref. 22 and references therein). Because the Lit-imposed death of T4-infected E. coli cells occurs at a defined point in the development of the phage, the Lit–Gol exclusion system is, in a sense, a prokaryotic example of programmed cell death.
At present, it is not clear how the combination of the Gol peptide and Lit activates proteolytic cleavage of EF-Tu. Activation of proteases most often involves the cleavage of inactive proenzymes or, as in the case of aspartate proteases, the dimerization of inactive monomers (23). We know of only one other example of a protease being activated by a peptide, the activation of the Ad2 protease involved in the maturation of adenovirus by an 11-residue peptide originating from a structural protein of the virion. This activation occurs through thiol-disulfide interchange between a cysteine residue in the peptide and the active site of the enzyme (24). The Gol peptide does contain a single cysteine (see Fig. 2), but a mutant peptide in which this cysteine has been replaced by alanine is still able to activate the cleavage of EF-Tu (S.E. & L.S., unpublished results). Hence, Gol must be activating proteolysis by a mechanism that does not involve disulfide exchange.
Clues to the mechanism of activation will come from studies of the effects of various amino acid changes in the Gol peptide on the activation of proteolysis. Although the effect of only two changes that prevent the activation of cleavage in vivo were investigated in the present study, it is interesting to note that both mutant peptides retained some residual activating ability in vitro. This observation is in agreement with what has been observed in vivo with all gol mutations known to date, in that the effect of the mutations is only partial and can be overcome by high concentrations of the Lit protein (5). Such behavior suggests that the amino acid changes may all be affecting the ability of the Gol peptide to bind its target (EF-Tu or Lit) rather than changing an amino acid with a critical role in the activation; for example, an amino acid that contributes to the formation of the protease active site. We cannot discount the possibility, however, that other, as yet unidentified amino acids in the peptide do perform a critical function in the activation.
The desirability of a regulatory system in which a peptide activates proteolysis seems obvious. A protease could lie dormant in the cell until a specific peptide appears, originating from an infectious agent, the degradation of another protein, or the expression of a developmentally regulated gene. The proteolysis of a specific target protein then occurs, initiating a particular cellular response. Although so far the sole known representative of this particular mechanism of activation operates during the exclusion of T-even phages by the defective prophage e14, it is conceivable that this same mechanism is used by cells in other situations.
Acknowledgments
We thank Robert Hausinger for useful discussions. This work was supported by a grant to L.S. from the National Science Foundation and to C.K. from the Biotechnology and Biological Sciences Research Council (U.K.). T.G. was funded by a University of East Anglia Research Based Studentship and a short-term European Molecular Biology Organization fellowship. S.E. was partially supported by a Cell and Molecular Biology fellowship and a research dissertation fellowship from Michigan State University (MSU). Y.-T.N.Y. and S.E. submitted parts of this work as partial fulfillment of the requirements for the Ph.D. degree from MSU.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: EF-Tu, elongation factor Tu.
A commentary on this article begins on page 2731.
References
- 1.Snyder L. Mol Microbiol. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 2.Yarmolinsky M. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]
- 3.Kao C, Snyder L. J Bacteriol. 1988;170:2056–2062. doi: 10.1128/jb.170.5.2056-2062.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hill C W, Gray J A, Brody H. J Bacteriol. 1989;171:4083–4084. doi: 10.1128/jb.171.7.4083-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergsland K J, Kao C, Yu Y-T N, Gulati R, Snyder L. J Mol Biol. 1990;213:477–494. doi: 10.1016/S0022-2836(05)80209-8. [DOI] [PubMed] [Google Scholar]
- 6.Champness W C, Snyder L. J Mol Biol. 1982;155:395–407. doi: 10.1016/0022-2836(82)90478-8. [DOI] [PubMed] [Google Scholar]
- 7.Yu Y-T N, Snyder L. Proc Natl Acad Sci USA. 1994;91:802–806. doi: 10.1073/pnas.91.2.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abel K, Jurnak F. Structure. 1996;4:229–238. doi: 10.1016/S0969-2126(96)00027-5. [DOI] [PubMed] [Google Scholar]
- 9.Hilgenfeld R. Curr Op Struct Biol. 1995;5:810–817. doi: 10.1016/0959-440x(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 10.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 11.Boon K, Vijgenboom E, Madsen L V, Talens A, Kraal B, Bosch L. Eur J Biochem. 1992;210:177–183. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- 12.Mester J R, de Graaf J M, Kraal B. FEBS Lett. 1993;321:149–152. doi: 10.1016/0014-5793(93)80097-e. [DOI] [PubMed] [Google Scholar]
- 13.Jensen M, Cool R H, Mortensen K K, Clark B F C, Parmeggiani A. Eur J Biochem. 1989;182:247–255. doi: 10.1111/j.1432-1033.1989.tb14824.x. [DOI] [PubMed] [Google Scholar]
- 14.Vallee B L, Auld D S. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 15.Arai K, Clark B F C, Duffy L, Jones M D, Kaziro Y, Laurson R A, L’Italien J, Miller D L, Nagarkatti S, Kanamura S, et al. Proc Natl Acad Sci USA. 1980;77:1326–1330. doi: 10.1073/pnas.77.3.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeidler W, Schirmer N K, Egle C, Ribeiro S, Kreutzer R, Sprinzl M. Eur J Biochem. 1996;239:265–271. doi: 10.1111/j.1432-1033.1996.0265u.x. [DOI] [PubMed] [Google Scholar]
- 17.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B F C, Nyborg J. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 18.Berchtold H, Reshetnikova L, Reiser C O A, Schirmer N K, Sprinzyl M, Hilgenfeld R. Nature (London) 1993;365:126–132. doi: 10.1038/365126a0. [DOI] [PubMed] [Google Scholar]
- 19.Sprinzl M. Trends Biochem Sci. 1994;19:245–250. doi: 10.1016/0968-0004(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 20.Laalami S, Sacerdot C, Vachon G, Mortensen K, Sperling-Petersen H U, Cenatiempo Y, Grunberg-Manago M. Biochimie. 1991;73:1557–1566. doi: 10.1016/0300-9084(91)90191-3. [DOI] [PubMed] [Google Scholar]
- 21.Schneider R J. Annu Rev Biochem. 1987;56:317–332. doi: 10.1146/annurev.bi.56.070187.001533. [DOI] [PubMed] [Google Scholar]
- 22.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, et al. Nature (London) 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 23.Navia M A, Fitzgerald P M D, McKeever B M, Leu C-T, Heimbach J C, Herber W K, Sigal I S, Darke P L, Springer J P. Nature (London) 1989;337:615–620. doi: 10.1038/337615a0. [DOI] [PubMed] [Google Scholar]
- 24.Webster A, Hay R T, Kemp G. Cell. 1993;72:97–104. doi: 10.1016/0092-8674(93)90053-s. [DOI] [PubMed] [Google Scholar]