Abstract
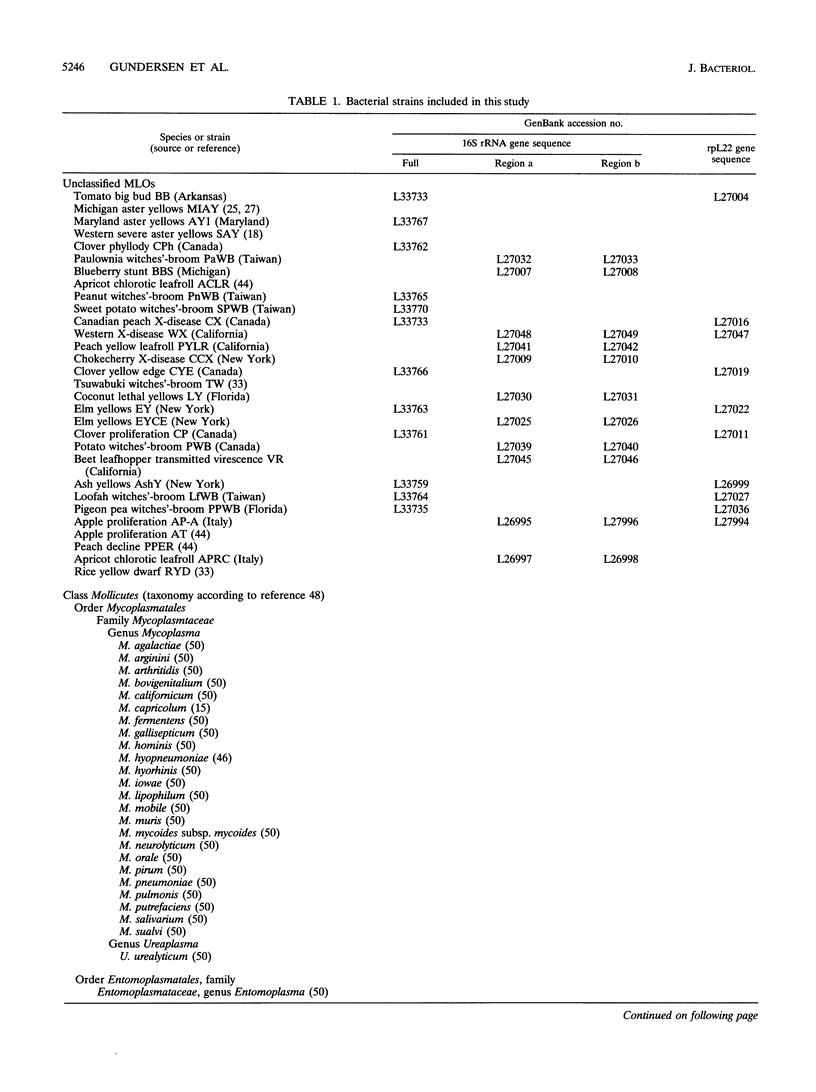
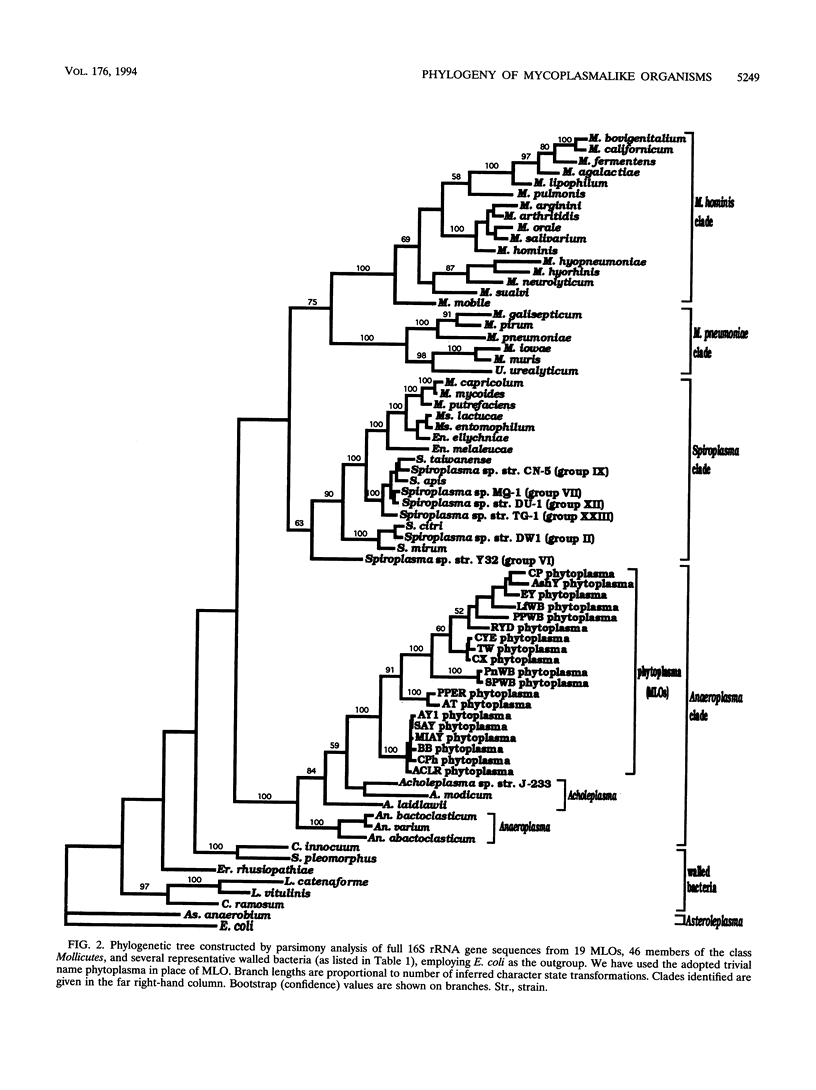
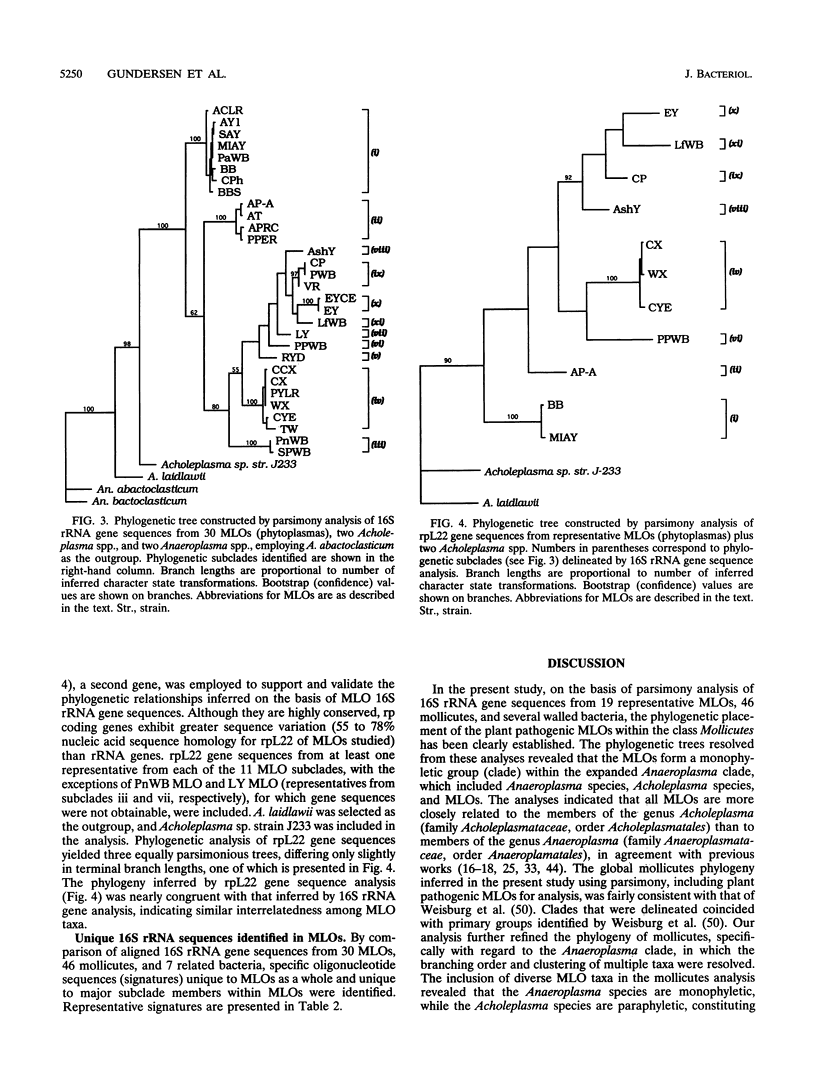
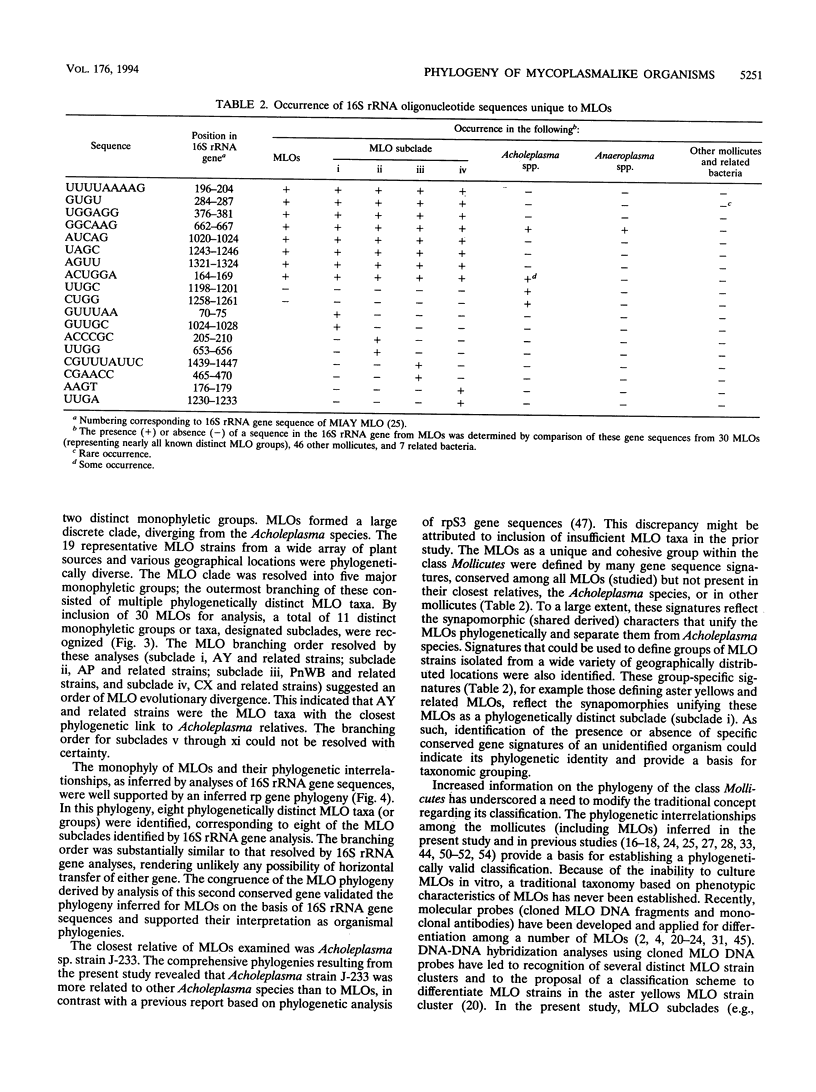
A global phylogenetic analysis using parsimony of 16S rRNA gene sequences from 46 mollicutes, 19 mycoplasmalike organisms (MLOs) (new trivial name, phytoplasmas), and several related bacteria placed the MLOs definitively among the members of the class Mollicutes and revealed that MLOs form a large discrete monophyletic clade, paraphyletic to the Acholeplasma species, within the Anaeroplasma clade. Within the MLO clade resolved in the global mollicutes phylogeny and a comprehensive MLO phylogeny derived by parsimony analyses of 16S rRNA gene sequences from 30 diverse MLOs representative of nearly all known distinct MLO groups, five major phylogenetic groups with a total of 11 distinct subclades (monophyletic groups or taxa) could be recognized. These MLO subclades (roman numerals) and designated type strains were as follows: i, Maryland aster yellows AY1; ii, apple proliferation AP-A; iii, peanut witches'-broom PnWB; iv, Canada peach X CX; v, rice yellow dwarf RYD; vi, pigeon pea witches'-broom PPWB; vii, palm lethal yellowing LY; viii, ash yellows AshY; ix, clover proliferation CP; x, elm yellows EY; and xi, loofah witches'-broom LfWB. The designations of subclades and their phylogenetic positions within the MLO clade were supported by a congruent phylogeny derived by parsimony analyses of ribosomal protein L22 gene sequences from most representative MLOs. On the basis of the phylogenies inferred in the present study, we propose that MLOs should be represented taxonomically at the minimal level of genus and that each phylogenetically distinct MLO subclade identified should represent at least a distinct species under this new genus.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Grau O., Laigret F., Carle P., Tully J. G., Rose D. L., Bové J. M. Identification of a plant-derived mollicute as a strain of an avian pathogen, Mycoplasma iowae, and its implications for mollicute taxonomy. Int J Syst Bacteriol. 1991 Oct;41(4):473–478. doi: 10.1099/00207713-41-4-473. [DOI] [PubMed] [Google Scholar]
- Iwami M., Muto A., Yamao F., Osawa S. Nucleotide sequence of the rrnB 16S ribosomal RNA gene from Mycoplasma capricolum. Mol Gen Genet. 1984;196(2):317–322. doi: 10.1007/BF00328065. [DOI] [PubMed] [Google Scholar]
- Kuske C. R., Kirkpatrick B. C. Phylogenetic relationships between the western aster yellows mycoplasmalike organism and other prokaryotes established by 16S rRNA gene sequence. Int J Syst Bacteriol. 1992 Apr;42(2):226–233. doi: 10.1099/00207713-42-2-226. [DOI] [PubMed] [Google Scholar]
- Lee I. M., Davis R. E., Hiruki C. Genetic Interrelatedness among Clover Proliferation Mycoplasmalike Organisms (MLOs) and Other MLOs Investigated by Nucleic Acid Hybridization and Restriction Fragment Length Polymorphism Analyses. Appl Environ Microbiol. 1991 Dec;57(12):3565–3569. doi: 10.1128/aem.57.12.3565-3569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. M., Gundersen D. E., Davis R. E., Chiykowski L. N. Identification and analysis of a genomic strain cluster of mycoplasmalike organisms associated with Canadian peach (eastern) X disease, western X disease, and clover yellow edge. J Bacteriol. 1992 Oct;174(20):6694–6698. doi: 10.1128/jb.174.20.6694-6698.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. O., Sears B. B. 16S rRNA sequence indicates that plant-pathogenic mycoplasmalike organisms are evolutionarily distinct from animal mycoplasmas. J Bacteriol. 1989 Nov;171(11):5901–5906. doi: 10.1128/jb.171.11.5901-5906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. O., Sears B. B. Evolutionary relationships of a plant-pathogenic mycoplasmalike organism and Acholeplasma laidlawii deduced from two ribosomal protein gene sequences. J Bacteriol. 1992 Apr;174(8):2606–2611. doi: 10.1128/jb.174.8.2606-2611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. O., Sears B. B. The genome size of a plant-pathogenic mycoplasmalike organism resembles those of animal mycoplasmas. J Bacteriol. 1991 Mar;173(6):2128–2130. doi: 10.1128/jb.173.6.2128-2130.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Kato S., Iwanami S., Murata N. Mycoplasmalike organisms (MLOs) and distinguish them from other MLOs. Appl Environ Microbiol. 1993 Apr;59(4):1206–1212. doi: 10.1128/aem.59.4.1206-1212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba S., Oyaizu H., Kato S., Iwanami S., Tsuchizaki T. Phylogenetic diversity of phytopathogenic mycoplasmalike organisms. Int J Syst Bacteriol. 1993 Jul;43(3):461–467. doi: 10.1099/00207713-43-3-461. [DOI] [PubMed] [Google Scholar]
- Neimark H. C., Lange C. S. Pulse-field electrophoresis indicates full-length Mycoplasma chromosomes range widely in size. Nucleic Acids Res. 1990 Sep 25;18(18):5443–5448. doi: 10.1093/nar/18.18.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J. Phylogenetic analysis using ribosomal RNA. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- Razin S., Hyman H. C., Nur I., Yogev D. DNA probes for detection and identification of mycoplasmas (Mollicutes). Isr J Med Sci. 1987 Jun;23(6):735–741. [PubMed] [Google Scholar]
- Rogers M. J., Simmons J., Walker R. T., Weisburg W. G., Woese C. R., Tanner R. S., Robinson I. M., Stahl D. A., Olsen G., Leach R. H. Construction of the mycoplasma evolutionary tree from 5S rRNA sequence data. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1160–1164. doi: 10.1073/pnas.82.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschke C., Ruland K., Herrmann R. Nucleotide sequence of the 16S rRNA of Mycoplasma hyopneumoniae. Nucleic Acids Res. 1987 May 11;15(9):3918–3918. doi: 10.1093/nar/15.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth K. F., Harrison N., Sears B. B. Phylogenetic relationships among members of the class Mollicutes deduced from rps3 gene sequences. Int J Syst Bacteriol. 1994 Jan;44(1):119–124. doi: 10.1099/00207713-44-1-119. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Tully J. G., Rose D. L., Petzel J. P., Oyaizu H., Yang D., Mandelco L., Sechrest J., Lawrence T. G., Van Etten J. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989 Dec;171(12):6455–6467. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Maniloff J., Zablen L. B. Phylogenetic analysis of the mycoplasmas. Proc Natl Acad Sci U S A. 1980 Jan;77(1):494–498. doi: 10.1073/pnas.77.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]




