Abstract
Interactions between ApCREB1a, ApCREB2, and ApC/EBP have been studied using conventional methods such as the yeast two-hybrid system. However, it is unclear whether these memory-related transcription factors actually interact in the native environment in neurons. To clarify this question, we have developed an Aplysia two-hybrid system and found, consistent with previous studies that ApCREB2 interacts with ApCREB1a and ApC/EBP, and that ApC/EBP forms homodimers. We have also found that ApCREB1a and ApC/EBP do not interact. Therefore, our study shows that formerly described interactions between the proteins actually occur in the Aplysia neurons and that interactions between these transcription factors are specific.
Because of its genetic robustness and ease of use, yeast has been the organism of choice for the development of two-hybrid systems (Sambrook and Russell 2001). However, some interactions of proteins may not occur in the yeast milieu because of the possible lack of associating factors, protein modifications (such as signal-induced phosphorylation), or because of lack of correct protein folding (Shioda et al. 2000). For example, the GAL4DB (DNA-binding domain) fusion SMAD4 (bait) and MSG1 (prey) interact in mammalian cells, but the bait–prey pair does not interact in yeast, which lacks the transforming growth factor β (Shioda et al. 2000). In this context, many groups have developed various types of mammalian cell two-hybrid screening systems to identify interacting proteins that are difficult to detect in the yeast system (Fearon et al. 1992; Luo et al. 1997; Shioda et al. 2000).
The Aplysia cell environment differs from the yeast cell environment in various aspects. For example, the Aplysia cell has high salt content and may contain neuron-specific modifying factors. Considering the importance of Aplysia as a model system for the study of memory, we have devised a two-hybrid system in Aplysia neurons.
Using the Aplysia two-hybrid system, we investigated the interaction among ApCREB1a, ApCREB2, and ApC/EBP, transcription factors involved in long-term facilitation in Aplysia. In previous studies using in vitro binding assays or the yeast two-hybrid system, the interactions among these transcription factors have been found to be mediated by the basic-leucine zipper (bZIP) domain (Landschulz et al. 1988), which has the dual function of dimerization and specific DNA-binding (Landschulz et al. 1989). ApCREB2 and ApC/EBP have been shown to interact through their bZIP domains (Bartsch et al. 1995). Also, the bZIP domain of ApCREB2 and mammalian CREB1 (homologous to ApCREB1a) interact in the yeast two-hybrid system (Bartsch et al. 1995). Rat C/EBP forms homodimers through the bZIP domain in cross-linking assays (Landschulz et al. 1989). To our knowledge, there has been no previous report of interaction between ApCREB1a and ApC/EBP (Bartsch et al. 1998). In our study, we used the Aplysia two-hybrid system to determine whether these specific bZIP-mediated interactions and the interaction between ApCREB1a and ApC/EBP actually occur in the Aplysia neuron.
We modified the Gal4-based yeast two-hybrid system (Fields and Song 1989) so that it would function in Aplysia neurons (see Fig. 1). The Gal4 DNA-binding domain (DB)-bait fusion DNA and Gal4 transcription activation domain (AD)-prey fusion DNA were subcloned into Aplysia expression vectors pNEX and pNEXδ (Kaang 1996), respectively.
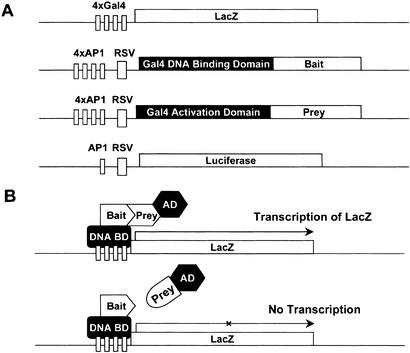
Figure 1.
Aplysia two-hybrid system. (A) Structure of DNA constructs. pNEX-Gal4DB group (bait) has the Gal4 DNA-binding domain fused to the bZIP domain of the transcription factor. pNEX-Gal4AD group (prey) contains the Gal4 transcription activation domain fused to the full-length transcription factor. 4xGal4-LacZ (reporter gene) contains four Gal4 DNA-binding sites in the promoter region of LacZ. pNEX2-luciferase (normalization gene) encodes the firefly luciferase gene. Rous sarcoma virus (RSV) long terminal repeat minimal promoter and four copies of the AP-1 enhancer, both of which are key elements of the pNEX vector, are denoted (Kaang et al. 1995). (B) A schematic diagram showing how the Aplysia two-hybrid system works. Gal4DB-bait fusion protein binds to the Gal4 DNA-binding sites of 4xGal4-LacZ. Only if there is interaction between the bait and prey, will the Gal4 DB and Gal4 AD be brought into proximity, which allows transcription of the reporter gene LacZ.
To test the proper functioning of the system, we compared the β-galactosidase activity after injection of full-length Gal4 with that of Gal4 AD and Gal4 DB (without any protein fused to either counterpart). Gal4 showed β-galactosidase activity (0.63 ± 0.16, number of injected ganglia, n = 5) 36.6 times that of the Gal4AD/Gal4DB pair (0.017 ± 0.004, n = 6; Fig. 2). We then assessed the β-galactosidase activity of Gal4DB-C/EBPbZIP and Gal4DB-CREB2bZIP, respectively, expressed without any Gal4AD-prey counterpart. Both showed a β-galactosidase activity that was even smaller than Gal4AD/Gal4DB (Gal4DB-C/EBPbZIP, 0.0030 ± 0.0017, n = 6; Gal4DB-CREB2bZIP, 0.0018 ± 0.0007, n = 7). These results show that neither the Gal4AD/Gal4DB pair alone, nor the Gal4DB-bait on its own, can activate the reporter gene. Therefore, the β-galactosidase activity of the Gal4AD/Gal4DB pair can be used as a baseline to determine whether β-galactosidase activity resulting from the interaction of the bait and prey is significant.
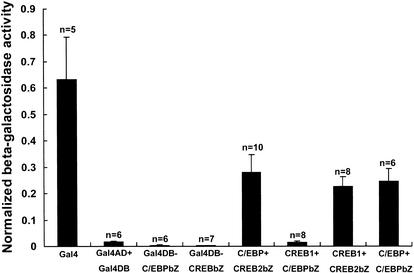
Figure 2.
Bar graph representing degree of interaction in terms of normalized β-galactosidase activity (β-galactosidase/luciferase activity). The height of each bar corresponds to the mean (normalized) β-galactosidase activity ± SEM. (bZ) The bZIP domain of each transcription factor. One-way ANOVA (analysis of variance) indicates a difference in degree of interaction (F = 14.94, df = 7, p < 0.0001). A comparison of means using the Student-Newman-Keuls test shows that interactions between full-length ApC/EBP and the bZIP domain of ApCREB2, full-length ApCREB1a and the bZIP domain of ApCREB2, and full-length ApC/EBP and the bZIP domain of ApC/EBP are significantly different from that between Gal4 AD and Gal4 DB with no protein fused (Gal4AD + Gal4DB). However, interaction between ApCREB1a and ApC/EBP is not significantly different from that between Gal4 AD and Gal4 DB.
We next assessed the β-galactosidase activity from interactions between ApCREB1a, ApCREB2, and ApC/EBP. We found interaction between full-length ApC/EBP and the bZIP domain of ApCREB2 (0.28 ± 0.07, n = 10), full-length ApCREB1a and the bZIP domain of ApCREB2 (0.23 ± 0.04, n = 8), and full-length ApC/EBP and the bZIP domain of ApC/EBP (0.25 ± 0.05, n = 6; one-way ANOVA, F = 14.94, df = 7, p < 0.0001, comparison of means by Student-Newman-Keuls test; Fig. 2). By contrast, we found no significant interaction between full-length ApCREB1a and the bZIP domain of ApC/EBP (0.015 ± 0.004, n = 8).
These results are consistent with previous reports (Landschulz et al. 1989; Bartsch et al. 1995). This implies that interactions between the transcription factors mentioned above actually occur in the Aplysia neuronal environment and that the bZIP regions mediate these interactions. Also, although both ApCREB1a and ApC/EBP have the bZIP domain, we have found no significant interaction between the two. This demonstrates both the selectivity of our two-hybrid system to respond to interacting pairs and the specificity of interactions between the transcription factors. However, it does not rule out the possibility that ApC/EBP could interact with ApCREB1a when regions other than the bZIP domain are intact, or the possibility that the two proteins interact indirectly.
In addition, we detected interactions between the ApCREB1a, ApCREB2, and ApC/EBP proteins in the intact ganglion by X-gal staining in situ (Fig. 3). Although cells were stained when injected with full-length ApC/EBP and the bZIP domain of ApCREB2, none were stained when injected with full-length ApCREB1a and the bZIP domain of ApC/EBP. However, when we observed hrGFP expression from coinjected pNEXδ-hrGFP in both ganglia, one cell (ApC/EBP and ApCREB2 bZIP) and 13 cells (ApCREB1a and ApC/EBP bZIP) from each ganglion expressed hrGFP (data not shown). This shows that the negative result in the latter was not caused by insufficient expression of the injected constructs. Thus the results from X-gal staining match those obtained by β-galactosidase assay.
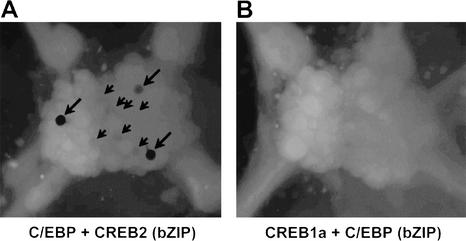
Figure 3.
X-gal staining of abdominal ganglia. Both ganglia were coinjected with pNEXδ-hrGFP to see whether the injected DNA constructs were properly expressed. We observed hrGFP expression in one cell (ApC/EBP and ApCREB2 bZIP) and 13 cells (ApCREB1a and ApC/EBP bZIP) in each ganglion (data not shown). (A) Ganglion injected with full-length ApC/EBP and the bZIP domain of ApCREB2. Three cells were strongly stained (large arrows), and some cells were weakly stained (small arrows). (B) Ganglion injected with full-length ApCREB1a and the bZIP domain of ApC/EBP. No cells were stained.
By developing the Aplysia two-hybrid system, we have created a tool that can reveal the components involved in the molecular mechanism of memory formation in Aplysia. For example, when a novel protein is identified in interaction screens conducted by in vitro methods or the yeast two-hybrid system, the Aplysia two-hybrid system should be used to confirm that the interaction actually occurs in Aplysia neuronal cells. Interactions between the new protein and other known memory-related proteins could also be assessed using the system. In this way, the Aplysia two-hybrid system would complement the weaknesses of in vitro methods and the yeast two-hybrid system, while benefiting from their ability to screen a vast number of proteins at one time.
Furthermore, the method can be extended to the study of the interaction of two proteins that are regulated in response to neurotransmitters. An example of this application would be to access the interaction of ApCREB1 and ApCREB2 by injecting the corresponding bait and prey DNA into the sensory neurons and assessing the reporter gene activity in response to or in absence of 5-HT (serotonin).
In summary, the Aplysia two-hybrid system allows quick and direct determination of interaction between memory-related transcription factors in their native protein environment.
MATERIALS AND METHODS
Plasmids
pAS99, pASCA, and pACT12.1 are the bZIP domains (or full length) of ApCREB2 and ApC/EBP (Alberini et al. 1994) subcloned in pAS1 and pACT2 vectors (Durfee et al. 1993) for yeast two-hybrid screening described in Bartsch et al. (1995). pNEX, pNEX2, and pNEXδ are vectors constitutively expressed in Aplysia neurons (Kaang 1996).
pNEX-Gal4DB Group (Bait)
To construct pNEX-Gal4DB, pNEX-Gal4DB-CREB2bZIP, and pNEX-Gal4DB-C/EBPbZIP, we inserted Gal4DB (from pAS1; Gal4 DNA-binding domain), Gal4DB-CREB2bZIP (from pAS99), and Gal4DB-C/EBPbZIP (from pASCA), respectively, into pNEX-Gal4 using XhoI-BamHI sites (regions of full-length Gal4 other than the Gal4 DB in pNEX-Gal4 were deleted).
pNEXδ-Gal4AD Group (Prey)
To construct pNEXδ-Gal4AD and pNEXδ-Gal4AD-C/EBP, we inserted Gal4AD (from pACT2; Gal4 transcription activation domain) and Gal4AD-CREB2 (from pACT12.1), respectively, into pNEXδ by cutting with HindIII-BamHI. pNEXδ-Gal4AD-CREB1a was constructed by inserting CREB1a (from pNEXδ-CREB1a by PCR) into pNEXδ-Gal4AD by cutting with BamHI-SacI.
4xGal4-LacZ (Reporter Gene)
The CRE region of CRE-LacZ (Kaang et al. 1993) was substituted by four Gal4-binding sites using NsiI. The four Gal4-binding sites are located upstream from β-galactosidase-encoding LacZ.
pNEX2-Luciferase (Normalization Vector)
The firefly luciferase gene was expressed from the weak expression vector pNEX2 (Kaang 1996).
pNEXδ-hrGFP
Green fluorescent protein (hrGFP) from phrGFP-1 (Stratagene) was cut with BamHI-KpnI and inserted into pNEXδ using the same sites.
pNEX-Gal4
pNEX-Gal4 was made by cutting the entire Gal4 gene from pMA210 using HindIII-BamHI and inserting the fragment into pNEX.
Microinjection, β-Galactosidase/Luciferase Activity Assay, and X-gal Staining
Microinjection, β-galactosidase/luciferase assay, and X-gal staining were performed primarily as described by Kaang (1996). Aplysia buccal, pedal, and abdominal ganglia were used for injection. For the β-galactosidase/luciferase assay, the concentration of DB-bait, AD-prey, reporter, and normalization gene in the microinjection solution (10 mM Tris-Cl, 100 mM NaCl, 0.1% fast green at pH 7.3) was 70, 70, 760, and 50 ng/μL, respectively (In the case of injection of single constructs such as pNEX-Gal4, the concentration of single construct, reporter, and normalization gene was 70, 760, and 50 ng/μL, respectively.) For X-gal staining, the concentration of DB-bait, AD-prey, reporter, and pNEXδ-hrGFP was 140, 140, 1520, and 100 ng/μL, respectively. After coinjection of the constructs into the 20–30 cells of each ganglion, it was incubated at 18°C for 24–48 h before β-galactosidase/luciferase assay or X-gal staining.
Acknowledgments
This work was supported by grants M1-0104-00-0140 under the NRL Program and M1-0108-00-0075 from MOST, and No. R01-1999-000-00147-0 from KOSEF. J.-A.L. was supported by a BK21 Fellowship from the Korean Ministry of Education.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kaang@snu.ac.kr; FAX 82-2-884-9577.
Article and publication are at http://www.learnmem.org/cgi/doi/10.1101/lm.55303.
REFERENCES
- Alberini CM, Ghirardi M, Metz R, Kandel ER. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Ghirardi M, Skehel PA, Karl KA, Herder SP, Chen M, Bailey CH, Kandel ER. Aplysia CREB2 represses long-term facilitation: Relief of repression converts transient facilitation into long-term functional and structural change. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn AE, Lee W-H, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Finkel T, Gillison ML, Kennedy SP, Casella JF, Tomaselli GF, Morrow JS, Dang CV. Karyoplasmic interaction selection strategy: A general strategy to detect protein-protein interactions in mammalian cells. Proc Natl Acad Sci. 1992;89:7958–7962. doi: 10.1073/pnas.89.17.7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Kaang B-K. Parameters influencing ectopic gene expression in Aplysia neurons. Neurosci Lett. 1996;221:29–32. doi: 10.1016/s0304-3940(96)13279-1. [DOI] [PubMed] [Google Scholar]
- Kaang B-K, Kandel ER, Grant SGN. Activation of cAMP-responsive genes by stimuli that produce long-term facilitation in Aplysia sensory neurons. Neuron. 1993;10:427–435. doi: 10.1016/0896-6273(93)90331-k. [DOI] [PubMed] [Google Scholar]
- Kaang B-K, Pfaffinger PJ, Grant SGN, Kandel ER, Furukawa Y. Overexpression of an Aplysia Shaker K+ channel gene modifies the electrical properties and synaptic efficacy of identified Aplysia neurons. Proc Natl Acad Sci. 1995;89:1133–1137. doi: 10.1073/pnas.89.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- ————— The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;143:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Luo Y, Batalao A, Zhou H, Zhu L. Mammalian two-hybrid system: A complementary approach to the yeast two-hybrid system. BioTechniques. 1997;22:350–352. doi: 10.2144/97222pf02. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shioda T, Andriole S, Yahata T, Isselbacher KJ. A green fluorescent protein-reporter mammalian two-hybrid system with extrachromosomal maintenance of a prey expression plasmid: Application to interaction screening. Proc Natl Acad Sci. 2000;97:5220–5224. doi: 10.1073/pnas.97.10.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]