Abstract
Human red blood cell membranes were used as a model system to determine if the systematic evolution of ligands by exponential enrichment (SELEX) methodology, an in vitro protocol for isolating high-affinity oligonucleotides that bind specifically to virtually any single protein, could be used with a complex mixture of potential targets. Ligands to multiple targets were generated simultaneously during the selection process, and the binding affinities of these ligands for their targets are comparable to those found in similar experiments against pure targets. A secondary selection scheme, deconvolution-SELEX, facilitates rapid isolation of the ligands to targets of special interest within the mixture. SELEX provides high-affinity compounds for multiple targets in a mixture and might allow a means for dissecting complex biological systems.
Systematic evolution of ligands by exponential enrichment (SELEX) (1) is an oligonucleotide-based combinatorial chemistry that has been used to isolate high-affinity ligands for a wide variety of protein and low molecular weight targets (for review, see refs. 2–6). Single-stranded oligonucleotides (either RNA or DNA) with the highest affinity for the target are isolated from a very large pool of random sequence molecules by reiterative rounds of selection and amplification. These ligands have dissociation constants in the picomolar to low nanomolar range for protein targets, whether they naturally bind nucleic acids or not, and in the high nanomolar to high micromolar range for low molecular weight targets. In addition, nucleic acid ligands display high specificity for their targets with the capacity to distinguish between homologous proteins (7–10) or nearly identical low molecular weight compounds (11–19). Thus, the capabilities of these ligands are well established and applications of SELEX to many research, diagnostic, and therapeutic purposes is ongoing (4, 5).
A selected pool with high affinity for a pure protein or low molecular weight target usually consists of relatively few sequences that can be grouped into a small number of families based on primary and secondary sequence similarities. These different families are often able to compete with each other for binding to the target, demonstrating that they bind to single or overlapping epitopes. In the case of proteins, the highest-affinity nucleic acid ligands often inhibit function, presumably by interactions that overlap the binding site of the natural ligand. Single-stranded nucleic acids form stable three-dimensional shapes (6) that can be selected—and often are—to bind most often to a dominant epitope on a protein.
Surprisingly, a single epitope also can dominate when nucleic acid libraries are presented with more complex targets such as viral particles (20) or 30S ribosomes (21). Similar results have been reported for peptide-based combinatorial chemistry libraries (22–25). The dominance of a single epitope after only a few rounds of selection and amplification is unexpected because targeting individual components of these larger entities would almost certainly result in ligands with affinities similar to those obtained against other pure targets; an example might be that the pure ribosomal protein S1 and 30S ribosomal particles containing S1 gave more or less the same ligands after SELEX (21). And yet it seems that if binding to one epitope in a mixture does not interfere physically with binding to others, we would expect the simultaneous selection of ligands to all of the high-affinity epitopes within the mixture. In that this has not been the case in experiments reported in the literature, the application of combinatorial chemistry methods to complex mixtures of targets appears to be hindered by an unexplainable problem of extreme epitope dominance.
Combinatorial chemistry libraries containing high-affinity ligands to many components within a complex mixture would be an effective tool for dissecting and comparing biological systems such as serum or urine samples from patients with a similar disease, whole cells, or even different tissues. With this in mind, we used human red blood cell (RBC) ghosts as a model to work out the methodologies for obtaining and manipulating a pool of ligands that recognizes many targets within a complex mixture. RBC membranes provided a reasonably complex mixture of potential targets (26, 27) that should remain unchanged over the course of the experiment. We first demonstrate that a pool of single-stranded DNA (ssDNA) molecules with high affinity for different targets on cell membranes can be isolated. We then demonstrate how techniques that are common to most any molecular biology laboratory can be used to match the oligodeoxynucleotide ligands with their target, and visa versa, providing all of the basic requirements for the broad application of SELEX to complex biological systems.
EXPERIMENTAL PROCEDURES
SELEX.
RBC ghosts were prepared by washing human erythrocytes in PBS (pH 7.4) before lysing them in 5 mM sodium phosphate, pH 8.0/1 mM EDTA at 0°C (28). The final membrane preparation was stored at 0°C in PBS (pH 7.4) after further washes in that solution. The concentration of RBC ghosts was estimated by counting the number in a small sample through a phase-contrast microscope, and the total protein concentration was measured by staining with Coomassie blue G-250 (Bio-Rad).
Approximately 10 pmol of synthetic template DNA (Operon Technologies, Alameda, CA) containing 30 random nucleotides flanked by fixed sequences complementary to the primers (5′-GGGAGCTCAGAATAAACGCTCAA-3′ and 5′-BBBGATCCGGGCCTCATGTCGAA-3′, where B is biotin) was amplified by PCR. All PCR mixtures contained 50 mM KCl, 10 mM Tris⋅HCl (pH 8.6), 2.5 mM MgCl2, BSA (170 mg/ml), all four dNTPs (each at 1 mM), 1 mM each primer, and Taq DNA polymerase (Boehringer Mannheim; 1,000 units/ml). A similar reaction containing 1 pmol of template, 0.1 mM dCTP, and 1.25 μM [α-32P]dCTP was used to produce radioactively labeled ssDNA for monitoring the binding affinity of the pool. Amplifications were carried out in a Perkin–Elmer/Cetus thermal cycler at 93°C for 30 sec, 50°C for 30 sec, and 72°C for 1 min, for 25 cycles. Nonbiotinylated ssDNA was size-purified from the larger biotinylated strand by electrophoresis in 8% polyacrylamide gels containing urea. The starting pool (50 pmol) and a trace amount of radioactively labeled ssDNA were denatured by heating at 70°C for 5 min in 200 ml of PBS (pH 7.3) and renatured at 0°C for 10 min. The DNA solution was prefiltered through nitrocellulose to counter-select DNA molecules with high affinity for the partitioning matrix. The filter was washed with 300 ml of PBS and the filtrate was divided into six aliquots. An equal volume of PBS containing total RBC ghost protein at 0, 0.11, 0.22, 0.43, 0.86, or 1.72 mg/ml was added to each aliquot. The mixture was incubated for 20 min at room temperature and then filtered through nitrocellulose. The filters were washed with 5 ml of PBS, and the amount of radioactively labeled ssDNA retained was calculated by measuring Cerenkov radiation in a scintillation counter. The ssDNA was isolated from the filter that retained 5–10 times the radioactivity bound to the background control filter and was amplified by PCR for the next round of selection.
Individual DNA molecules were isolated from the round 25 pool by PCR amplification with primers that introduce BamHI and HindIII restriction sites at the 5′ and 3′ ends of the DNA. Restriction endonuclease-digested PCR products were ligated into pUC18 and introduced into Escherichia coli SURE (Stratagene) by electroporation. Plasmids were isolated from single bacterial colonies, and the nucleotide sequences in the inserted DNAs were determined by standard dideoxynucleotide methods. The sequences were searched for patterns in their primary and in their possible secondary sequences by inspection.
Nitrocellulose Filter Binding Assays.
Dissociation constants for single RBC ghost ligands were determined by Scatchard analysis (29, 30). Synthetic oligodeoxynucleotides (10 pmol) were radioactively labeled at their 5′ end. Reaction mixtures contained 50 pmol of [γ-32P]ATP and 10 units T4 polynucleotide kinase (Boehringer Mannheim) in 70 mM glycine⋅NaOH, pH 9.5/10 mM MgCl2/5 mM DTT/0.1% Nonidet P-40, and reactions were carried out at 37°C for 30 min. Binding reactions contained fixed amounts of RBC ghosts and various concentrations of radioactively labeled ssDNA in PBS (pH 7.3). The reactions were incubated at room temperature for 30 min before filtration through nitrocellulose (Millipore) that had been prewashed with 5 ml of PBS. The filters were immediately washed with 5 ml of PBS and then air-dried. The amount of radioactivity retained by each filter was measured by scintillation counting. Standard filter binding assays were used for comparisons between random and evolved nucleic acid ligand pools (1).
Cross-Linking of Synthetic Oligodeoxynucleotides.
Oligodeoxynucleotides were synthesized (Operon) with a primary aliphatic amine (with a six-carbon spacer group) on the 5′ end of each molecule. The conjugation reactions between the photoreactive molecule and the amino group on the ssDNA contained 5 mM N-hydroxysulfosuccinimidyl-4-azidobenzoate (sulfo-HSAB; Pierce) and 0.01 mM oligodeoxynucleotide in a buffer of 100 mM triethylamine (pH 9.5). The reactions were incubated at room temperature for 15 min and then at 37°C for 15 min. The oligodeoxynucleotides were separated from the unconjugated sulfo-HSAB by centrifugation through columns containing Sephadex G-50 (Pharmacia) according to the suppliers instructions. The oligodeoxynucleotides were then radioactively labeled at their 3′ end with [α-32P]dideoxyadenosine 5′-triphosphate (3,000 Ci/mmol; 1 Ci = 37 GBq) by terminal deoxynucleotidyltransferase in buffer containing 100 mM sodium cacodylate (pH 7.2), 0.2 mM 2-mercaptoethanol, and 2 mM CoCl2 at 37°C for 1 hr and purified by electrophoresis in 8% polyacrylamide gels containing urea.
Typical binding reactions for cross-linking contained 10 nM radioactively labeled photoreactive oligodeoxynucleotide, 10 μM random-sequence oligodeoxynucleotide (30 nucleotides long; Operon), and 7.7 × 106 RBC ghosts in PBS (pH 7.3). Reactions were incubated at room temperature for 30 min before irradiation for 100 pulses at 308 nm by a XeCl excimer laser (Lumonics Ex-748) (31). Similar reactions in which the radioactively labeled ssDNA molecules lacking the photoreactive group were cross-linked to the ghosts were irradiated with a 254-nm Mineral Light for 5 min. Reaction products were analyzed by SDS/PAGE (4–12% polyacrylamide) and autoradiography.
Deconvolution-SELEX.
Approximately 1 pmol of round 25 pool DNA was amplified by PCR as described above except the nonbiotinylated primer sense strand (Operon) contained the primary aliphatic amine group (with a six-carbon spacer group) at the 5′ end and the PCR contained all four dNTPs (each at 50 μM) and 1 μM [α-32P]dCTP at a specific activity of 6,000 Ci/mmol. The nonbiotinylated strands were purified as described above. The cross-linking reagent from sulfo-HSAB was conjugated to the amino group at the 5′ ends, and the pool of ssDNA molecules was cross-linked to the RBC ghosts as described above. The SDS/PAGE-separated products were transferred to nitrocellulose by electrophoresis. Autoradiography of the nitrocellulose blot was used to detect the cross-linking products. The appropriate bands of nitrocellulose were excised and placed directly in PCR mixtures for amplification. Individual DNA molecules were isolated from the round 4 pools and their nucleotide sequences were analyzed as described above.
Protein Purification.
A version of c56t was synthsized (Operon) with three biotin groups at the 3′ end and the aliphatic amine required for conjugation of the photoreactive group on the 5′ end. The sulfosuccinimidyl (4-azidophenyldithio) propionate conjugation reaction was carried out as described above. Binding reactions (1-ml total volume) contained 100 nM cross-linkable biotinylated c56t, 0.1 μM random-sequence DNA (30 nucleotides long), and 7.7 × 108 RBC ghosts in PBS (pH 7.3) and were irradiated as described above. The RBC ghosts were pelleted by centrifugation, and the membrane proteins were solubilized in a solution containing 10 mM Tris⋅HCl (pH 7.5), 200 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS at 4°C for 2 hr. Enough magnetic beads with streptavidin covalently attached (Dynal) to bind 200 pmol of biotinylated oligodeoxynucleotide was prepared according to the suppliers instructions and added to the solution. Binding of the biotinylated DNA molecules took place in 10 mM Tris⋅HCl, pH 7.5/1 M NaCl/2 mM EDTA/0.1% Triton X-100/0.01% SDS at 4°C for 2 hr. The beads were washed sequentially with 10 ml of 10 mM Tris⋅HCl, pH 7.5/2 mM EDTA/0.1% Triton X-100/0.01% SDS containing 1 M, 0.5 M, and then 0.1 M NaCl. The disulfide bond in the sulfosuccinimidyl (4-azidophenyldithio) propionate group was reduced by incubation of the beads in 10 mM Tris⋅HCl, pH 7.5/0.1 M NaCl/2 mM EDTA/0.1% Triton X-100/0.01% SDS/200 mM DTT at room temperature for 1 hr. The proteins and nucleic acids were precipitated in 0.3 M NaOAc and 2 vol of ethanol, pelleted by centrifugation, and analyzed by SDS/PAGE (4–12% polyacrylamide) with staining of the proteins by silver.
RESULTS
Selection of ssDNA Molecules That Bind RBC Ghosts.
Selection of ssDNA ligands aimed at human RBC ghosts was carried out by using essentially the same methodologies as originally described for pure protein targets (1). We used ssDNA because we had not yet decided what modified RNA library was likely to be most useful for later experiments (32–34). A pool of 1013 ssDNA molecules containing a 30-base random region flanked by fixed sequences required for the hybridization of primers was incubated with the RBC ghosts. Tighter-binding sequences were partitioned from the rest of the pool by nitrocellulose filtration, and that subset of selected sequences was amplified by PCR. The pool showed slow but steady improvement in binding affinity during the 25 rounds of selection and amplification (data not shown). The concentration of RBC ghosts in the binding reactions was reduced to increase the selection stringency as the affinity of the ssDNA pool for the membranes increased.
The round 25 pool, which binds approximately 10-fold better than the starting pool (data not shown), was cloned and the nucleotide sequences of 69 clones were determined.¶ The most interesting feature of the round 25 pool was its high sequence complexity compared with that of pools selected for binding to pure targets. When SELEX is performed over 10–15 rounds for a single protein target, ligand families are easily found (9, 10). Usually clones with identical sequences to other clones are observed and these repeats are an indication that the SELEX protocol is nearing its final round (2, 3).
Forty-one of the round 25 sequences could be grouped in one of six distinct sequence families based on common primary sequence motifs; these family relationships are not robustly determined but are based on the presence of at least two sequences that are not obvious derivatives of the same progenitor clone. The remaining clones had no significant sequence similarities to any of the others. These clones may be nonbinding background sequences that have not yet been purged from the pool during the selection process, or they might be single representatives of other sequence families that are less prevalent in the final pool than those identified. Very few repeated sequences were found; three within the most represented family (motif I) and two that were not members of any identified family.
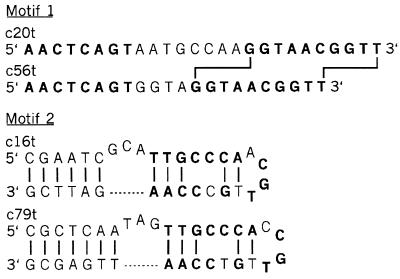
Motifs I and II, which compose approximately 16% and 10% of the entire pool, respectively, were chosen for further analysis. Oligodeoxynucleotides that consist of the conserved regions of two individual clones from each of these sequence families were synthesized (Fig. 1). These ssDNA molecules ranged from 22 to 35 nucleotides long, with the boundaries being chosen by inspection of the conserved regions of each sequence family. There was sufficient sequence variation among the motif II clones to propose a secondary structure. This was not the case for the motif I members, although they could form a short helix with the nucleotides at the extreme 5′ and 3′ ends. The dissociation constants for these truncates and the RBC membranes were determined by Scatchard analysis of a series of binding reactions in which the concentration of RBC ghosts was held constant while that of the nucleic acids was varied (29, 30). The motif I truncates (c20t and c56t) differ in the number of bases that separate two conserved sequence elements (eight and four, respectively), yet binding analysis of each measured approximately equal dissociation constants of 1.2 nM and 1.6 nM for 1.6 × 103 and 1.3 × 103 binding sites per ghost, respectively (data not shown). Similar analyses measured dissociation constants of approximately 1.8 nM and 2.0 nM for 6.5 × 103 and 6.0 × 103 binding sites for the two motif II truncates (c16t and c79t, respectively). Therefore, these two motifs specify high-affinity binding to the RBC ghosts but appear to recognize targets that differ a fewfold in concentration on the membranes.
Figure 1.
Deoxynucleotide sequences of the motif I and motif II truncates used in the binding and cross-linking experiments (see text). Invarient residues within each motif are shown in boldface type. The shift in the spacing of the 3′ conserved element between the motif I ligands is indicated by lines connecting the sequences. The proposed secondary structures of the motif II ligands are shown with base-pairing interactions denoted by lines connecting the nucleotides.
Identification of Targets.
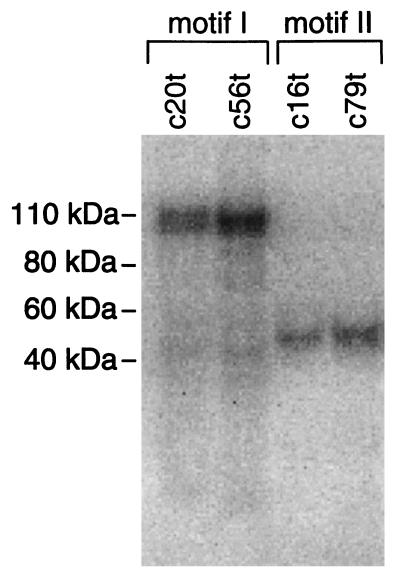
Both the sequence complexity of the round 25 pool and the Scatchard analysis of the binding of individual motif truncates suggest that ligands for multiple targets on the RBC membranes were selected. Photoaffinity cross-linking experiments were performed with the four motif I and II truncates to more thoroughly test whether these individual sequence motifs recognize different targets on the ghosts (Fig. 2). Each truncate was synthesized with a primary aliphatic amine (with a six-carbon spacer group) on the 5′ end. The phenyl azide photoreactive molecule from N-hydroxysulfosuccinimidyl-4-azidobenzoate (sulfo-HSAB) was conjugated to the 5′ end of each synthetic DNA through this amino group. Additionally, the DNA molecules were radioactively labeled on the 3′ end. The photoreactive truncates were allowed to bind to the ghosts in the presence of a thousandfold molar excess of nonradioactive random-sequence ssDNA before irradiation with a 308-nm excimer laser. Cross-links between the ssDNA molecules and RBC membrane proteins were detected by SDS/PAGE and autoradiography. The motif I truncates cross-linked to a protein dimer band with an apparent molecular weight centered around 105 kDa when complexed to the oligodeoxynucleotides. In contrast, the photoreactive motif II truncates labeled a protein that migrates slightly above 40 kDa when associated with ssDNA molecules. The identical protein bands were labeled by c56t lacking the phenyl azide group by cross-linking through thymidine with short-wavelength UV (254 nm) irradiation. However, two control synthetic ssDNAs of the same length and base composition as c56t, but with the primary sequences randomly shuffled, formed no detectable cross-links (data not shown).
Figure 2.
Autoradiograph of the SDS/PAGE analysis of the cross-linking products formed between the truncated ssDNA ligands indicated above each lane and their targets on the RBC membranes.
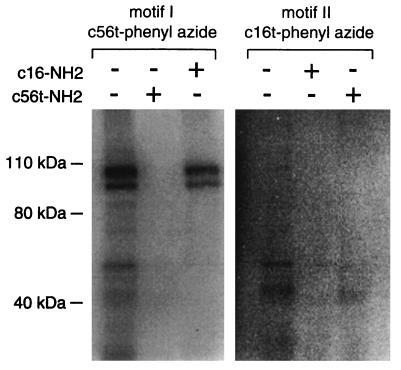
The binding specificities of a motif I truncate (c56t) and a motif II truncate (c16t) were further tested by a cross-competition experiment (Fig. 3). Addition of a thousandfold molar excess of the same sequence lacking the phenyl azide group abolished cross-linking of both c56t and c16t to their appropriate target. However, addition of a thousandfold molar excess of the other sequence to the binding reaction reduced the amount of nonspecific cross-linking products but did not diminish the specific cross-link. Therefore, these different sequence families from the round 25 pool recognize different targets with high affinity and specificity.
Figure 3.
Autoradiograph of the SDS/PAGE analysis of the cross-linking products formed between c56t and c16t and their targets on the RBC membranes in the presence or absence of competitor ligand. The radioactive ligand present in each group of three lanes and the presence (+) or absence (−) of a thousandfold excess of nonradioactively labeled ligand is indicated above. The specific cross-linking products for each ligand is denoted by an arrow.
Deconvolution-SELEX.
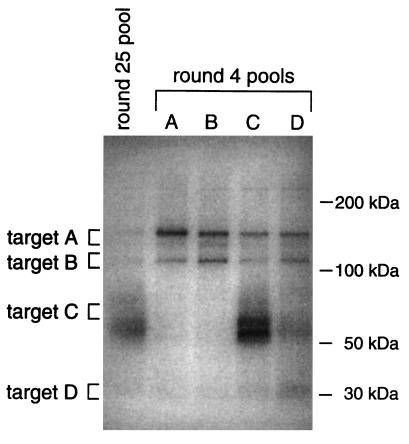
The utility of a selected pool of nucleic acids to dissect complex systems is contingent upon the ability to quickly identify which sequences are ligands for components of interest within the targeted mixture. Therefore, a secondary selection was developed to deconvolute the complex round 25 pool. PCR amplification of the random starting pool and the round 25 pool in the presence of a primer with an aliphatic amine at its 5′ end allowed subsequent conjugation of the photoreactive phenyl azide group from sulfo-HSAB to these pools. Binding reactions containing RBC ghosts, radioactively labeled versions of these pools, and a thousandfold molar excess of nonradioactive random-sequence ssDNA molecules lacking the hybridization site for the primers were irradiated at 308 nm with an excimer laser and the resulting cross-linking products were detected by SDS/PAGE and autoradiography (Fig. 4). No protein bands were radioactively labeled by the random starting pool (data not shown). However, multiple cross-linking products were detected in the reaction that contained the round 25 pool, indicating the minimum number of targets that are recognized by the selected pool. As with the c56t cross-linking reaction described above, essentially the same results were seen when the random and round 25 pools lacking the photoreactive agent were cross-linked to the ghosts by irradiation with short-wavelength UV (data not shown). These results with the selected pool once again demonstrate that ligands to multiple targets on the RBC membranes were selected and provided a means by which the selected pool could be deconvoluted.
Figure 4.
Autoradiograph of the SDS/PAGE analysis of the cross-linking product formed by the radioactively labeled round 25 starting pool and each of the round 4 deconvolution-SELEX pools (A–D) with the RBC membranes. The approximate region of the gel excised for each target is indicated by brackets.
To isolate the ssDNA molecules that recognize specific targets on the membranes, the RBC proteins and their associated ssDNA molecules were first electrophoretically transferred from the SDS/polyacrylamide gel described above to nitrocellulose. This was expected to remove uncross-linked ssDNA that remained in the gel. Four radioactively labeled bands (targets A–D) that varied in molecular weight and in radioactive intensity were excised from the filter and the associated ssDNA molecules were amplified by PCR in the presence of the aliphatic amine-containing primer. The phenyl azide cross-linking reagent was conjugated to the resulting ssDNA pools, which were again cross-linked to the ghosts. Four rounds of this deconvolution-SELEX scheme resulted in pools (A–D) with clearly visible biases upon cross-linking to the RBC membranes (Fig. 4).
The four round 4 pools were cloned and the nucleotide sequences of several clones from each pool (13 from pool A, 14 from pool B, 16 from pool C, and 17 from pool D) were determined.¶ As expected from the residual cross-linking of each of the pools to the other targets, there was some overlap between the sequences in the pools. Clearly, however, distinct ligands were selected to each target, and it is expected that further rounds of selection and amplification would result in more distinct sequence compositions. The ability of the sequences that predominate in each pool to recognize the appropriate target was confirmed by cross-linking of individual clones to the RBC ghosts (data not shown). Each of these clones cross-linked somewhat to target C in addition to the anticipated target even at a relatively high (10,000-fold molar excess) nonspecific competitor ssDNA concentration, possibly due to the presence of nucleotides in addition to the binding motif in the full-length molecules.
In addition to eliminating prevalent sequences that do not bind the excised target, the deconvolution-SELEX procedure was successful in isolating rare sequences that do recognize that target. Although motif I, the most prevalent sequence family in the original pool selected for binding to the entire RBC membrane, was again dominant in pools A and B, motif II, the third most represented sequence family, was not found in any of the pools. Most importantly, two new sequence motifs were present in pool D that had not been seen in the 69 clones isolated from the original round 25 pool, and several “orphan” sequences from round 25 were now well represented. Therefore, the original pool selected for binding to the membranes is deeper in sequence complexity and target recognition capacity than was measured by the nucleotide sequencing analysis.
Ligand-Mediated Target Purification.
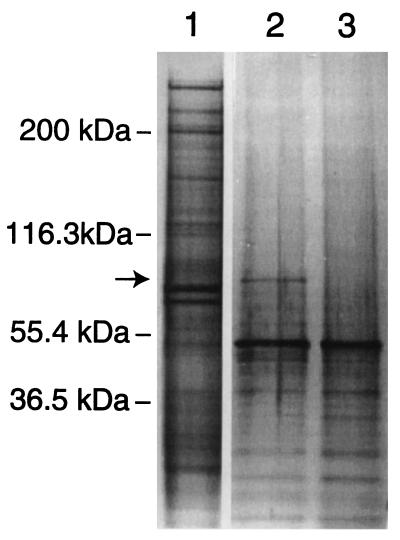
One would like to be able to purify any protein target for which a high-affinity nucleic acid ligand has been identified. A one-step partial purification of the protein recognized by the motif I truncate c56t was performed to demonstrate a general scheme by which the selected nucleic acid ligands can be used to mediate isolation of their target. A version of c56t containing the aliphatic amine required for conjugation of the photoreactive phenyl azide cross-linking group at the 5′ end and three biotin molecules at the 3′ end was synthesized. In this experiment, the phenyl azide from sulfosuccinimidyl (4-azidophenyldithio) propionate, a slightly different form of the cross-linking reagent in which the spacer arm contains a disulfide bond, was attached to the ssDNA molecules. Binding reactions containing or omitting this photoreactive ssDNA, a thousandfold molar excess of non-cross-linkable random-sequence ssDNA, and RBC ghosts were irradiated at 308 nm as described. The proteins were then extracted from the membrane by treatment with the detergents Triton X-100 and SDS. Magnetic beads with strepavidin covalently attached were added to the solution of solubilized membrane proteins, and the c56t molecules were allowed to bind to the beads via the biotin groups attached to their 3′ ends. The beads were thoroughly washed before the protein target cross-linked to the c56t molecules was released by reduction of the disulfide bond and analyzed by SDS/PAGE. Staining of the proteins in the polyacrylamide gel with silver revealed the presence of a very distinct band migrating at approximately 97 kDa in the lane containing the product of the c56t binding reaction that is absent in the lane containing the product of the control reaction lacking the DNA (Fig. 5). This band was eliminated by addition of a thousandfold molar excess of c56t lacking both the cross-linking and biotin groups (data not shown). The apparent molecular weight of the c56t target, its concentration in the RBC membrane as determined by the Scatchard analysis described above, and its migration as a dimer of approximately 200 kDa on a nonreducing SDS gel (data not shown) suggest that the target of motif I is the transferrin receptor monomer (CD71). The protein could be sequenced if the identity was important.
Figure 5.
Silver-stained denaturing polyacrylamide gel used to analyze the ligand mediated target purification procedure. Lanes: 1, untreated RBC membranes; 2, purification with c56t; 3, purification minus c56t. The target band isolated in the reaction containing the c56t ligand is indicated by an arrow.
DISCUSSION
This SELEX experiment against human RBC ghosts clearly demonstrates that ligands to multiple targets are isolated when a complex mixture is targeted by nucleic acid combinatorial libraries. In essence, the experiment is really a set of multiple simultaneous selections, in which the competition of ligands for binding at one target site is independent of the same competitive process at a different target site. For example, in the first round of selection, there were approximately 10 ssDNA molecules for every copy of the target recognized by the motif I clones. In subsequent rounds, reduction of the concentration of RBC ghosts and increases in the number of molecules that bind to the target resulted in more stringent competition between these ligands for the motif I binding site (35). To a certain extent, this process must be unaffected by similar competitions for other binding sites that are physically separated on the membranes. However, families of ligands are lost when the concentration of their target falls below the value of their Kd for the binding site. Therefore, we would expect that earlier round pools are more complex and contain ligands with a wider range of affinities for a broader spectrum of targets, whereas later round pools become narrower in scope as the decreasing target concentrations select for the highest-affinity ligands.
Both the sequence complexity of the round 25 pool and the measurement of the binding characteristics of two consensus sequence motifs suggested that the pool contained ligands for more than one target. However, the photoaffinity cross-linking of these different DNA molecules to their targets was the first direct evidence that the pool contained nucleic acid families that recognize different RBC proteins. The cross-competition experiment between the two motifs further demonstrated that these ligands are specific for their targets. These cross-linking experiments also provided an essential connection between the library of selected ligands and the complex mixture of targets that was exploited in deconvolution-SELEX to match individuals from the two sets. This procedure led to the identification of the ligands for the four target mixtures that were chosen, and more importantly, amplified sequences that had not been identified in the sequence analysis of the original selected pool. This further demonstrates that the selected pool is rich in sequence complexity and suggests that ligands to almost any protein of interest might be isolated. The opportunity to isolate rare ligands might be further enhanced by starting at an earlier round when the pool has an even higher sequence complexity.
Combinatorial libraries provide a powerful tool for dissecting complex biological systems. Libraries selected for binding to mixtures of targets potentially contain ligands to many epitopes that are inaccessible to traditional antibody technology. Unlike antibodies, these libraries are not biased by natural mechanisms such as immune tolerance. In addition, appropriate counterselection schemes can efficiently yield ligands that recognize differences between two mixtures. Many approaches can be envisioned to study the appearance and disappearance of various types of cellular targets such as developmental markers or tumor-specific antigens, and selections against cellular or nuclear extracts would extend this analysis to intracellular targets. The ligands isolated in these experiments can then be used to identify and study their cellular targets, and many may become important medical diagnostic and therapeutic agents (4).
Several encouraging reports with random peptide libraries have demonstrated the potential application of combinatorial libraries to the study of biological systems (22–25). However, the small numbers of ligands isolated in these studies suggests that peptide libraries have insufficient binding affinity to fully probe the complete array of targets available in complex mixtures. Low-affinity binding limits the analysis to abundant targets because ligands will be selected for only those present at concentrations near to or greater than the value of the Kd (35). In addition, peptides require circularization, dimerization, or presentation in the context of a larger protein to achieve sufficient structural stability for even modest affinity binding to a target (36–41). These structural requirements limit the number of potential ligands that can be sampled in an experiment. In contrast, relatively short nucleic acid ligands form stable structures that display high affinities for their targets. These features permit the use of very large starting pools to probe for the presence of much rarer targets. In this study, the c56t truncate consists of only 22 nucleotides, yet has a Kd value of 1.6 nM for a target that is relatively rare on the red blood cell membrane. In addition to direct counterselection, nucleic acid libraries also permit the use of subtractive hybridization technologies to obtain differences between selected pools. This feature may allow more rapid comparisons between larger numbers of cell or tissue types by eliminating the need for individual experiments with direct counterselection against unwanted targets for every comparison. Instead, libraries selected for binding to different mixtures could later be counterselected against each other by nucleic acid hybridization. Overall, the experiments reported herein and those previously reported illustrate the great potential for combinatorial chemistries in dissecting complex biological systems.
Acknowledgments
We thank Steve Ringquist, Shauna Simons, Hang F. G. Chen, Rachel North, and all of our colleagues at the University of Colorado, Boulder, and NeXstar Pharmaceuticals, Inc., for their contributions to this work. This work was supported by National Institutes of Health Grants GM 28685 and GM 19963 to L.G. and by a Damon Runyon–Walter Winchell Cancer Research Fund Fellowship, DRG-1139, to K.N.M.
ABBREVIATIONS
- SELEX
systematic evolution of ligands by exponential enrichment
- RBC
red blood cell
- ssDNA
single stranded DNA
- sulfo-HSAB
N-hydroxysulfosuccinimidyl-4-azidobenzoate
Footnotes
All sequences are available from L.G.
References
- 1.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 2.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 3.Fitzwater T, Polisky B. In: Methods in Enzymology. Abelson J, Simon M I, editors. San Diego: Academic; 1996. pp. 275–301. [DOI] [PubMed] [Google Scholar]
- 4.Gold L. J Biol Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 5.Gold L, Brown D, He Y-Y, Shtatland T, Singer B S, Wu Y. Proc Natl Acad Sci USA. 1997;94:59–64. doi: 10.1073/pnas.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eaton B E, Gold L, Zichi D A. Chem Biol. 1995;2:633–638. doi: 10.1016/1074-5521(95)90023-3. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C, MacDougal S, Gold L. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Gold L. Biochemistry. 1994;33:8746–8756. doi: 10.1021/bi00195a016. [DOI] [PubMed] [Google Scholar]
- 9.Jellinek D, Lynott C K, Rifkin D B, Janjic N. Proc Natl Acad Sci USA. 1993;90:11227–11231. doi: 10.1073/pnas.90.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jellinek D, Green L S, Bell C, Janjic N. Biochemistry. 1994;33:10450–10456. doi: 10.1021/bi00200a028. [DOI] [PubMed] [Google Scholar]
- 11.Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 12.Famulok M, Szostak J W. J Am Chem Soc. 1992;114:3990–3991. [Google Scholar]
- 13.Sassanfar M, Szostak J W. Nature (London) 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 14.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 15.Connell G J, Illangesekare M, Yarus M. Biochemistry. 1994;32:5497–5502. doi: 10.1021/bi00072a002. [DOI] [PubMed] [Google Scholar]
- 16.Lauhon C T, Szostak J W. J Am Chem Soc. 1995;117:1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- 17.Jiang F, Kumar R A, Jones R A, Patel D J. Nature (London) 1996;382:183–186. doi: 10.1038/382183a0. [DOI] [PubMed] [Google Scholar]
- 18.Nolte A, Klubmann S, Bald R, Erdmann V A, Furste J P. Nat Biotechnol. 1996;14:1116–1119. doi: 10.1038/nbt0996-1116. [DOI] [PubMed] [Google Scholar]
- 19.Klubmann S, Nolte A, Bald A, Erdmann V A, Furste J P. Nat Biotechnol. 1996;14:1112–1115. doi: 10.1038/nbt0996-1112. [DOI] [PubMed] [Google Scholar]
- 20.Pan W, Craven R C, Qiu Q, Wilson C B, Wills J W, Golovine S, Wang J F. Proc Natl Acad Sci USA. 1995;92:11509–11513. doi: 10.1073/pnas.92.25.11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ringquist S, Jones T, Snyder E E, Gibson T, Boni I, Gold L. Biochemistry. 1995;34:3640–3648. doi: 10.1021/bi00011a019. [DOI] [PubMed] [Google Scholar]
- 22.Kruif J, Terstappen L, Boel E, Logtenberg T. Proc Natl Acad Sci USA. 1995;92:3938–3942. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barry M A, Dower W J, Johnston S A. Nat Med. 1996;2:299–305. doi: 10.1038/nm0396-299. [DOI] [PubMed] [Google Scholar]
- 24.Pasqualini R, Ruoslahti E. Nature (London) 1996;380:364–366. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 25.Van Ewijk W, de Kruif J, Germeraad W T V, Berendes P, Ropke C, Platenburg P P, Logtenberg T. Proc Natl Acad Sci USA. 1997;94:3903–3908. doi: 10.1073/pnas.94.8.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennet V. Annu Rev Biochem. 1985;54:273–304. doi: 10.1146/annurev.bi.54.070185.001421. [DOI] [PubMed] [Google Scholar]
- 27.Telen M J. Blood. 1995;85:299–306. [PubMed] [Google Scholar]
- 28.Yu J, Steck T L. J Biol Chem. 1975;250:9170–9175. [PubMed] [Google Scholar]
- 29.Scatchard G. Ann NY Acad Sci. 1949;51:660–667. [Google Scholar]
- 30.Robb R J, Mayer P C, Garlick R. J Immunol Methods. 1985;81:15–30. doi: 10.1016/0022-1759(85)90118-8. [DOI] [PubMed] [Google Scholar]
- 31.Jensen K B, Atkinson B L, Willis M C, Koch T H, Gold L. Proc Natl Acad Sci USA. 1995;92:12220–12224. doi: 10.1073/pnas.92.26.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jellinek D, Green L, Bell C, Lynott K, Gill N, Vargeese C, Kirschenheuler G, McGee D, Abesinghe P, Pieken W, et al. Biochemistry. 1995;34:11363–11372. doi: 10.1021/bi00036a009. [DOI] [PubMed] [Google Scholar]
- 33.Green L, Jellinek D, Beebe L, Bell C, Feistner B, Gill S, Jucker F, Janjic N. Chem Biol. 1995;2:683–695. doi: 10.1016/1074-5521(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y, Nieuwlandt D, Magallanez A, Feistner B, Jayasena S. Nucleic Acids Res. 1996;24:3407–3414. doi: 10.1093/nar/24.17.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Irvine D, Tuerk C, Gold L. J Mol Biol. 1991;222:739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- 36.McCafferty J, Griffiths A D, Winter G, Chriswell D J. Nature (London) 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 37.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 38.Janda K D. Proc Natl Acad Sci USA. 1994;91:10779–10785. doi: 10.1073/pnas.91.23.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladner R C. Trends Biotechnol. 1995;13:426–430. doi: 10.1016/S0167-7799(00)88997-0. [DOI] [PubMed] [Google Scholar]
- 40.Wrighton N C, Farrell F X, Chang R, Kashyap A K, Barbone F P, Mulcahy L S, Johnson D L, Barrett R W, Jolliffe L K, Dower W J. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 41.Livnah O, Stura E A, Johnson D L, Middleton S A, Mulcahy L S, Wrighton N C, Dower W J, Jolliffe L K, Wilson I A. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]